Abstract
Pandemic influenza A H1N1 (pH1N1) virus emerged in 2009. In the subsequent 4 years, it acquired several genetic changes in its hemagglutinin (HA). Mutations may be expected while virus is adapting to the human host or upon evasion from adaptive immune responses. However, pH1N1 has not displayed any major antigenic changes so far. We examined the effect of the amino acid substitutions found to be most frequently occurring in the pH1N1 HA protein before 1 April 2012 on the receptor-binding properties of the virus by using recombinant soluble HA trimers. Two changes (S186P and S188T) were shown to increase the receptor-binding avidity of HA, whereas two others (A137T and A200T) decreased binding avidity. Construction of an HA protein tree revealed the worldwide emergence of several HA variants during the past few influenza seasons. Strikingly, two major variants harbor combinations of substitutions (S186P/A137T and S188T/A200T, respectively) with opposite individual effects on binding. Stepwise reconstruction of the HA proteins of these variants demonstrated that the mutations that increase receptor-binding avidity are compensated for by the acquisition of subsequent mutations. The combination of these substitutions restored the receptor-binding properties (avidity and specificity) of these HA variants to those of the parental virus. The results strongly suggest that the HA of pH1N1 was already optimally adapted to the human host upon its emergence in April 2009. Moreover, these results are in agreement with a recent model for antigenic drift, in which influenza A virus mutants with high and low receptor-binding avidity alternate.
INTRODUCTION
The spread of new animal influenza A viruses (IAVs) in the human population requires specific adaptations in several viral proteins. Clearly, the hemagglutinin (HA) receptor-binding and fusion protein needs to adapt to the new sialic acid (SIA) receptor repertoire present on epithelial cells in the human upper airway (1–3). However, optimal adaptation does not simply translate into the HA protein acquiring high-avidity binding to human receptors. Strong binding by HA of decoy receptors, for example, receptors present on soluble proteins in the mucus, is probably detrimental for virus replication. Also, efficient release of newly assembled viral particles requires a delicate balance between binding of HA to host cell receptors and neuraminidase (NA)-mediated cleavage of this interaction (4–6).
The emergence of the new pandemic H1N1 (pH1N1) virus (7, 8) in 2009 has resulted in the accumulation of sequences of large numbers of pH1N1 virus isolates over a number of consecutive influenza seasons. Thus, for the first time in history, the evolution of a newly emerged IAV spreading in the human population can be followed in great detail. Apart from the possible selection of adaptive changes in HA that result in optimized receptor-binding properties, HA is also under selective pressure by the host immune response. IAV can escape from antibody recognition by the accumulation of amino acid substitutions in the antigenic sites of HA, a process called “antigenic drift” (9), yet testing of thousands of viral isolates did not reveal any considerable changes in the antigenic properties of pH1N1 viruses up to 2012 (10). A systematic analysis of the evolution of the receptor-binding properties of pH1N1 viruses has not yet been performed.
Evidence linking the IAV antigenic drift with changes in HA receptor-binding avidity has recently been presented (11). Whereas single amino acid substitutions are not expected to provide sufficient antigenic change to allow escape from a polyclonal antiserum directed against multiple antigenic epitopes, single amino acid changes can lead to changes in the receptor-binding avidity of the HA protein. As a result of the increased receptor-binding avidity, the neutralizing capacity of immune serum is reduced. Although such mutations in HA are proposed to be selected mainly due to their effect on receptor-binding avidity, they may also slightly affect the antigenic properties of HA, as the receptor-binding site (RBS) partly overlaps the antigenic sites. In naive individuals, however, the increased binding avidity of HA may reduce viral fitness, for instance, by decreased release of newly assembled virions or by enhanced binding to decoy receptors. As a result, subsequent compensatory mutations that reduce binding avidity but also further modify antigenic epitopes on HA are selected. Thus, Yewdell and coworkers have proposed a model for antigenic drift in which influenza A virus mutants with high and low receptor-binding avidity alternate (11).
Here we analyzed all pH1N1 HA sequences deposited in the NCBI influenza virus database before 1 October 2012 and tested the effect of the most frequently occurring amino acid substitutions on HA receptor binding. To this end, recombinant soluble trimeric HA proteins were produced and analyzed using solid-phase binding and hemagglutination assays, as well as glycan array analysis (12–14). The expression of recombinant soluble HA proteins allows the rapid production of HAs encoded by primary isolates and circumvents the need for producing and culturing complete viruses that may incorporate additional changes during propagation in eggs or MDCK cells. In addition to analyzing the effect of single amino acid substitutions, we also identified and tested combinations of substitutions that frequently cosegregate on specific branches of an HA protein tree, especially during the latest flu seasons. Remarkably, single amino acid substitutions displaying opposite effects on binding (increased binding by S186P and S188T, decreased binding by A137T and A200T) occurred in combinations (S186P or S188T together with A137T or A200T, respectively) that had no net effect on binding. Thus, in the first 3 years of pH1N1 evolution, mutations in HA that increase HA receptor-binding avidity, followed by the selection of compensatory mutations that restore binding to the original levels, have been selected. The results strongly suggest that the HA of pH1N1 was already optimally adapted to the human host upon its emergence in April 2009. Furthermore, our results are in agreement with the model of Hensley and coworkers (11), in which transient increases in binding avidity precede antigenic variation.
MATERIALS AND METHODS
Genes, expression vectors, protein expression, and purification.
Codon-optimized HA-encoding cDNA of A/California/07/2009 (Cal07) H1N1 (GenScript) was cloned into the pCD5 expression vector as described previously (13, 15). The resulting expression vector encodes an HA protein containing a heterologous signal peptide, a C-terminal trimerization domain (GCN4), and a streptavidin tag (Streptag-II) and lacking the transmembrane and cytoplasmic domains. All mutant H1 proteins were created by site-directed mutagenesis with a QuikChange site-directed mutagenesis kit (Stratagene, CA). The HA proteins were expressed in HEK293S GnT1(−) cells, purified from the cell culture supernatants using streptavidin beads, and quantified as described previously (13, 15).
Viruses.
A wild-type recombinant virus of strain A/California/04/2009 (Cal04) and an HA substitution mutant (Cal04 T200A) (16) were grown in MDCKII cells in Opti-MEM/GlutaMAX (Gibco) medium containing 2% bovine serum albumin and 1 μg/ml trypsin-TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone) (Sigma-Aldrich). Viruses were inoculated at a multiplicity of infection of 0.01 and harvested after 48 h in four independent batches. Viral particles were quantified by a quantitative reverse transcription-PCR (qPCR) using primers specific for the NP segment, as described previously (17). Briefly, viral RNA was extracted in triplicate for each virus batch using a viral RNA extraction kit (Qiagen). qPCR was performed on 2-fold dilution series of the viral RNA preparations.
HA receptor-binding assays.
Hemagglutination assays were performed with 0.5% human erythrocytes. Hemagglutination assays were performed with HAs precomplexed with mouse monoclonal antibodies against the streptavidin tag and a second polyclonal goat anti-mouse antiserum as described previously (13, 15) or with recombinant virus. Twofold dilutions of precomplexed HA, starting at a concentration of 100 μg/ml HA, were tested in 96-well format. The number of particles present in three independently grown stocks of each of the two recombinant viruses was determined by quantitative PCR as described previously (17). Hemagglutination titers were determined using 1.25-fold dilutions starting with equal amounts of particles. A solid-phase HA-binding assay using fetuin as a ligand and antibody-complexed HA was also performed as described previously (13, 15). HA binding was detected by reading the optical density at 450 nm. Different concentrations of HA were analyzed in the fetuin-binding assay. The data point shown (at 40 μg/ml HA) corresponds to the linear part of the curve.
Glycan array analyses.
Microarrays were printed as described previously (12). The glycan array analysis of the HA proteins was performed as previously described (13). Briefly, 200 μg/ml recombinant HA was precomplexed with a horseradish peroxidase-linked anti-streptavidin tag antibody and an Alexa Fluor 488 antimouse antibody (4:2:1 molar ratio) for 30 min at 0°C, prior to incubation for 90 min on the microarray slide under a microscope cover glass in a humidified chamber at room temperature. After repeated washes with phosphate-buffered saline (PBS) with 0.05% Tween, PBS, and deionized water, the slides were immediately subjected to imaging.
Phylogenetic analysis.
All 7,132 available full-length pH1N1 HA protein sequences were downloaded from the NCBI influenza virus database (18) on 1 October 2012 and used for analysis. All sequences from pH1N1 viruses isolated in seasons IV to VI (349 sequences, after identical sequences were removed) and 200 sequences from selected isolates from seasons I, II, and III each (excluding identical sequences) were aligned and a phylogenetic tree was constructed using the neighbor-joining algorithm in the Unipro UGENE package (19). Aided by the alignment, amino acid substitutions were mapped to branch points in the tree. Terminal branches were grouped into 82 clusters, and amino acid substitutions leading to the earliest branch point of a cluster were considered to define the signature sequence for that cluster. An alignment of all 7,132 sequences was subsequently used to manually assign each individual sequence to 1 of the 82 clusters on the basis of the presence of the signature sequence. A list of GenBank accession numbers of all sequences included in each cluster is provided in Table S1 in the supplemental material. For each cluster, a single sequence with no further amino acid substitutions in addition to the signature sequence was selected for the construction of the phylogenetic tree shown in Fig. 3 (the tree was constructed by use of the neighbor-joining algorithm in the UGENE package).
Fig 3.
Phylogenetic tree based on HA of pH1N1. A phylogenetic tree (obtained by neighbor joining) was constructed using full-length viral HA sequences (GenBank accession numbers are listed in Table S2 in the supplemental material) identical to the signature sequence of the cluster (see Materials and Methods) to which it belongs. Cluster numbers describe three properties. The first number (1 to 4) indicates the flu season (seasons I, II, III, and IV to VI, respectively) in which the cluster was the most prominent (as a percentage of the total number of sequences in a season). The decimal number ranks the clusters in order of size. The clusters dominant in season III (given a prefix of 3) and seasons IV to VI (given a prefix of 4) are colored red and blue, respectively. The number after the underscore indicates the number of sequences present in a cluster (all 7,132 sequences were assigned to a cluster). Substitutions defining the branches are indicated. The substitutions for the Cal04 and Cal07 reference strains are indicated in red and were analyzed for their effect on receptor binding.
Modeling.
The three-dimensional crystal structure of the HA head domain (20) of strain A/California/04/2009 (Protein Data Bank accession number 3UBN) in binary complex with an α2-6-linked trisaccharide [NeuAca(2-6)-Galβ(1-4)-GlcNAc] was used for analysis of the potential effects of introduction of amino acid substitutions at positions 186/137 or 188/200. The SWISS-MODEL program (21) was used to predict the structures of mutants with the T200A (see Fig. 7A and C), S188T (see Fig. 7B), and T200A/S186P/A137T (see Fig. 7D) substitutions. Subsequent energy minimizations were not necessary, as inspection of the modeled structure by GROMOS software revealed no unfavorable energy interactions. None of the predicted structures displayed considerable changes in the average position of the C-α backbone residues when superpositioned on the original structure of the protein with Protein Data Bank accession number 3UBE (the root mean square deviation of superpositioned C-α backbone residues surrounding the receptor-binding site was less than 0.2 Å). Structures were analyzed for the detection of lost or newly formed hydrogen bonds using the Swiss-PdbViewer application (21), as indicated in Fig. 7.
Fig 7.
Structural model of the amino acid substitutions in clade A and B HA proteins. The two pairs of amino acid substitutions that were shown to compensate for the effects on HA binding of each other are circled in red (panels A and B show clade B mutations S188T/A200T; panels C and D show clade A mutations) in the wild type (A, C) and double-substitution mutants (B, D). The receptor trisaccharide NeuAca(2-6)-Galβ(1-4)-GlcNAc is colored in purple (SIA), yellow (Gal), and blue (GlcNAc). The three structural elements that interact with the receptor are indicated in ribbon presentation (white, 130 and 220 loops; purple, 190 helix). Important backbone and side chain structures are also displayed. Two water molecules involved in a hydrogen bond network (green dotted lines) between residue S186, the 220 loop, and the sialic acid are indicated in red. Substitution S186P disrupts this network by clashes (red dotted lines in panel D) between the hydrophobic proline backbone and both water molecules. Substitution A137T leads to the formation of a new hydrogen bond between the threonine side chain hydroxyl group and the sialic acid. Remarkably, formation of this new bond decreases binding avidity, whereas the disruption of the water-supported network by S186P increases binding.
RESULTS
Receptor binding of HA proteins with single substitutions.
In order to get insight into the amino acid substitutions in the pH1N1 HA protein after the introduction of this virus into the human population, all 7,132 full-length HA protein sequences available from the NCBI influenza virus database on 1 October 2012 were aligned and amino acid substitutions were counted. Figure 1 displays the most frequently observed substitutions per half-year flu season (season I was from 1 April 2009 to 30 September 2009; the data for seasons IV to VI were grouped together because of the small number of sequences available from seasons V and VI; no sequences from viruses isolated after 1 April 2012 were available). Fifteen substitutions (relative to the sequence of the A/California/07/2009 [Cal07] reference strain) occurred at a frequency of >5% in the last three flu seasons (seasons IV to VI). The effects on receptor binding of six frequently occurring substitutions in the globular head of HA, which were already frequently detected during season III (D100N, A137T, S186P, S188T, A200T, and S206T), were studied by using a recombinant protein approach in the background of HA from Cal07 (13). The receptor-binding properties of the recombinant soluble HA trimers expressed in HEK293S GnTI(−) cells were examined by analyzing their ability to bind the glycoprotein fetuin, which carries three sialylated N-linked glycans, in a solid-phase enzyme-linked immunosorbent assay. In addition, the HA proteins were analyzed for their ability to agglutinate human erythrocytes (Fig. 2). In agreement with previous results (13), most HA proteins displayed relatively low fetuin binding and hemagglutination titers, which were nevertheless well above background levels. Substitution S206T, which was already very prevalent during the first flu season (74% of isolates in season I; 99.4% in seasons IV to VI; Fig. 1), did not affect HA receptor binding. As all other substitutions occurred in the background of the S206T substitution (Fig. 1; described in Fig. 3 below), the effect of the other substitutions was tested in this background. Substitutions S186P and S188T increased fetuin binding, whereas only S186P gave rise to increased hemagglutination activity. In contrast, substitutions A137T and A200T led to decreased fetuin binding and hemagglutination, whereas D100N had no effect on HA receptor binding. As an additional reference, the receptor binding of the HA protein of A/California/04/2009 (Cal04) (13), which differs from Cal07 only at position 200 (A200T), is also shown in Fig. 2. Again, the A200T substitution negatively affected receptor binding (compare Cal07 and Cal04 in Fig. 2), while receptor binding was not affected by the identity of the residue at position 206 (compare Cal07 S206T/A200T and Cal04 in Fig. 2). Also, when several other substitutions described above were analyzed in the background of S206, very similar results were obtained (data not shown).
Fig 1.
Sequence variation in the HA protein of pH1N1. Amino acid substitutions at every position of HA of pH1N1 (using all 7,132 HA sequences) were counted per half-year flu season (season I, 1 April 2009 to 30 September 2009, etc.). The most frequent substitutions were plotted (y axis, percent occurrence) for each season. The data for seasons IV to VI were grouped together because of the low number of sequences available for seasons V and VI. The locations of the signal peptide (SP), HA stem, and HA head domains are indicated at the bottom (the drawing is not to scale). The most frequent mutations in the head domain of HA in season III that still prevailed in seasons IV to VI are shaded in black.
Fig 2.
Receptor-binding avidity of HA proteins with single amino acid substitutions. The effect of the most frequently occurring amino acid substitutions on the binding avidity of soluble HA trimers of pH1N1 was determined by solid-phase binding assay (40 μg/ml HA on fetuin) (OD450, optical density at 450 nm) (A) and hemagglutination (human erythrocytes) (B). HA from strain A/California/07/2009 (Cal07) was used as the template for the introduction of the substitutions indicated on the x axis. (C) The effect of a single amino acid substitution (T200A) on the binding avidity of virus particles was determined by hemagglutination using strain A/California/04/2009 (Cal04) and a recombinant virus of this strain carrying substitution T200A in HA. Equal numbers of viral particles were used on the basis of quantification by quantitative PCR. The average result of four experiments (each performed in triplicate) with independently grown viral stocks is shown. The hemagglutination of the two viruses was significantly different (P < 0.05) (B). Error bars show standard deviations.
Using reverse genetics, we next determined whether the relatively small differences in receptor binding detected with the soluble HA trimers would also be observed if hemagglutination was carried out with viruses containing the same mutation. Similar to the findings for HA trimers carrying the A200T substitution, which were invariably 2-fold less efficient in hemagglutination, a pH1N1 virus containing substitution A200T was also found to be significantly less efficient in hemagglutination (1.6-fold, P < 0.05; Fig. 2B). Thus, small differences in receptor-binding avidity detected by using recombinant soluble HA trimers can also be observed when hemagglutination is performed with virus particles.
Phylogenetic analysis of pH1N1.
A phylogenetic tree of pH1N1 HA was constructed in order to determine the evolutionary pathways along which the substitutions described above have been introduced into the virus population. Previously, phylogenetic analysis of pH1N1 detected only limited variation in the HA protein in the first 2 seasons (22). In later studies, which were mostly restricted to analysis of sets of sequences obtained from a specific country or region, segregation into specific clades was described (23–28). To avoid the effects of sampling bias in time and place, a consensus phylogenetic tree of the HA protein was constructed on the basis of all 7,132 sequences. Individual sequences were grouped into clusters on the basis of signature sequences determined for each cluster. Substitutions defining the signature sequences were mapped to branch points in the HA phylogenetic tree (Fig. 3). The number of sequences in each cluster was determined, and the relative frequency at which a cluster was identified in a flu season was plotted (Fig. 4). The major clusters present in season III (indicated in red in Fig. 3) and seasons IV to VI (indicated in blue in Fig. 3) formed four clades (clades A to D) in the phylogenetic tree. Amino acid substitution A200T (which decreased binding avidity) was selected several times along different branches (as was the case for several other substitutions, e.g., E377K). A200T was already detected in one of the first isolates recovered in 2009 (reference strain A/California/04/2009) but became dominant (present in 52% of all virus isolates obtained after 30 September 2010) only after its introduction along another branch in flu season III (clade B in Fig. 3). Interestingly, the phylogenetic analysis shows that A200T was obtained by clade B viruses after the acquisition of substitution S188T, which was shown to enhance avidity, as described above. Other viruses in clade B obtained the D100N rather than the A200T substitution after having gained S188T. Also in clade A, substitutions which had opposite effects on receptor-binding avidity (S186P and A137T) were found to cosegregate. However, for clade A viruses, too few sequences were available to unequivocally determine the temporal order of their acquisition.
Fig 4.
Distribution of HA pH1N1 clusters per flu season. Results for clusters that contained more than 5% of the total number of sequences in any of the seasons are plotted (the y axis indicates the percentage of sequences in a cluster relative to the total number of sequences in a season; e.g., cluster 1.1 harbors 47% of the total number of sequences in season I [green bar], 31% of the sequences in season II [red bar], but only 2% of the sequences in season III [yellow bar]). Clusters are sorted by cluster number on the x axis (numbering is according to that used for the phylogenetic tree and is explained in the legend to Fig. 3). Clusters belonging to clade A or B (Fig. 3) are indicated.
Receptor binding of mutant HA proteins with multiple substitutions.
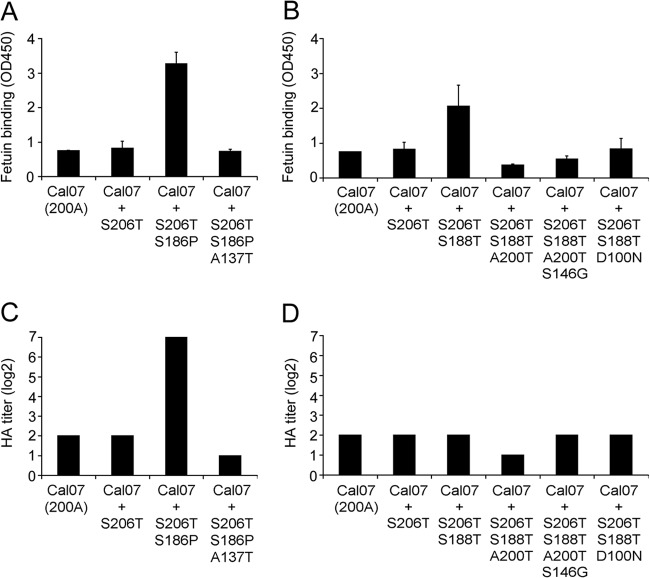
To evaluate the development of HA receptor-binding characteristics along the evolutionary branches present in clades A and B, mutations were sequentially introduced into the HA head domain putatively involved in receptor binding (indicated in red in Fig. 3). Analysis of the receptor-binding avidity of the resulting recombinant proteins (Fig. 5) shows that the dramatic increase in binding avidity caused by substitution S186P (clade A) was counteracted by substitution A137T. A similar result was observed for clade B. The increased receptor binding caused by a single amino acid substitution (S188T) was followed by subsequent substitutions (A200T or D100N) that reduced HA receptor binding to values similar to (D100N) or slightly lower than (A200T) those observed for pH1N1 strain Cal07. The strain in the branch in clade A containing substitutions S188T/A200T subsequently acquired an additional substitution (S146G) which restored hemagglutination to the level of that for the pH1N1 Cal07 strain.
Fig 5.
Receptor-binding avidity of HA proteins of clades A and B with multiple amino acid substitutions. The effect of amino acid substitutions consecutively occurring during the evolution of clades A (A, C) and B (B, D) on the binding avidity of soluble HA trimers of pH1N1 was determined by a solid-phase binding assay (40 μg/ml HA on fetuin) (A, B) and hemagglutination (human erythrocytes) (C, D). HA from strain A/California/07/2009 (Cal07) was used as the template for the introduction of the substitutions indicated on the x axis.
Glycan array analysis.
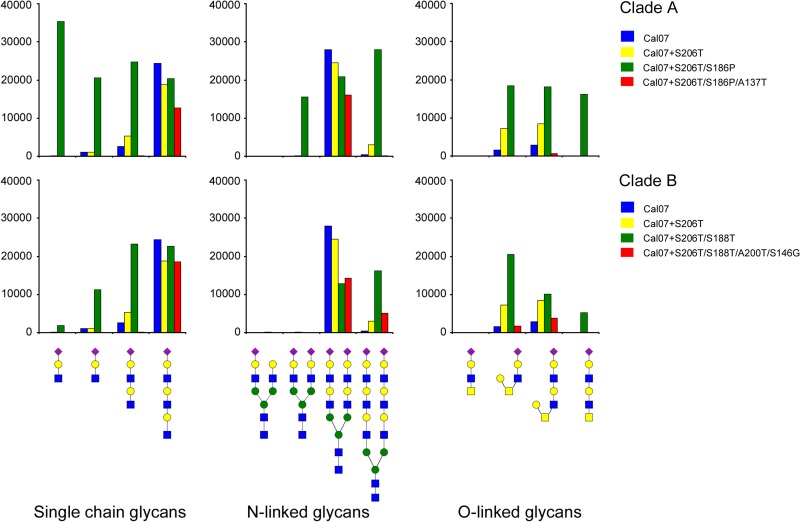
The results presented above demonstrate that amino acid substitutions that are introduced along the branches of clade B and, possibly, clade A lead to a temporary increase in binding avidity, followed by the acquisition of compensatory mutations that restore binding to the original levels. Slight differences in the magnitude of the effects were observed when the results of hemagglutination and the solid-phase binding assay were compared. Whereas fetuin is known to carry three N-linked glycans, erythrocytes have a much more complex glycosylation pattern. To determine to what extent the substitutions in clades A and B affect receptor specificity, we made use of an extended version of the custom glycan array previously described (20). Several poly-LacNAc glycans were added to this extended version, as these specific structures have been shown to be preferentially bound by human and swine H1 hemagglutinins (13, 29, 30). Trimeric HAs of pH1N1 strain Cal07 bound efficiently only to poly-LacNAc structures terminated by a 2-6-linked sialic acid (Fig. 6, blue bars). The addition of substitution S206T (Cal07 S206T) had little effect, except for allowing some binding to two O-linked glycans to be gained (Fig. 6, yellow bars). In contrast, introduction of substitution S186P (Cal07 S206T/S186P, clade A) or S188T (Cal07 S206T/S188T, clade B), which resulted in increased fetuin binding and/or hemagglutination titers, largely expanded the set of glycans that could be bound (Fig. 6, green bars). The HA protein carrying the S206T and S186P substitutions appeared to be an especially efficient binder of short single-LacNAc-containing glycans, although this protein also efficiently bound to short O-linked glycans. In complete agreement with the results obtained by hemagglutination and fetuin-binding assays, the addition of successive substitutions (Cal07 S206T/S188T/A200T/S146G and Cal07 S206T/S186P/A137T) restored the glycan binding profile (Fig. 6, red bars) to the pattern that was observed with the initial pH1N1 Cal07 strain (Fig. 6, blue bars). We conclude that binding specificity, similar to binding avidity, is only temporarily altered by amino acid substitutions due to the rapid acquisition of compensatory substitutions which restore binding to the levels and in the patterns already observed for the first isolates of 2009.
Fig 6.
Receptor-binding specificity of HA proteins of clades A and B with multiple amino acid substitutions. Precomplexed HA trimers were applied to a custom-made glycan array. The effect of amino acid substitutions that were introduced into wild-type (Cal07) HA in a stepwise reconstruction of clades A and B was examined. The structures of glycans (arranged in separate panels for N-linked glycans, O-linked glycans, and single-chain glycans attached by an ethyl linker to the array) that were bound (relative fluorescence units are indicated on the y axes) are shown below the panels. Binding was strictly specific for glycans carrying α2-6-linked sialic acids (indicated by purple diamonds in the glycan structure) attached to a β1-4-linked galactose (yellow circles). Blue squares, N-acetylglucosamine; yellow squares, N-acetylgalactosamine; green circles, mannose.
NA coevolution.
It has been reported that IAV neuraminidase activity and receptor binding by HA require a delicate balance for optimal replication (4–6). Thus, changes in one of these proteins may induce or be the consequence of changes in the other protein. Of the 7,132 HA sequences that we analyzed, 4,864 were matched by a full-length NA sequence and were screened for the presence of coevolutionary patterns between HA and NA in clades A and B (Table 1). HA of clade B was associated with NA substitutions N44S, V241I, and N396K. Substitutions V241I and N396K were tightly linked to the introduction of substitution S188T in HA (cluster 4.7). Ninety-two percent of the sequences in cluster 4.7 and up to 100% of the sequences in clusters that evolved from cluster 4.7 were associated with V241I/N396K in NA, whereas this combination was rarely found in any other cluster. The introduction of A200T in HA clade A (cluster 3.2) was tightly associated (92%) with the introduction of substitution N44S in NA. Clusters evolving from cluster 3.2 were up to 100% associated with N44S/V241I/N396K in NA. This combination of mutations in NA was found only 5 times outside clade A (possibly as a consequence of reassortment with clade A viruses). Clade A was 100% associated with substitution V394I in NA. This substitution was not strongly associated with any other cluster.
Table 1.
Coevolution of substitution patterns in HA and NA
| Clustera | Substitution(s) in HA | No. of sequences in each cluster | No. (%) of sequences with the indicated mutation in NA |
||
|---|---|---|---|---|---|
| N44S | V241I | N396K | |||
| 3.8 | S454N | 14 | 1 (7) | 4 (29) | 3 (21) |
| 4.15 | S454N, E377K | 7 | 0 (0) | 1 (14) | 2 (29) |
| 4.7 | S454N, E377K, S188T | 39 | 2 (5) | 36 (92) | 37 (95) |
| 4.2 | S454N, E377K, S188T, D100N | 77 | 0 (0) | 74 (96) | 75 (97) |
| 3.2 | S454N, E377K, S188T, A200T | 55 | 49 (92) | 50 (94) | 51 (96) |
| 4.9 | S454N, E377K, S188T, A200T, I463T | 19 | 19 (100) | 19 (100) | 19 (100) |
| 4.1 | S454N, E377K, S188T, A200T, S146G | 86 | 82 (95) | 84 (98) | 84 (98) |
| 4.5 | S454N, E377K, S188T, A200T, S146G, L-7M | 33 | 33 (100) | 32 (97) | 33 |
| 4.1 | S454N, E377K, S188T, A200T, S146G, N263D | 12 | 12 (100) | 12 (100) | 12 (100) |
Modeling of substitutions in the pH1N1 structure.
Substitution pairs S188T/A200T and S186P/A137T were modeled into the crystal structure of pH1N1 (strain Cal04) in complex with the trisaccharide NeuAca(2-6)-Galβ(1-4)-GlcNAc (20). In Fig. 7, the 130 loop, 190 helix, and 220 loop which make up the RBS are shown. Residues 188 and 200, substitution of which caused enhanced (S188T) or decreased (A200T) binding avidity, are located at the surface of HA on the backside of the 190 α helix that is involved in receptor binding. Residue 190D in the helix forms a hydrogen bond with the GlcNAc of the sialylated ligand and is crucial for the recognition of α2-6-linked sialic acid-containing receptors. The side chain and backbone hydroxyl groups of residue S188 form a hydrogen network (including a water molecule) with the backbone of residues 189A and 190D (Fig. 7A). Addition of a solvent-exposed hydrophobic methyl group caused by substitution S188T (Fig. 7B) likely affects this hydrogen network and presumably caused the observed increase in binding avidity. Substitution A200T (Fig. 7B) resulted in the creation of a hydrogen bond between the hydroxyl group in the side chain of 200T and the amino group in the side chain of 191Q. We conclude that changing the interactions with the 190 α helix at opposing sides of crucial residue 190D keeps the interactions with the sialoside receptor in balance. Residues 186 and 137 are located directly in the receptor-binding site. Serine 186 interacts via a hydrogen bond network (including water molecules) with the sialic acid moiety and the 220 loop (Fig. 7C). Substitution S186P disrupted this network completely (Fig. 7D). Substitution A137T led to the formation of a hydrogen bond between the side chain hydroxyl group of 137T with the sialic acid moiety. We conclude that the absence and the presence of direct interactions with the sialic acid moiety maintain receptor binding by the S186P/A137T mutant HA protein at a level comparable to that of the first pH1N1 isolates in 2009.
DISCUSSION
The zoonotic transfer of IAV from swine to human led to the new H1N1 pandemic in April 2009. This offered an excellent opportunity to follow the early evolution of the viral HA protein in its new host. Two selective forces are expected to drive the evolution of this protein: optimizing HA binding to the new receptor repertoire and escaping from adaptive immune responses. Phylogenetic analysis of the HA protein (Fig. 3) showed that several clades have become prominent during the latest flu seasons. Making use of recombinant soluble HA trimers, we analyzed the cumulative effects of consecutive single amino acid substitutions in the HA head domain during the evolution of two of the major clades. Remarkably, for both clades, a transient increase in binding avidity with a broadening of receptor specificity was observed. This was rapidly followed by the accumulation of fully compensatory substitutions that restored the binding avidity and specificity to the initial levels.
We identified several substitutions in the HA protein of pH1N1 that affected binding of recombinant soluble HA trimers to sialosides. Modeling of the mutations in a structure model of pH1N1 HA showed that changes in receptor binding are likely explained by the effect of these mutations on hydrogen bond formation either with the 190 helix or with the sialylated glycan directly. Importantly, mutations in HA that affected receptor binding, as detected with the recombinant protein approach, were also shown to affect receptor binding in the context of complete virus particles (13, 16; this study). Several of the substitutions that affected receptor binding in vitro were shown by others to affect the pathogenicity or transmission of pH1N1 viruses in animal models (16, 31), demonstrating that changes in receptor binding identified with our recombinant protein approach are of biological significance in vivo, but the specific impact on viral phenotypes of the HA substitutions studied here, with the exception of A200T, remains to be determined.
The rapid accumulation of mutations that compensate for the increase in receptor-binding avidity caused by preceding mutations suggests that the binding properties of the HA of the earliest isolates of pH1N1 were already optimal for infection of the human host and have likely evolved as such in the swine host. NeuAca2-6 polylactosamine-containing glycans are the receptors that are the most efficiently bound by pH1N1 HA. Glycan analysis of primary swine respiratory epithelial cells (29) and the human respiratory tract (3) has shown that these glycans are abundantly present in both hosts. Optimal adaptation of HA binding to human receptors in the swine host is therefore plausible. In a previous study, we showed that two mutations in HA of pH1N1 (strain Cal04), including T200A, that result in enhanced binding (13) also decrease transmission in a ferret model (16). Decreased transmission may explain the selection of compensatory mutations that restore the binding properties of HA to their original values.
The optimal receptor-binding properties of a particular HA in a specific host are also determined by the specific activity of the accompanying NA protein. pH1N1 resulted from reassortment between a triple-reassortant swine H1N1 virus (contributing HA) and a Eurasian H1N1 swine virus (contributing NA) (32, 33). The resulting combination of an HA exhibiting low avidity for glycan receptors and weak NA enzymatic activity was shown to provide the functional balance between HA and NA required for efficient replication and transmission of pH1N1 in humans (6). In agreement with the notion that the receptor-binding properties of HA need to be balanced by the enzymatic activity of the NA protein to achieve optimal replication and transmission, our analysis indicates the presence of coevolutionary patterns of substitutions in the HA and NA proteins of pH1N1. Future studies will be needed to evaluate whether the NA substitutions coselected with specific HAs are associated with changes in the specific activity of NA.
Why are mutations that increase receptor binding selected if they are rapidly followed by compensatory mutations that decrease receptor binding again? Our observation may be explained by the model of Hensley et al. (11). In this model, on the basis of results obtained with alternating passages of IAV in immune and naive mice, IAV initially escapes from neutralizing antibody pressure by the selection of mutations that enhance binding avidity. Single amino acid substitutions usually induce only very limited antigenic change in HA, and their selection is therefore not readily driven by a polyclonal antibody response. However, mutations that enhance receptor-binding avidity may improve escape from a polyclonal antibody response and therefore lead to their selection in an increasingly immune population. Mutations that restore binding properties are subsequently selected in naive individuals. The accumulation of mutations affecting receptor binding ultimately leads to a more extensive antigenic change due to the overlap of antigenic sites and the receptor-binding site. In agreement with this model, we have revealed multiple occurrences of mutation pairs (S188T/T200A, S188T/D100N, and, possibly, S186P/A137T) that increase and subsequently decrease the receptor-binding avidity of HA during the early evolution of pH1N1. Only a single event of a sequential increase and a subsequent decrease of binding avidity was detected along each branch, and thus, extensive antigenic change is not yet expected. Indeed, an extensive study of the antigenic properties of a large set of pH1N1 viruses isolated from March to August 2011 (season V) revealed only small antigenic differences compared to the original Cal07 virus (10). Furthermore, these differences could not be attributed to any specific branch of an evolutionary tree of the season V viruses, which corresponded well to the phylogenetic tree shown in Fig. 3.
The evolution of HA appears to be driven by a complex interplay between HA, NA, host cell receptors, and innate and adaptive immune responses. This complex interplay collectively determines the pathogenicity and transmission of IAV and the ability of the virus to maintain itself in or to gain access to a certain host population. In the current genomics era, it will be of interest to link the enormous amounts of genomics data on IAVs to the phenotypic properties of the viral proteins in order to gain new insights into the evolution of these pathogens and to be able to better predict their biological properties.
Supplementary Material
ACKNOWLEDGMENTS
This work has been financially supported by an NWO-ALW grant from the Netherlands Organization for Scientific Research (NWO) to C. A. M. de Haan and by NIH grant AI058113 to J. C. Paulson. R. P. de Vries is a recipient of a Rubicon grant from NWO. Several glycans used for HA binding assays were provided by the Consortium for Functional Glycomics (http://www.functionalglycomics.org/), funded by NIGMS grant GM62116. This work has also been partially supported by the Center for Research on Influenza Pathogenesis (CRIP) and the NIAID-funded Center of Excellence for Influenza Research and Surveillance (CEIRS; contract HHSN266200700010C) to A. Garcia-Sastre.
Footnotes
Published ahead of print 9 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01955-13.
REFERENCES
- 1.Glaser L, Stevens J, Zamarin D, Wilson IA, Garcia-Sastre A, Tumpey TM, Basler CF, Taubenberger JK, Palese P. 2005. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J. Virol. 79:11533–11536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 74:8502–8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walther T, Karamanska R, Chan RW, Chan MC, Jia N, Air G, Hopton C, Wong MP, Dell A, Malik Peiris JS, Haslam SM, Nicholls JM. 2013. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 9:e1003223. 10.1371/journal.ppat.1003223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, Kawaoka Y. 2000. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J. Virol. 74:6015–6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner R, Matrosovich M, Klenk HD. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 12:159–166 [DOI] [PubMed] [Google Scholar]
- 6.Xu R, Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, Wilson IA. 2012. Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. J. Virol. 86:9221–9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605–2615 [DOI] [PubMed] [Google Scholar]
- 8.Neumann G, Noda T, Kawaoka Y. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Air GM, Laver WG, Webster RG. 1987. Antigenic variation in influenza viruses. Contrib. Microbiol. Immunol. 8:20–59 [PubMed] [Google Scholar]
- 10.Klimov AI, Garten R, Russell C, Barr IG, Besselaar TG, Daniels R, Engelhardt OG, Grohmann G, Itamura S, Kelso A, McCauley J, Odagiri T, Smith D, Tashiro M, Xu X, Webby R, Wang D, Ye Z, Yuelong S, Zhang W, Cox N. 2012. WHO recommendations for the viruses to be used in the 2012 Southern Hemisphere influenza vaccine: epidemiology, antigenic and genetic characteristics of influenza A(H1N1)pdm09, A(H3N2) and B influenza viruses collected from February to September 2011. Vaccine 30:6461–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hensley SE, Das SR, Bailey AL, Schmidt LM, Hickman HD, Jayaraman A, Viswanathan K, Raman R, Sasisekharan R, Bennink JR, Yewdell JW. 2009. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 326:734–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. 2004. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U. S. A. 101:17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Vries RP, de Vries E, Moore KS, Rigter A, Rottier PJ, de Haan CA. 2011. Only two residues are responsible for the dramatic difference in receptor binding between swine and new pandemic H1 hemagglutinin. J. Biol. Chem. 286:5868–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevens J, Blixt O, Paulson JC, Wilson IA. 2006. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat. Rev. Microbiol. 4:857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Vries RP, de Vries E, Bosch BJ, de Groot RJ, Rottier PJ, de Haan CA. 2010. The influenza A virus hemagglutinin glycosylation state affects receptor-binding specificity. Virology 403:17–25 [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Romero C, de Vries E, Belicha-Villanueva A, Mena I, Tscherne DM, Gillespie VL, Albrecht RA, de Haan CA, Garcia-Sastre A. 2013. Substitutions T200A and E227A in the hemagglutinin of pandemic 2009 influenza A virus increase lethality but decrease transmission. J. Virol. 87:6507–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Widjaja I, de Vries E, Tscherne DM, Garcia-Sastre A, Rottier PJ, de Haan CA. 2010. Inhibition of the ubiquitin-proteasome system affects influenza A virus infection at a postfusion step. J. Virol. 84:9625–9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okonechnikov K, Golosova O, Fursov M. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167 [DOI] [PubMed] [Google Scholar]
- 20.Xu R, McBride R, Nycholat CM, Paulson JC, Wilson IA. 2012. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J. Virol. 86:982–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- 22.Galiano M, Agapow PM, Thompson C, Platt S, Underwood A, Ellis J, Myers R, Green J, Zambon M. 2011. Evolutionary pathways of the pandemic influenza A (H1N1) 2009 in the UK. PLoS One 6:e23779. 10.1371/journal.pone.0023779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barr IG, Cui L, Komadina N, Lee RT, Lin RT, Deng Y, Caldwell N, Shaw R, Maurer-Stroh S. 2010. A new pandemic influenza A(H1N1) genetic variant predominated in the winter 2010 influenza season in Australia, New Zealand and Singapore. Euro Surveill. 15(42):pii=19692 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19692 [DOI] [PubMed] [Google Scholar]
- 24.Dapat IC, Dapat C, Baranovich T, Suzuki Y, Kondo H, Shobugawa Y, Saito R, Suzuki H. 2012. Genetic characterization of human influenza viruses in the pandemic (2009-2010) and post-pandemic (2010-2011) periods in Japan. PLoS One 7:e36455. 10.1371/journal.pone.0036455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreira JL, Borborema SE, Brigido LF, Oliveira MI, Paiva TM, Santos CL. 2011. Sequence analysis of the 2009 pandemic influenza A H1N1 virus haemagglutinin gene from 2009-2010 Brazilian clinical samples. Mem. Inst. Oswaldo Cruz 106:613–616 [DOI] [PubMed] [Google Scholar]
- 26.Kao CL, Chan TC, Tsai CH, Chu KY, Chuang SF, Lee CC, Li ZR, Wu KW, Chang LY, Shen YH, Huang LM, Lee PI, Yang C, Compans R, Rouse BT, King CC. 2012. Emerged HA and NA mutants of the pandemic influenza H1N1 viruses with increasing epidemiological significance in Taipei and Kaohsiung, Taiwan, 2009-10. PLoS One 7:e31162. 10.1371/journal.pone.0031162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ledesma J, Pozo F, Reina G, Blasco M, Rodriguez G, Montes M, Lopez-Miragaya I, Salvador C, Reina J, Ortiz de Lejarazu R, Egido P, Lopez Barba J, Delgado C, Cuevas MT, Casas I. 2012. Genetic diversity of influenza A(H1N1)2009 virus circulating during the season 2010-2011 in Spain. J. Clin. Virol. 53:16–21 [DOI] [PubMed] [Google Scholar]
- 28.Strengell M, Ikonen N, Ziegler T, Julkunen I. 2011. Minor changes in the hemagglutinin of influenza A(H1N1)2009 virus alter its antigenic properties. PLoS One 6:e25848. 10.1371/journal.pone.0025848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bateman AC, Karamanska R, Busch MG, Dell A, Olsen CW, Haslam SM. 2010. Glycan analysis and influenza A virus infection of primary swine respiratory epithelial cells: the importance of NeuAc{alpha}-2-6 glycans. J. Biol. Chem. 285:34016–34026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, Sasisekharan V, Sasisekharan R. 2008. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat. Biotechnol. 26:107–113 [DOI] [PubMed] [Google Scholar]
- 31.Ye J, Sorrell EM, Cai Y, Shao H, Xu K, Pena L, Hickman D, Song H, Angel M, Medina RA, Manicassamy B, Garcia-Sastre A, Perez DR. 2010. Variations in the hemagglutinin of the 2009 H1N1 pandemic virus: potential for strains with altered virulence phenotype? PLoS Pathog. 6:e1001145. 10.1371/journal.ppat.1001145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.