Abstract
A decisive step in a virus infection cycle is the recognition of a specific receptor present on the host cell surface, subsequently leading to the delivery of the viral genome into the cell interior. Until now, the early stages of infection have not been thoroughly investigated for any virus infecting hyperthermophilic archaea. Here, we present the first study focusing on the primary interactions between the archaeal rod-shaped virus Sulfolobus islandicus rod-shaped virus 2 (SIRV2) (family Rudiviridae) and its hyperthermoacidophilic host, S. islandicus. We show that SIRV2 adsorption is very rapid, with the majority of virions being irreversibly bound to the host cell within 1 min. We utilized transmission electron microscopy and whole-cell electron cryotomography to demonstrate that SIRV2 virions specifically recognize the tips of pilus-like filaments, which are highly abundant on the host cell surface. Following the initial binding, the viral particles are found attached to the sides of the filaments, suggesting a movement along these appendages toward the cell surface. Finally, we also show that SIRV2 establishes superinfection exclusion, a phenomenon not previously described for archaeal viruses.
INTRODUCTION
Viruses infecting Archaea constitute an integral, yet unique part of the virosphere. In particular, a significant portion of the viruses infecting hyperthermophilic archaea display morphotypes—bottle shaped, lemon shaped, droplet shaped, etc.—not known to be associated with the other two cellular domains, Bacteria and Eukarya (1–3). Furthermore, the distinctiveness of archaeal viruses extends to their genome sequences (4, 5) and the structure of proteins that they encode (1). The ways these viruses interact with their hosts are therefore also likely to be unique. However, until now, the studies on archaeal viruses were mostly confined to biochemical and genetic characterization of their virions, and the knowledge on virus-host interplay in Archaea is minuscule compared to the wealth of data available on bacterial and eukaryotic systems. In particular, insights are lacking into the entry process of hyperthermophilic archaeal viruses.
Recognition of a suitable host cell is an essential first step in the infection cycle of any virus. This is typically achieved by specific interactions between a receptor-binding protein exposed on the virion and a receptor present on the host cell surface, which subsequently leads to cell envelope penetration, accompanied by internalization of the viral genome (6). A variety of cell surface structures are known to be targeted by viruses. For example, in the case of bacterial viruses, nearly all components of the cell envelope are known to serve as receptors (7), including lipopolysaccharide (8–10), pili (11–13), flagella (14–17), (lipo-)teichoic acids (18–20), peptidoglycan (21), or various integral membrane proteins (22–24). The only archaeal virus for which a potential cellular receptor has been identified is ϕCh1, infecting the hyperhalophilic host Natrialba magadii (25). Notably, ϕCh1 is a member of the viral order Caudovirales (26), sharing clear evolutionary history with tailed double-stranded DNA (dsDNA) bacteriophages (27). Similarly to bacterial viruses, ϕCh1 utilizes tail fibers to bind to its cellular receptor. Galactose moieties were found to be important for ϕCh1 adsorption; however, the exact nature of the receptor remains to be identified (25).
Sulfolobus islandicus rod-shaped virus 2 (SIRV2) (28) and its host, Sulfolobus islandicus LAL14/1 (29), represent a valuable model system to study virus-host interactions in Archaea (30). SIRV2 is a member of the family Rudiviridae, within the recently established order Ligamenvirales (31). It has a nonenveloped, stiff, rod-shaped virion composed of four structural proteins encasing a linear dsDNA genome of 35 kb (28). Both termini of the virion are decorated with three fibers composed of the minor structural protein P1070 (32) and thought to be involved in host recognition. At the end of the infection cycle, SIRV2 induces the formation of large pyramidal structures on the surface of infected cells that serve as portals for the release of progeny viruses (33–35). A similar egress mechanism has been also demonstrated for the unrelated icosahedral archaeal virus STIV (36, 37), indicating that mechanisms underlying virus-host interactions in a particular virus-host system are sometimes applicable to a wider range of archaeal viruses. In contrast to the egress mechanism, which has been characterized to some detail, almost nothing is known about the entry process of SIRV2. Here, we investigate the SIRV2-S. islandicus interaction, focusing on the early stages of infection.
MATERIALS AND METHODS
Strain cultivation and virus purification.
Sulfolobus islandicus strain LAL14/1 (29) was used as a host for SIRV2 in all experiments. The cells were cultivated with aeration (150 rpm) in a water bath shaker, Innova 3100 (Eppendorf), filled with silicon oil, in 50-ml flasks at 75°C, pH 3.5. The rich medium was prepared as described previously (38). SIRV2 was purified by CsCl density gradient centrifugation and the virus titer determined by plaque assay as described previously (28).
Adsorption assay.
For adsorption assays, LAL14/1 cells (optical density at 600 nm [OD600] = 0.15; 108 CFU/ml) were infected using a multiplicity of infection (MOI) of 0.1. At defined time intervals, a sample of infected culture was removed and the adsorption stopped by immediate centrifugation (10,000 × g, 5 min, room temperature [RT]). The number of remaining PFU was determined by the plaque assay and compared to the amount of virus present in a cell-free control incubated at 75°C. The adsorption rate constant was calculated as described previously (39).
Receptor saturation assay.
For the receptor saturation assay, a constant number of LAL14/1 cells (grown to a cell density of 108 CFU/ml) were infected using MOIs between 0.1 and 370. At 30 min postinfection, the cells were removed by centrifugation (10,000 × g, 5 min, RT) and the number of nonadsorbed viral particles in the supernatants was determined using the plaque assay and compared to the amount of virus present in a cell-free control incubated at 75°C.
Superinfection assay.
LAL14/1 cells were infected at an MOI of 10 for 1 h. Cells were washed twice with fresh rich medium in two rounds of gentle centrifugation (Jouan BR4i, rotor AB 50.10A [Thermo Scientific]; 3,500 rpm, 10 min, 20°C). Infected cells were subjected to a second round of infection using an MOI of 0.1, and the number of unadsorbed particles remaining in the supernatant was determined as described above.
Filament purification.
S. islandicus filaments were purified as described previously (40) with minor modifications. Briefly, 3 liters of LAL14/1 cells were grown to an OD600 of 0.3, collected (Avanti J-26XP, rotor JLA 16.250 [Beckman Coulter]; 3,500 rpm, 15 min, 15°C) and resuspended in 15 ml of Brock's basal salt medium. Filaments were mechanically sheared from the cells by vortexing at maximum speed for 15 min. Cells and debris were removed by two steps of centrifugation (Jouan BR4i rotor AB 50.10A; Thermo Scientific): 8,000 rpm for 30 min at 15°C, followed by 6,000 rpm for 1 h at 15°C. The filaments were pelleted (Beckman rotor 70.1Ti at 65,000 rpm, 1 h, 15°C) and further purified on a CsCl gradient (Beckman rotor SW60, 55,000 rpm, 48 h, 15°C). Gradient fractions containing filaments were collected and processed for transmission electron microscopy (TEM).
Interaction of SIRV2 with cellular appendages.
SIRV2 virions were incubated with either S. islandicus LAL14/1 cells (MOI = 10) or purified filaments in a thermoblock (Fisher Scientific) at 75°C for 1 to 2 or 5 to 10 min, respectively, and immediately prepared for TEM.
TEM.
For conventional negative-stain TEM, 10 μl of sample was added on Formvar-coated grids (Eloïse Instruments Service SARL) for 2 min, air dried, and stained with 2% uranyl acetate (EuroMedex) for 30 s. Samples were imaged in a JEOL 1200EX-II transmission electron microscope at 80 kV.
Electron cryotomography.
A suspension of LAL14/1 cells infected with SIRV2 was mixed (1 min postinfection) with an equal amount of 10 nm gold fiducial markers (Aurion). Three microliters of this mixture was added to a glow-discharged R2/2 Quantifoil grid and rapidly plunged into liquid ethane.
Samples were transferred into a FEI TITAN Krios transmission electron microscope at liquid nitrogen temperature. The microscope was equipped with a field emission gun operated at 300 kV. Zero-loss filtered tilt series were collected on a 4x4k Gatan charge-coupled-device (CCD) camera in a range of −60° to + 60° in steps of 1.5° or 2° and a defocus of 8 to 9 μm. The magnification was chosen to give a pixel size of 0.477 nm in the final image. Tomograms were generated with the IMOD software (41) and denoised by nonlinear anisotropic diffusion (NAD) (42). Tomograms were segmented and surface rendered using Amira (Mercury systems).
RESULTS AND DISCUSSION
SIRV2 adsorption is very rapid.
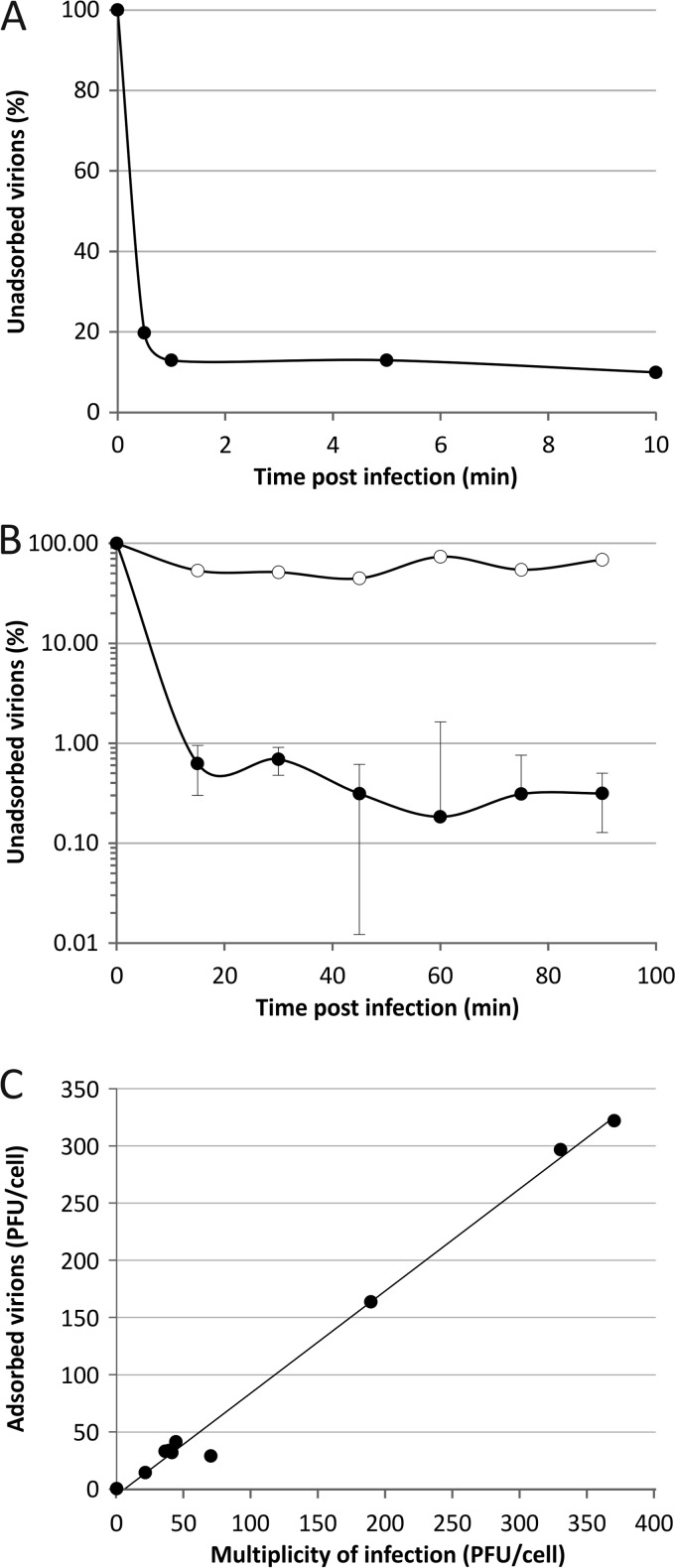
To gain insights into the initial stages of SIRV2 entry, we followed the kinetics of SIRV2 adsorption to LAL14/1 cells. The adsorption was very efficient, with ∼80% of virions being bound to cells within the first 30 s of infection (Fig. 1A). Further incubation of the virus in the presence of the host cells resulted in additional virion binding; ∼99% of virions were bound within 20 to 30 min postinfection (p.i.). All adsorption assays were conducted under the conditions optimal for the growth of S. islandicus cells, i.e., at high temperature (75°C) and in acidic pH (pH 3.5). The possibility that the observed effects were due to high-temperature- and/or acid-induced virion inactivation rather than adsorption was eliminated by performing a cell-free control in which the same amount of SIRV2 as used for the infection was incubated at 75°C in the LAL14/1 growth medium.
Fig 1.
Adsorption of SIRV2 to cells of S. islandicus LAL14/1. (A) Kinetics of SIRV2 adsorption. Cells were infected with SIRV2 using an MOI of 0.1 at 75°C. The number of unbound virus particles was determined at different time points postinfection as described in Materials and Methods. (B) SIRV2-mediated superinfection exclusion. Cells were infected at an MOI of 10 for 1 h, washed twice with rich medium, and subjected to a second round of infection using an MOI of 0.1. The number of unadsorbed particles remaining in the supernatant was determined. The kinetics of adsorption to noninfected, control cells is shown by closed circles, while open circles represent adsorption to preinfected cells. All experiments were conducted in triplicate, and error bars represent standard deviations. When error bars are not visible, the deviation was below 5%. (C) Receptor saturation. Cells were infected with SIRV2 using MOIs ranging from 0.1 to 370. At 30 min postinfection, the number of unadsorbed viral particles present in the supernatant was determined by plaque assay and compared to that of the cell-free control.
Upon the first encounter of a susceptible host, many bacterial viruses initially bind reversibly to the structures on the cell surface and only then commit to the infection by attaching to the cell irreversibly, a stage subsequently followed by delivery of the viral genome into the cell interior (8, 21, 43–45). To test the reversibility of the SIRV2 adsorption, we investigated whether viral particles could be washed off from the host cell surface. However, no virus particles could be released at any of the time points tested (starting with 1 min postinfection), suggesting that SIRV2 binding very quickly becomes irreversible and the reversible step, if it occurs, is transient.
Such a rapid adsorption rate (calculated as 2 × 10−8 ml min−1 at 1 min p.i.) is surprising, given that viruses of halophilic archaea—the only group of archaeal viruses for which adsorption has been studied—often bind to their hosts extremely slowly (46). For example, only 30% of salterprovirus His1 (47, 48) and Haloarcula hispanica tailed virus 1 (HHTV-1; Siphoviridae) (46) virions adsorb in 3 h. The fast adsorption of SIRV2 is consistent with recent transcriptomics data revealing that transcription of the early SIRV2 genes starts within 1 min of infection (49). Despite the fact that SIRV2 adsorption is quick, the intracellular phase of the viral cycle is fairly long (∼10 to 15 h). We hypothesize that the duration of both stages has been fine-tuned during SIRV2's evolution to minimize the time spent by the virus in the hostile extracellular environment, i.e., high temperature and acidic pH.
SIRV2 establishes superinfection exclusion.
In the case of many bacterial virus-host systems, virus infection of a cell renders it resistant to subsequent infections by related viruses—a phenomenon known as superinfection exclusion (50). Although relatively widespread among bacterial viruses, to the best of our knowledge, superinfection exclusion has not been described for any archaeal virus. We investigated whether SIRV2 infection modulates the susceptibility of its host to subsequent infections. For this purpose, LAL14/1 cells were preinfected with SIRV2 at an MOI of 10 for 1 h, to ensure that infection was established in all cells. After removing the unadsorbed viral particles, the cells were challenged with a second course of infection at an MOI of 0.1. We observed a dramatic decrease in the amount of virions bound to preinfected cells compared to the noninfected control cells (Fig. 1B). Infected cells were no longer able to efficiently adsorb the virus even after 90 min of incubation. This result suggests that upon infection SIRV2 establishes superinfection exclusion; the exact mechanism underlying this phenomenon remains to be elucidated.
SIRV2 receptor is highly abundant.
The abundance and nature of cell surface molecules that serve as receptors for virions are specific for each virus-host system. The receptor saturation assay is a classical experiment used to determine the approximate number of receptors present on the host cell surface (39). For this purpose, S. islandicus cells were infected with SIRV2 at various MOI values ranging from 0.1 to 370 and the number of free particles remaining in the supernatant was determined and compared to the initial number of virions added into the cell-free control (Fig. 1C). Remarkably, even at the highest MOI tested, 95% of virions were bound 30 min p.i., indicating that the receptor mediating the primary interaction between SIRV2 and LAL14/1 is highly abundant, consistently with the high adsorption rate.
SIRV2 binds to long filaments on S. islandicus cells.
To gain more-detailed insight into the interaction of SIRV2 with LAL14/1 cells, we followed the initial stages of infection by transmission electron microscopy and whole-cell electron cryotomography (cryo-ET). The surface of noninfected LAL14/1 cells was covered with pilus-like filaments. The number of filaments varied between individual cells. With negative staining, the filament diameter was close to 10 nm but the length was highly variable. In cryo-ET, the filaments appeared thinner, with a diameter of 5 nm. Negative staining involves dehydration and flattening of the sample on the carbon support film, which may cause the filaments to appear thicker in projection. In contrast, for cryo-ET the filaments are rapidly frozen in their native, fully hydrated state. Cryo-ET thus shows the actual in situ structure and dimensions of the filaments. The exact nature of the filaments remains to be determined.
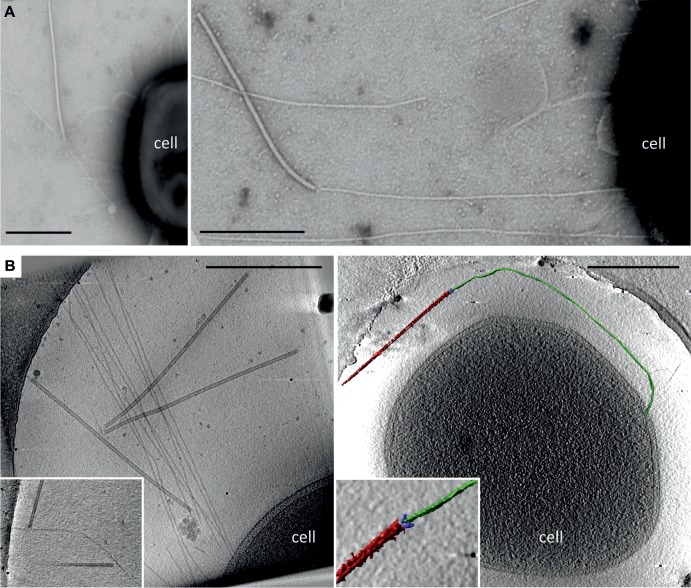
It has been previously observed that SIRV2 virions copurify with filamentous structures (51). To verify whether the structures on the surface of S. islandicus cells might be involved in SIRV2 binding, the cells were infected at an MOI of 10 and observed by TEM and cryo-ET. Indeed, an interaction between SIRV2 virions and the cellular filaments was observed (Fig. 2). The interaction involved the terminal fibers of the SIRV2 virion. Typically, a single filament accommodated several SIRV2 virions (Fig. 2B). This is consistent with the receptor saturation experiment, which indicated that nearly 370 particles could adsorb per one LAL14/1 cell.
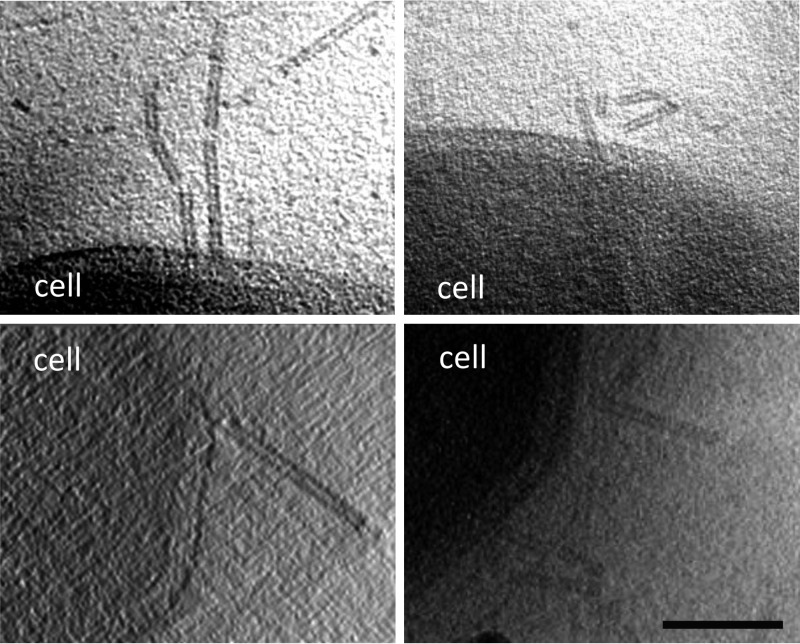
Fig 2.
Electron micrographs of SIRV2 interaction with S. islandicus LAL14/1 cells. Samples were collected 1 min postinfection and negatively stained for TEM (A) or plunge-frozen for electron cryotomography (cryo-ET) (B). The virions interact both at the filament tips (right panels) and along the length of the filaments (left panels). The inset in the lower left panel depicts two virions bound to the sides of a single filament. The lower right panel shows a segmented tomographic volume of the SIRV2 virion (red) attached to the tip of an S. islandicus filament (green). The three terminal virion fibers that appear to mediate the interaction are shown in blue (the inset depicts a magnified view of the interaction between the virion fibers and the tip of the filament). A complete tomogram of the cell depicted in the lower right panel can be found in the supplemental material. Scale bars, 500 nm.
SIRV2 specifically interacts with the tips of the cellular filaments.
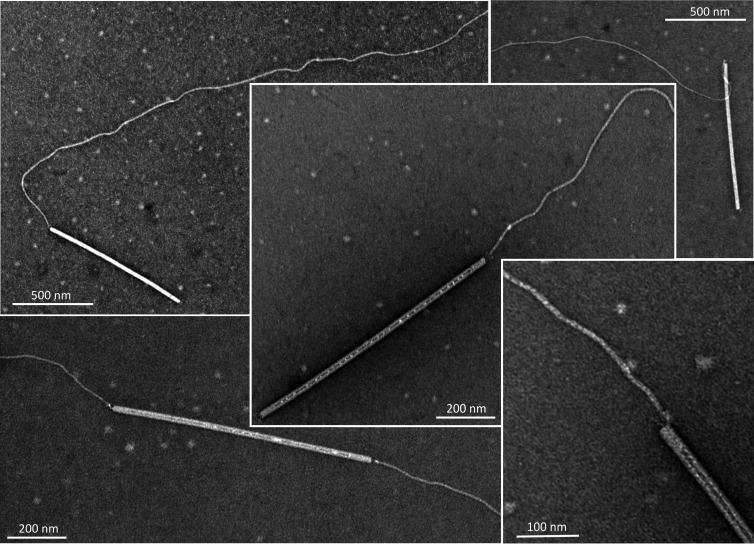
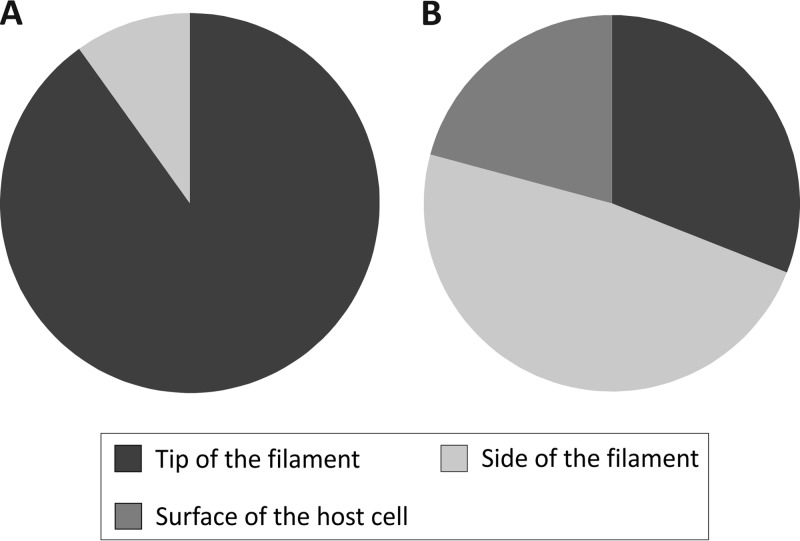
To verify that the filaments indeed represent SIRV2 receptors, the fibers were removed from noninfected LAL14/1 cells by vortexing and purified on CsCl density gradient (see Materials and Methods). The filament preparations were then tested for SIRV2 binding. TEM analysis revealed that virions preferentially interact with the tip of the filaments (Fig. 3); of the 202 observed interactions (in 4 independent experiments), only 9.9% (20 interactions) occurred along the length of a filament (Fig. 4A). In some cases, the two termini of a rod-shaped virion were bound to two different filaments (Fig. 3), indicating that the two ends of the virion are functionally equivalent. Unexpectedly, this preference was not observed with filaments attached to LAL14/1 cells, where the virions typically bound not only to the tip but also to the sides of the filaments (Fig. 2) and were also observed at the cell surface. Of the 629 observed cases of virus-host interactions (in 6 independent experiments), 31% (195) occurred with the tips and 48.2% (303) with the sides of the LAL14/1 filaments, while the remaining 20.8% (131) of virions were found to interact directly with the cell surface (Fig. 4B).
Fig 3.
Transmission electron micrographs of SIRV2 interaction with purified cellular filaments. The filaments were removed from S. islandicus LAL14/1 cells as described in Materials and Methods.
Fig 4.
Interactions between SIRV2 and purified filaments (A) or S. islandicus LAL14/1 cells (B). SIRV2 virions were incubated with purified filaments for 5 to 10 min or LAL14/1 cells for 1 to 2 min at 75°C and prepared for TEM (see Materials and Methods). Binding of viral particles to the tips, the sides of LAL14/1 filaments, or the cell surface was counted in electron micrographs of negatively stained samples.
Concluding remarks.
Collectively, our data provide valuable insight into the entry process of SIRV2 and suggest the following sequence of events. SIRV2 binds to the tip of the filament with its three terminal fibers (Fig. 2 and 3) and subsequently progresses along the filaments toward the cell surface. Interestingly, in one case we observed virions bound to the tip of a 12.5-μm-long filament, which raises the question of how the virus overcomes such a long distance to reach the cell body. Once the SIRV2 virion reaches the cell surface, it disassembles, presumably as the viral DNA is delivered to the cell interior. Although the last step remains enigmatic, partially disassembled virions that we observed by cryo-ET at the cell surface postinfection (Fig. 5) are consistent with such a process. Many bacterial viruses utilize filamentous cellular appendages, such as pili or flagella, as primary receptors (7). Superficially, the adsorption of SIRV2 to LAL14/1 filaments resembles the interaction of filamentous Ff inoviruses with F-pili. The pIII protein of Ff phages binds to the tip of the F-pilus; subsequent retraction of the pilus brings the virion close to the cell surface, where upon binding the secondary receptor, TolA, the viral genome is translocated into the cytoplasm (12). A similar F-pilus retraction-driven entry has been also described for certain single-stranded RNA (ssRNA) phages of the family Leviviridae (13). However, the apparent translocation of SIRV2 virions along the LAL14/1 filaments, as judged from the differential binding of the virions to purified filaments (Fig. 4A) and filaments attached to LAL14/1 cells (Fig. 4B), implies that the mechanism of SIRV2 entry might differ from that employed by pilus-specific bacterial viruses. Indeed, no retracting pili have been identified in archaea so far, which is also consistent with the apparent lack of genes encoding typical retraction ATPases in the archaeal pilus operons (52, 53). A retraction-independent mechanism is utilized by flagellotrophic bacteriophages, which instead harness the energy of flagellar rotation to move along the flagellum toward the cell surface (14, 16, 17). Notably, the flagella (called archaella in archaea) of Sulfolobus are considerably thicker (∼14 nm in diameter [54]) than the LAL14/1 filaments to which SIRV2 binds. Whether the mechanism of SIRV2 translocation along the filaments is related to that of flagellotrophic bacteriophages is under investigation. The work described here provides the basis for future studies, which should illuminate the mechanistic details of SIRV2 cell entry.
Fig 5.
A tomographic slice through S. islandicus LAL14/1 cells 1 min after infection with SIRV2 reveals partially disassembled SIRV2 virions at the cell surface. Scale bar, 100 nm.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Agence Nationale de la Recherche (ANR) program BLANC, project EXAVIR. E.R.J.Q. was supported by a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche of France and the Université Pierre et Marie Curie, Paris, France.
Footnotes
Published ahead of print 2 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02742-13.
REFERENCES
- 1.Krupovic M, White MF, Forterre P, Prangishvili D. 2012. Postcards from the edge: structural genomics of archaeal viruses. Adv. Virus Res. 82:33–62 [DOI] [PubMed] [Google Scholar]
- 2.Pina M, Bize A, Forterre P, Prangishvili D. 2011. The archeoviruses. FEMS Microbiol. Rev. 35:1035–1054 [DOI] [PubMed] [Google Scholar]
- 3.Prangishvili D. 2013. The wonderful world of archaeal viruses. Annu. Rev. Microbiol. 67:565–585 [DOI] [PubMed] [Google Scholar]
- 4.Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. 2011. Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol. Mol. Biol. Rev. 75:610–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prangishvili D, Garrett RA, Koonin EV. 2006. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 117:52–67 [DOI] [PubMed] [Google Scholar]
- 6.Poranen MM, Daugelavicius R, Bamford DH. 2002. Common principles in viral entry. Annu. Rev. Microbiol. 56:521–538 [DOI] [PubMed] [Google Scholar]
- 7.Vinga I, São-José C, Tavares P, Santos MA. 2006. Bacteriophage entry in the host cell, p 163–203 In Wêgrzyn G. (ed), Modern bacteriophage biology and biotechnology. Research Signpost, Kerala, India [Google Scholar]
- 8.Daugelavičius R, Cvirkaitė V, Gaidelytė A, Bakienė E, Gabrėnaitė-Verkhovskaya R, Bamford DH. 2005. Penetration of enveloped double-stranded RNA bacteriophages phi13 and phi6 into Pseudomonas syringae cells. J. Virol. 79:5017–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inagaki M, Kawaura T, Wakashima H, Kato M, Nishikawa S, Kashimura N. 2003. Different contributions of the outer and inner R-core residues of lipopolysaccharide to the recognition by spike H and G proteins of bacteriophage phiX174. FEMS Microbiol. Lett. 226:221–227 [DOI] [PubMed] [Google Scholar]
- 10.Molineux IJ. 2001. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 40:1–8 [DOI] [PubMed] [Google Scholar]
- 11.Bamford DH, Palva ET, Lounatmaa K. 1976. Ultrastructure and life cycle of the lipid-containing bacteriophage phi 6. J. Gen. Virol. 32:249–259 [DOI] [PubMed] [Google Scholar]
- 12.Rakonjac J, Bennett NJ, Spagnuolo J, Gagic D, Russel M. 2011. Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 13:51–76 [PubMed] [Google Scholar]
- 13.Shiba T, Miyake T. 1975. New type of infectious complex of E. coli RNA phage. Nature 254:157–158 [DOI] [PubMed] [Google Scholar]
- 14.Yen JY, Broadway KM, Scharf BE. 2012. Minimum requirements of flagellation and motility for infection of Agrobacterium sp. strain H13-3 by flagellotropic bacteriophage 7-7-1. Appl. Environ. Microbiol. 78:7216–7222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi Y, Shin H, Lee JH, Ryu S. 2013. Identification and characterization of a novel flagellum-dependent Salmonella-infecting bacteriophage, iEPS5. Appl. Environ. Microbiol. 79:4829–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerrero-Ferreira RC, Viollier PH, Ely B, Poindexter JS, Georgieva M, Jensen GJ, Wright ER. 2011. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 108:9963–9968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuel AD, Pitta TP, Ryu WS, Danese PN, Leung EC, Berg HC. 1999. Flagellar determinants of bacterial sensitivity to chi-phage. Proc. Natl. Acad. Sci. U. S. A. 96:9863–9866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young FE, Smith C, Reilly BE. 1969. Chromosomal location of genes regulating resistance to bacteriophage in Bacillus subtilis. J. Bacteriol. 98:1087–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia G, Corrigan RM, Winstel V, Goerke C, Grundling A, Peschel A. 2011. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J. Bacteriol. 193:4006–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raisanen L, Schubert K, Jaakonsaari T, Alatossava T. 2004. Characterization of lipoteichoic acids as Lactobacillus delbrueckii phage receptor components. J. Bacteriol. 186:5529–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaidelytė A, Cvirkaite-Krupovic V, Daugelavičius R, Bamford JK, Bamford DH. 2006. The entry mechanism of membrane-containing phage Bam35 infecting Bacillus thuringiensis. J. Bacteriol. 188:5925–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakutytė L, Baptista C, São-José C, Daugelavičius R, Carballido-Lopez R, Tavares P. 2011. Bacteriophage infection in rod-shaped gram-positive bacteria: evidence for a preferential polar route for phage SPP1 entry in Bacillus subtilis. J. Bacteriol. 193:4893–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letellier L, Boulanger P, Plancon L, Jacquot P, Santamaria M. 2004. Main features on tailed phage, host recognition and DNA uptake. Front. Biosci. 9:1228–1339 [DOI] [PubMed] [Google Scholar]
- 24.Randall-Hazelbauer L, Schwartz M. 1973. Isolation of the bacteriophage lambda receptor from Escherichia coli. J. Bacteriol. 116:1436–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein R, Rossler N, Iro M, Scholz H, Witte A. 2012. Haloarchaeal myovirus phiCh1 harbours a phase variation system for the production of protein variants with distinct cell surface adhesion specificities. Mol. Microbiol. 83:137–150 [DOI] [PubMed] [Google Scholar]
- 26.Klein R, Baranyi U, Rossler N, Greineder B, Scholz H, Witte A. 2002. Natrialba magadii virus phiCh1: first complete nucleotide sequence and functional organization of a virus infecting a haloalkaliphilic archaeon. Mol. Microbiol. 45:851–863 [DOI] [PubMed] [Google Scholar]
- 27.Krupovic M, Forterre P, Bamford DH. 2010. Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J. Mol. Biol. 397:144–160 [DOI] [PubMed] [Google Scholar]
- 28.Prangishvili D, Arnold HP, Gotz D, Ziese U, Holz I, Kristjansson JK, Zillig W. 1999. A novel virus family, the Rudiviridae: structure, virus-host interactions and genome variability of the Sulfolobus viruses SIRV1 and SIRV2. Genetics 152:1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaubert C, Danioux C, Oberto J, Cortez D, Bize A, Krupovic M, She Q, Forterre P, Prangishvili D, Sezonov G. 2013. Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open Biol. 3:130010. 10.1098/rsob.130010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prangishvili D, Koonin EV, Krupovic M. 2013. Genomics and biology of Rudiviruses, a model for the study of virus-host interactions in Archaea. Biochem. Soc. Trans. 41:443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prangishvili D, Krupovic M. 2012. A new proposed taxon for double-stranded DNA viruses, the order “Ligamenvirales”. Arch. Virol. 157:791–795 [DOI] [PubMed] [Google Scholar]
- 32.Steinmetz NF, Bize A, Findlay KC, Lomonossoff GP, Manchester M, Evans DJ, Prangishvili D. 2008. Site-specific and spatially controlled addressability of a new viral nanobuilding block: Sulfolobus islandicus rod-shaped virus 2. Adv. Funct. Mater. 18:3478–3486 [Google Scholar]
- 33.Bize A, Karlsson EA, Ekefjard K, Quax TE, Pina M, Prevost MC, Forterre P, Tenaillon O, Bernander R, Prangishvili D. 2009. A unique virus release mechanism in the Archaea. Proc. Natl. Acad. Sci. U. S. A. 106:11306–11311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quax TE, Krupovic M, Lucas S, Forterre P, Prangishvili D. 2010. The Sulfolobus rod-shaped virus 2 encodes a prominent structural component of the unique virion release system in Archaea. Virology 404:1–4 [DOI] [PubMed] [Google Scholar]
- 35.Quax TE, Lucas S, Reimann J, Pehau-Arnaudet G, Prevost MC, Forterre P, Albers SV, Prangishvili D. 2011. Simple and elegant design of a virion egress structure in Archaea. Proc. Natl. Acad. Sci. U. S. A. 108:3354–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brumfield SK, Ortmann AC, Ruigrok V, Suci P, Douglas T, Young MJ. 2009. Particle assembly and ultrastructural features associated with replication of the lytic archaeal virus Sulfolobus turreted icosahedral virus. J. Virol. 83:5964–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snyder JC, Brumfield SK, Peng N, She Q, Young MJ. 2011. Sulfolobus turreted icosahedral virus c92 protein responsible for the formation of pyramid-like cellular lysis structures. J. Virol. 85:6287–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, Lanzendorfer M, Kristjansson JK. 1994. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst. Appl. Microbiol. 16:609–628 [Google Scholar]
- 39.Adams MH. 1959. Bacteriophages. Interscience Publishers, Inc., New York, NY [Google Scholar]
- 40.Lassak K, Neiner T, Ghosh A, Klingl A, Wirth R, Albers SV. 2012. Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 83:110–124 [DOI] [PubMed] [Google Scholar]
- 41.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 42.Frangakis AS, Hegerl R. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 135:239–250 [DOI] [PubMed] [Google Scholar]
- 43.Cvirkaite-Krupovic V, Krupovic M, Daugelavičius R, Bamford DH. 2010. Calcium ion-dependent entry of the membrane-containing bacteriophage PM2 into its Pseudoalteromonas host. Virology 405:120–128 [DOI] [PubMed] [Google Scholar]
- 44.Hu B, Margolin W, Molineux IJ, Liu J. 2013. The bacteriophage T7 virion undergoes extensive structural remodeling during infection. Science 339:576–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jakutytė L, Lurz R, Baptista C, Carballido-Lopez R, São-José C, Tavares P, Daugelavičius R. 2012. First steps of bacteriophage SPP1 entry into Bacillus subtilis. Virology 422:425–434 [DOI] [PubMed] [Google Scholar]
- 46.Kukkaro P, Bamford DH. 2009. Virus-host interactions in environments with a wide range of ionic strengths. Environ. Microbiol. Rep. 1:71–77 [DOI] [PubMed] [Google Scholar]
- 47.Pietilä MK, Atanasova NS, Oksanen HM, Bamford DH. 2013. Modified coat protein forms the flexible spindle-shaped virion of haloarchaeal virus His1. Environ. Microbiol. 15:1674–1686 [DOI] [PubMed] [Google Scholar]
- 48.Bath C, Dyall-Smith ML. 1998. His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J. Virol. 72:9392–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quax TE, Voet M, Sismeiro O, Dillies MA, Jagla B, Coppee JY, Sezonov G, Forterre P, van der Oost J, Lavigne R, Prangishvili D. 2013. Massive activation of archaeal defense genes during viral infection. J. Virol. 87:8419–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu MJ, Henning U. 1994. Superinfection exclusion by T-even-type coliphages. Trends Microbiol. 2:137–139 [DOI] [PubMed] [Google Scholar]
- 51.Zillig W, Prangishvilli D, Schleper C, Elferink M, Holz I, Albers S, Janekovic D, Gotz D. 1996. Viruses, plasmids and other genetic elements of thermophilic and hyperthermophilic Archaea. FEMS Microbiol. Rev. 18:225–236 [DOI] [PubMed] [Google Scholar]
- 52.Albers SV, Driessen AJ. 2005. Analysis of ATPases of putative secretion operons in the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology 151:763–773 [DOI] [PubMed] [Google Scholar]
- 53.Albers SV, Pohlschroder M. 2009. Diversity of archaeal type IV pilin-like structures. Extremophiles 13:403–410 [DOI] [PubMed] [Google Scholar]
- 54.Albers SV, Meyer BH. 2011. The archaeal cell envelope. Nat. Rev. Microbiol. 9:414–426 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.