Abstract
Measles virus (MV) infection causes an acute childhood disease that can include infection of the central nervous system and can rarely progress to severe neurological disease for which there is no specific treatment. We generated potent antiviral peptide inhibitors of MV entry and spreading and MV-induced cell fusion. Dimers of MV-specific peptides derived from the C-terminal heptad repeat region of the MV fusion protein, conjugated to cholesterol, efficiently protect SLAM transgenic mice from fatal MV infection. Fusion inhibitors hold promise for the prophylaxis of MV infection in unvaccinated and immunocompromised people, as well as potential for the treatment of grave neurological complications of measles.
INTRODUCTION
Measles virus (MV) is one of the most infectious microorganisms known and continues to cause extensive morbidity and mortality worldwide. Despite the availability of a vaccine and the measles initiative launched by WHO, UNICEF, and their partners to increase vaccine coverage, MV has not been eradicated and has caused 140,000 deaths globally as recently as 2010 (1), making it one of the top causes of death among vaccine-preventable diseases. The measles incidence in North America has increased in recent years, with hundreds of confirmed cases in 2011. European eradication of MV is also far behind the expected deadlines, and numerous outbreaks have occurred during the last few years (2, 3). While vaccination is a priority for the control of measles, it alone may not be sufficient (2, 4) and should be complemented by the use of antiviral therapy to restrict virus dissemination (4).
MV infection causes an acute febrile respiratory illness with a skin rash and may cause acute, profound suppression of the immune system. The neurological sequelae of measles can occur within days to years after acute MV infection, often resulting in severe disability and death (5, 6). Acute postinfectious encephalomyelitis occurs primarily in older children and adults during or shortly after acute measles, and subacute sclerosing panencephalitis (SSPE) is a late neurodegenerative complication associated with the persistent infection of brain cells (7).
MV belongs to the Paramyxoviridae family, and its lipid envelope carries the two glycoproteins directly involved in viral entry and pathogenesis: a fusion protein (F) and a receptor-binding protein (H). The MV fusion (F) protein, like other paramyxovirus F proteins belonging to the group of “class I” fusion proteins (8–14), is synthesized as a precursor protein that is proteolytically processed posttranslationally to form a trimer of disulfide-linked heterodimers. This cleavage event exposes a new N terminus, called the fusion peptide, which is essential for membrane fusion activity. To initiate infection, the receptor-binding protein (H) binds to cellular surface receptors and activates the viral F protein to undergo the required conformational changes leading to fusion. The F protein passes through a transient extended intermediate form and inserts its fusion peptide into the target cell membrane before refolding upon itself to attain its postfusion conformation in a series of steps that drive membrane merger (8, 9, 15, 16).
Several cell surface receptors have been shown to interact with MV: CD46 in laboratory MV strains, CD150 (or SLAM) in both wild-type (WT) and laboratory MV strains, and nectin-4, which promotes viral egress from the respiratory tract (17, 18). It is not yet known how MV enters the brain. It has been suggested that specific MV H receptors may not be necessary for the MV-induced central nervous system (CNS) manifestations, since the virus seems to spread without budding, implicating direct cell-to-cell and transsynaptic transmission (19–21). The presence of F protein and alterations of its fusion phenotype have been associated with severe CNS infections (22); if F-mediated membrane fusion is blocked, viral spread between neurons is halted (23). These findings suggest that it may be possible to halt CNS infection by targeting the F protein and its function.
Peptides derived from either N- or C-terminal heptad repeat (HR) regions (HRN and HRC, respectively) of paramyxovirus F proteins can interfere with the structural rearrangements required for viral fusion during infection (24–28). The current paradigm for the mechanism of HR-derived peptide action is that HRC peptides bind to the postulated extended intermediate state of F, after the fusion peptide has been inserted into the target membrane, and prevent the transition to the postfusion conformation (15). The efficacy of peptide inhibition depends on both the strength of the interaction of the peptide with the target fusion protein and the temporal window of access to the target sequence (29, 30). We used structure-based design to improve the strength of the peptide-target interaction, and we conjugated a cholesterol group to the peptides to increase the inhibitor concentration at the location of receptor binding (30, 31); in this way, we obtained a potent fusion inhibitor that prevented and treated lethal Nipah virus (NiV) encephalitis in vivo (30). Recently, we showed that peptide efficacy against NiV, human parainfluenza virus type 3 (HPIV3), and human immunodeficiency virus type 1 (HIV-1) can also be improved by combining cholesterol conjugation with dimerization of the HRC peptide (32). Here, we report that cholesterol-conjugated dimeric HRC peptides derived from MV F can effectively inhibit MV fusion, block viral spread, and prevent MV infection both ex vivo in brain explants and in vivo in an established animal model of MV encephalitis.
MATERIALS AND METHODS
Plasmids and reagents.
The genes for the WT MV G954 H and F proteins were codon optimized, synthesized, and subcloned into the mammalian expression vector pCAGGS.
Peptide synthesis.
All peptides were produced by standard 9-fluorenylmethoxy carbonyl solid-phase methods. The cholesterol moieties were attached to the peptides via chemoselective reaction between the thiol group of an extra cysteine residue added C terminally to the sequence and a bromoacetyl derivative of cholesterol or a bis-maleimide (bis-MAL)-functionalized cholesterol core as previously described (30–32).
Transient expression of genes for H and F proteins.
Transfections of 293T cells were performed according to the Lipofectamine 2000 manufacturer's protocols (Invitrogen).
Cells and viruses.
293T (human kidney epithelial) cells and Vero-SLAM (African green monkey kidney) cells were grown in Dulbecco's modified Eagle's medium (GIBCO; Invitrogen) supplemented with 10% fetal bovine serum (FBS) and antibiotics in 5% CO2. The Vero-SLAM culture medium was supplemented with Geneticin. WT MV strain G954 (genotype B3.2) was isolated in Gambia in 1993 (33). Recombinant MV IC323 expressing enhanced green fluorescent protein (EGFP), MV-IC323-EGFP (34), was kindly provided by Y. Yanagi (Kyushu University, Fukuoka, Japan). Both virus strains were propagated and titrated on Vero-SLAM cells.
Viral entry assay.
Vero-SLAM monolayer cells were incubated with WT MV G954 in the presence of various concentrations of peptides. After 90 min, 2× minimal essential medium containing 10% FBS was mixed with 1% Avicel and added to the dishes. The plates were then incubated at 37°C for 72 h. After removal of the medium overlay, the cells were immunostained for plaque detection. The plaques in the control (no peptide) and experimental wells were counted under a dissecting stereoscope.
Viral spread assay.
Vero-SLAM monolayer cells were incubated with WT MV-IC323-EGFP. After 90 min, the virus was aspirated and medium with various concentrations of peptides was added. After 72 h, fluorescence was read with a SpectraMax M5 plate reader (35).
β-Gal complementation-based fusion assay.
The β-galactosidase (β-Gal) complementation-based fusion assay was performed as described previously (36, 37). Briefly, 293T cells transiently transfected with SLAM and the omega reporter subunit were incubated with cells coexpressing MV glycoproteins H and F and the alpha reporter subunit (30).
Preparation of brain explants, peptide treatment, and MV infection.
Organotypic slices were obtained from SLAM transgenic (TG) murine hippocampi as previously described (38). Briefly, the hippocampi were isolated from the brains of 7- to 10-day-old mice and cut with a McIlwain tissue chopper (WPI-Europe) to obtain 350-μm-thick progressive slices. The brain slices were then dissociated in cold Hanks balanced salt solution buffer and deposited on Millipore cell culture insert membranes (Millicell cell culture insert, 30 mm, hydrophilic polytetrafluoroethylene). They were subsequently cultured in GlutaMAX minimal essential medium supplemented with 25% horse serum, 5 g/liter of glucose, 1% HEPES, and 1 mg/ml of human recombinant insulin at 37°C in 5% CO2 in a humidified atmosphere. The medium was changed 24 h later and then every 2 days. Infection of organotypic brain explants was performed after 2 days of culture by placing 104 PFU of MV-IC323-EGFP on the center of the hippocampal slices. Medium containing HRC peptides at the indicated concentrations was added 5 h before infection. Time-lapse progression of the infection was followed with an inverted fluorescence microscope (Zeiss) and camera (AxioCam; Zeiss).
Immunofluorescence.
Hippocampal slices infected with MV-IC323-EGFP for 3 days as described above were stained with the indicated antibodies (Abs) and analyzed by confocal microscopy. Briefly, after being blocked and permeabilized in phosphate-buffered saline (PBS)–4% FBS–0.3% Triton X-100, sections were sequentially incubated with a primary Ab overnight at 4°C and with a secondary Ab for 2 h at room temperature (RT). The primary Abs used were anti-glial fibrillary acidic protein (GFAP) rabbit polyclonal serum (Z0334; Dako), anti-microtubule-associated protein 2 (MAP-2) rabbit polyclonal IgG (H300/sc-20172; Santa Cruz), and an anti-MV F-specific monoclonal Ab (clone Y503) (39).
For the biodistribution study, 7-day-old suckling SLAM TG mice were injected subcutaneously (s.c.) with a monomeric or dimeric peptide. After 8 h, their brains were harvested and frozen with cold isopentane on dry ice. The cryosections were dried for 30 min and fixed in a 4% formalin solution. After multiple washes in PBS, saturation was performed with PBS–4% FBS (30 min at RT) before incubation with a specific rabbit anti-MV HRC Ab (overnight at 4°C) in PBS–4% FBS. After multiple washes, tissue sections were incubated with the secondary goat anti-rabbit Ab conjugated with Alexa 488 in PBS–4% FBS (2 h, RT). The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). After multiple washes in PBS, mounting was performed with Fluoroprep (bioMérieux).
Brains of infected suckling mice were harvested when neurological signs of infection were observed. Brain hemispheres were separated and fixed in a 4% formalin solution for 48 h at 4°C, embedded in a 30% sucrose solution overnight, and frozen with cold isopentane on dry ice. The cryosections were dried for 30 min and fixed with a 4% formalin solution. After multiple washes in PBS, the tissue sections were saturated and permeabilized in PBS–4% FBS–0.3% Triton X-100 (30 min at RT) and then incubated with an anti-nucleocapsid Ab conjugated with biotin (clone 120-biotin) (40, 41) in PBS–4% fetal calf serum–0.3% Triton X-100 (overnight at RT). After multiple washes, tissue sections were incubated with Alexa 647-conjugated streptavidin in PBS–4% FBS (at RT for 2 h). The nuclei were counterstained with DAPI. After multiple washes in PBS, mounting was performed with Fluoroprep (bioMérieux).
Brain sections were analyzed with an Axioplan 2 imaging microscope (Zeiss) and a confocal spectral SP5 microscope (Leica) on the imagery platform PLATIM (IFR128 BioSciences Lyon-Gerland).
Viral load quantification by reverse transcription-qPCR.
Brain explants were harvested 3 days after infection. RNA extraction and reverse transcription-quantitative PCR (qPCR) were performed as previously described (42). The MV N-specific forward and reverse primers used were GTG ATC AAA GTG AGA ATG AGC and GCT GAC CTT CGA CTG TCC T, respectively, as described before (43, 44), and the glyceraldehyde 3-phosphate dehydrogenase forward and reverse primers used were GCAT GGC CTT CCG TGT CC and TGT CAT CAT ACT TGG CAG GTT TCT, respectively. Calculations were performed as previously described (42).
Biodistribution analysis.
Suckling SLAM TG mice (three per group) were injected s.c. with 6 mg/kg of MV HRC2, MV HRC4, or the vehicle in 50 μl of water (Aguettant). After 8 h, their brains were collected, frozen in isopentane, and kept at −80°C until cryosection and staining. In further experiments, to obtain more material for analysis, 3- to 5-week-old mice (3 animals per group) received MV HRC4 (6 mg/kg) in 200 μl of water (Aguettant) s.c. or intranasally (i.n.) in 20 μl of diluent. At each time point, blood was collected by intracardiac puncture in EDTA Vacutainer tubes and sera were conserved at −20°C until their use in enzyme-linked immunosorbent assays (ELISAs). Organs were collected from each animal and conserved at −80°C. The protocol was approved by the Regional Ethical Committee (CECCAPP protocol ENS-2012-041).
ELISA.
Each organ was weighed and mixed in PBS (1:1, wt/vol) with an Ultra-Turrax homogenizer. Samples were then treated with acetonitrile–1% trifluoroacetic acid (1:4, vol/vol) for 1 h on a rotor at 4°C and then centrifuged for 10 min at 8,000 rpm. Supernatant fluids were collected and tested in an ELISA for determination of the peptide concentration. MaxiSorp 96-well plates (Nunc) were coated overnight with purified rabbit anti-MV F HRC Abs (5 μg/ml) in carbonate/bicarbonate buffer, pH 7.4. Plates were washed twice with PBS and then incubated with 3% BSA in PBS (blocking buffer) for 30 min. The blocking buffer was replaced with 2 dilutions of each sample in PBS–3% BSA in duplicate and incubated for 90 min at RT. After multiple washes in PBS, the peptide was detected with a horseradish peroxidase (HRP)-conjugated rabbit custom-made anti-MV F HRC Ab (1:1,500) in blocking buffer for 2 h at RT. HRP activity was measured by reading absorbance at 492 nm on the Sigmafast o-phenylenediamine dihydrochloride substrate system (Sigma-Aldrich, France). Standard curves were established for each peptide under the same ELISA conditions as for the test samples, and the detection limit was determined to be 0.15 nM.
Infection of mice.
SLAM TG mice (45) were bred at the institute's animal facility (PBES, ENS-Lyon) and used as heterozygotes for SLAM transgenes. One-week-old mice were infected i.n. by the application of 10 μl of MV G954 to both nares (500 PFU of MV/mouse). To evaluate the effect of HRC peptides, the SLAM TG mice were treated with peptide i.n. at 6 mg/kg 24 h before and on the day of infection and the treatment was then continued with daily s.c. injections of peptide at 4 mg/kg up to 7 days postinfection. The control mice received the same number of administrations/injections of the diluent. All animals were observed daily for clinical signs (neurological symptoms, ataxia, lethargy) and euthanized once clinical signs were observed. The protocol was approved by the Regional Ethical Committee (CECCAPP protocol ENS-2011-003).
Statistical analysis.
Data are expressed as means and standard deviations. Statistical analyses were performed with the F test, the unpaired t test, the Mantel-Cox test, and GraphPad Prism software.
RESULTS
Design of MV F-derived fusion inhibitors.
The antiviral activity of peptides derived from the HRC region of the fusion protein differs considerably among enveloped viruses (15, 32, 46, 47). Potency, especially for weak inhibitors, can be increased by peptide engineering strategies. On the basis of our previous experience with optimization of the antiviral properties of HRC-specific peptides (30, 32), we applied cholesterol conjugation and dimerization strategies to the design of MV F-derived fusion inhibitors.
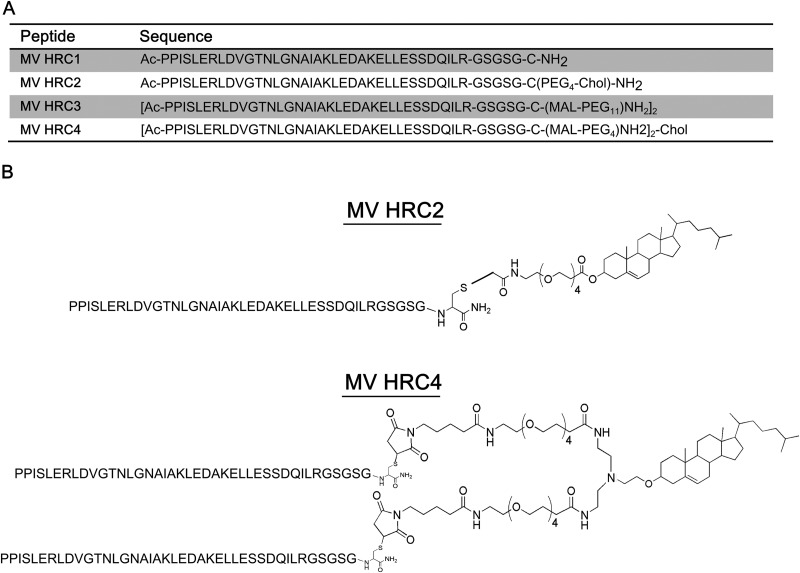
The peptide sequence was derived from the HRC region (residues 450 to 485) of MV F, from a previously identified MV HRC peptide by Lambert et al. (24), and extended with a C-terminal GSGSG linker/spacer sequence and a cysteine residue to allow conjugation to cholesterol via thiol-reactive reagents (Fig. 1). This core amino acid sequence was used for the subsequent modifications. The control peptide sequence (MV HRC1) featured the cysteine residue alkylated with iodoacetamide. Similar to the previously reported NiV, HPIV3, and HIV inhibitors (32), reaction of the core sequence with a bromoacetyl derivative of cholesterol featuring a four-unit polyethylene glycol spacer (PEG4) produced the cholesterol-conjugated monomer (MV HRC2), while reaction with bis-MAL-functionalized PEG11 produced the HRC dimer (MV HRC3) and reaction with bis-MAL-functionalized PEG4 produced the cholesterol-conjugated dimer (MV HRC4) (32).
Fig 1.
Sequences and structures of the MV HRC peptides. (A)The peptides consist of the MV HRC (amino acids 450 to 485 of MV F) with a GSGSG spacer, either dimerized (HRC3 and HRC4) or monomeric (HRC1 and HRC2). Specific peptides are linked to a PEG spacer (HRC2, HRC3, and HRC4) and modified by cholesterol (Chol) conjugation (HRC2 and HRC4). Ac, acetyl. (B) Schematic representation of MV peptides HRC2 and HRC4.
Inhibition of WT MV entry by MV F-derived peptides.
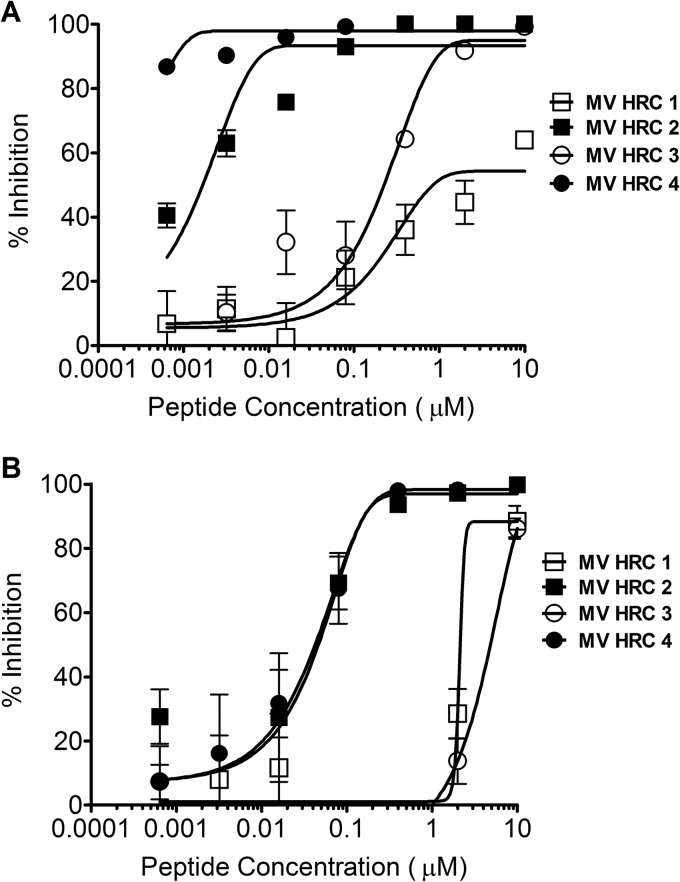
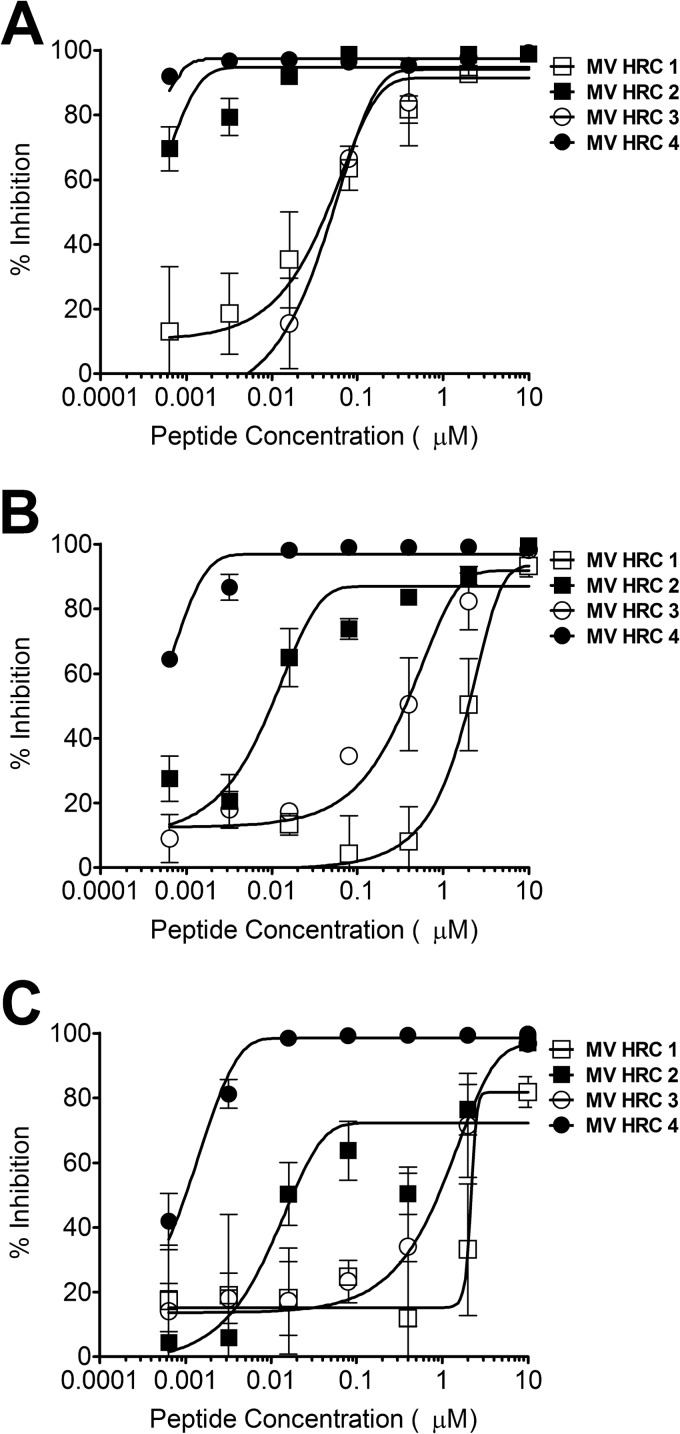
The inhibitory activities of MV HRC1, HRC2, HRC3, and HRC4 (Fig. 1A and B) against WT MV G954 were assessed in plaque reduction assays (Fig. 2A). Both peptides containing the cholesterol moiety (MV HRC2 and HRC4) performed better than their unconjugated counterparts (MV HRC1 and HRC3). While the 50% inhibitory concentrations (IC50s) of the unconjugated peptides were ∼7,000 nM (MV HRC1) and ∼300 nM (MV HRC3), the cholesterol-conjugated peptide IC50s were only ∼2 nM (MV HRC2) and <1 nM (MV HRC4) and the IC90s were 26.6 and 2.7 nM, respectively (Table 1). The dimeric peptide with cholesterol outperformed the other peptides over a wide range of concentrations and was significantly more efficient than the corresponding monomer (P = 0.0016, F test). Dimerization and cholesterol conjugation independently improve the overall efficacy of the entry inhibitor against MV (Fig. 2A), and the effects of cholesterol and of dimerization are additive. Similar results were obtained with the MV-IC323-EGFP WT strain (data not shown).
Fig 2.
Inhibition of MV entry and spread. (A) Inhibition of MV entry by MV HRC peptides. Vero-SLAM cell monolayers were infected with WT MV G954 in the presence of the indicated concentrations of MV HRC peptides. Viral entry was assessed by plaque reduction assay (multiplicity of infection [MOI], 2 × 10−4). Results are presented as percent reductions (y axis) in the number of plaques compared to that in the absence of treatment, as a function of the compound concentration (x axis). Each point represents the mean of four experiments (± the standard error). MV HRC4 is significantly more efficient at inhibiting entry than MV HRC2 is (P = 0.0016, F test). (B) Inhibition of MV spread by MV HRC peptides. Vero-SLAM cell monolayers were infected with WT MV-IC323-EGFP. After 90 min, medium with the indicated peptide concentrations was added. Viral spread was assessed by using a readout of EGFP fluorescence after 72 h (MOI, 2 × 10−4). Results are presented as percent reductions in fluorescence (y axis) compared to that in the absence of treatment, as a function of the compound concentration (x axis). Each point represents the mean of a representative experiment (± the standard deviation).
Table 1.
IC50s and IC90s obtained with different in vitro assays
| Assay | IC50a/IC90b (nM) |
P value for MV HRC2 vs MV HRC4 | |||
|---|---|---|---|---|---|
| MV HRC1 | MV HRC2 | MV HRC3 | MV HRC4 | ||
| Plaque reduction | 6,941/>10,000 | 2.03/26.6 | 285/2,837 | <1/2.7 | 0.0016 |
| Spreading | ∼2,132 | 56.91 | ∼2,286 | 56.71 | |
| Fusion at 2 h | 58.44 | <1 | 68.57 | <1 | |
| Fusion at 4 h | 1,992 | 15.57 | 381.5 | <1 | |
| Fusion at 6 h | 2,088/>10,000 | 580/2,259 | 2,305/2,768 | 1.05/3.2 | <0.0001 |
| Ex vivo culture | ND | 9.38 | ND | 10.07 | |
The IC50s of MV HRC1, HRC2, HRC3, and HRC4 were determined by entry, spreading, fusion, and ex vivo assays.
The IC90 was calculated when the IC50 could not be determined. Statistical comparisons of MV HRC2 and HRC4 fits were performed with the F test.
Spread of virus is curtailed by the dimeric cholesterol-conjugated peptides.
For clinical utility, an antiviral agent should prevent multiple rounds of infection (48). A multicycle replication assay we have used in the past (35) was adapted to assess the effect of the cholesterol-conjugated peptides on viral infection in Vero-SLAM cells, with an MV carrying the gene for EGFP, MV-IC323-EGFP (34). Vero-SLAM cells were infected with MV-IC323-EGFP, and after a 90-min adsorption period to permit viral entry, the medium was replaced with fresh medium containing the concentrations of peptides indicated on the x axis in Fig. 2B. The fluorescence level, which reflects the amount of infection by the EGFP-producing virus, was quantitated after 72 h as previously described (35). Under these conditions, when viral entry had already occurred, only the cholesterol-conjugated peptides were effective inhibitors. The unconjugated peptides prevented 90% of viral spread only at the highest concentration and thus failed to completely block the spread of infection. At 3 days postinfection, neither HRC1 nor HRC3 reduced MV infection (Fig. 2B); the fluorescence in the presence of these peptides was the same as that in untreated control wells. At the same time point, the IC50 of the MV HRC2 peptide was ∼57 nM, similar to that of the MV HRC4 dimeric peptide (∼57 nM under the same conditions) (Table 1), indicating that the conjugated peptides prevent MV from spreading.
Cell-to-cell fusion is inhibited by MV F-derived peptides.
While the two unconjugated peptides completely lost their inhibitory efficacy versus viral spreading as described above (Fig. 2B), they inhibited simple viral entry (Fig. 2A). The cholesterol-conjugated peptides inhibited both spread and entry at 1,000 nM. To investigate the reasons for this difference, we performed a quantitative fusion assay based on β-Gal complementation, in which the expression of β-Gal results from the fusion of cells expressing viral envelope glycoproteins (MV G954 H and F) with cells expressing the MV receptor SLAM. The indicated concentrations of MV HRC peptides were present during the entire fusion process, allowing peptides to act at the stages of triggering/activation of the fusion protein, as well as during subsequent fusion. Fusion was quantitated by measuring β-Gal complementation after 2, 4, and 6 h as done previously (37) (Fig. 3). Fusion was inhibited by all of the peptides after 2 h; however, the dimeric cholesterol-conjugated peptide (MV HRC4) remained fusion inhibitory at the 6-h time point.
Fig 3.
Inhibition of cell fusion. Fusion of MV H- and F-coexpressing cells with SLAM-bearing cells in the presence of the indicated compound concentrations was quantitated at 2 h (A), 4 h (B), and 6 h (C) with a β-Gal complementation assay. Results are presented as percent reductions in luminescence (y axis) compared to that in the absence of treatment, as a function of the compound concentration (x axis). Each point is the mean result of three experiments (± the standard error). MV HRC4 is significantly more efficient at inhibiting fusion than MV HRC2 is (P < 0.0001, F test).
Cholesterol-conjugated, MV-derived peptides inhibit MV infection in brain explants.
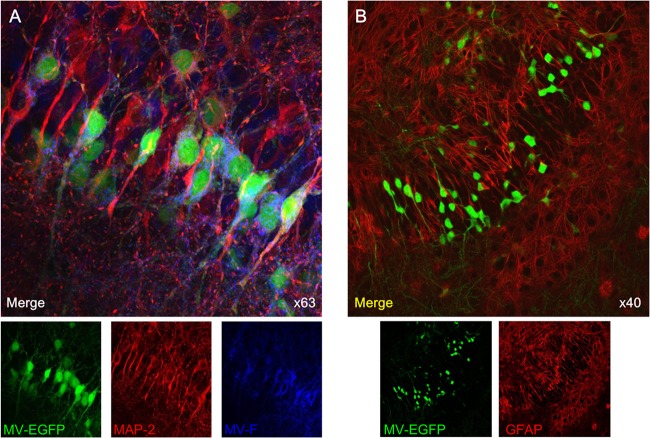
To analyze the effect of MV-derived peptides on brain MV infection, hippocampal slices from neonatal SLAM TG mice (41, 45, 49, 50) were used to establish an ex vivo infection model. Infection with MV-IC323-EGFP is monitored in this model by fluorescence microscopy (Fig. 4 and 5). In agreement with the data from infected human brain tissue (7), neurons are the main target of MV infection in these TG murine organotypic cultures (Fig. 4A). MV infection was not detected in astrocytes (Fig. 4B). The virus did not induce syncytia, as shown by time-lapse analysis (see Movie S1 in the supplemental material). Staining of the ex vivo cultures with anti-MV F protein Abs showed MV F expression during virus replication in neurons (Fig. 4A), confirming that F expression correlates with spreading (51) and that F is available as a target for MV F-specific peptides.
Fig 4.
MV infection of SLAM TG organotypic brain explants. Hippocampal slices obtained from SLAM TG murine brains were infected with MV-IC323-EGFP at 104 PFU/slice of for 3 days, stained with the indicated Abs, and analyzed by confocal microscopy. (A) MV-infected slices (green) were stained with neuron-specific MAP-2 (red)- and MV F-specific (blue) Abs. (B) MV-infected slices (green for EGFP) were stained with an astrocyte-specific GFAP (red) Ab and did not show any colocalization.
Fig 5.
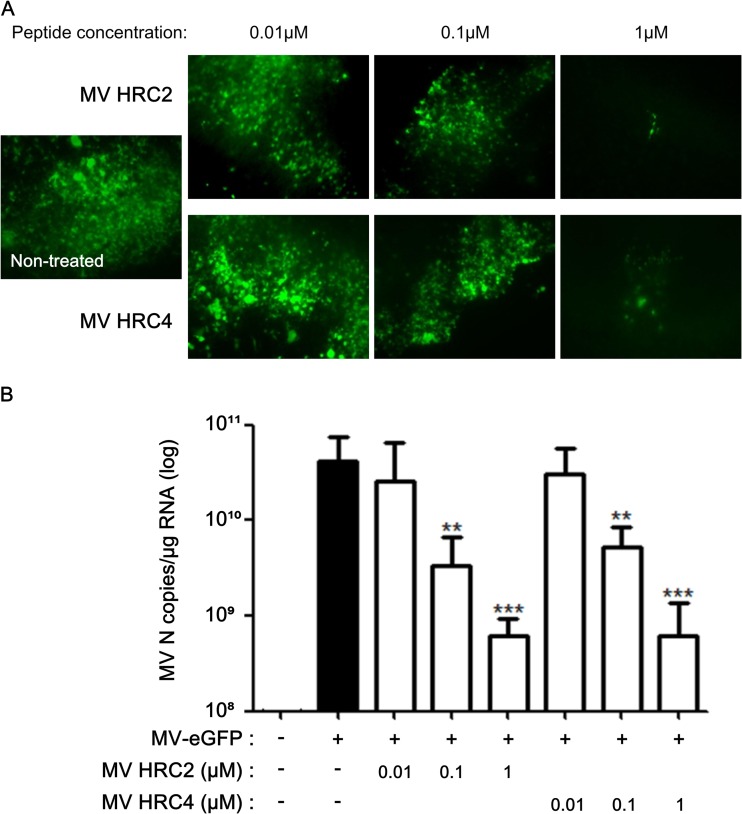
MV HRC peptides inhibit MV infection of organotypic brain explants. Hippocampal slices from SLAM TG murine brains were treated with the indicated concentrations of either HRC2 or HRC4 for 5 h or left untreated (control) and infected with MV-IC323-EGFP at 104 PFU/slice for 3 days. (A) Cultures from MV-infected explants (green) were observed by immunofluorescence microscopy. (B) Total RNA was harvested from organotypic slices at 3 days postinfection, and the level of MV N gene expression was quantified by reverse transcription-qPCR. Results are expressed as means ± standard deviations of triplicate cultures (**, P < 0.01; ***, P < 0.001 [unpaired t test]).
The brain explants were infected with MV-IC323-EGFP in the presence of the various peptide concentrations to test antiviral potency ex vivo. Viral replication was reduced in the presence of MV HRC2 and HRC4 (Fig. 5). Only a few cells were positive for EGFP fluorescence in brain explants treated with the highest peptide concentration (Fig. 5A), and the expression of the gene for MV nucleoprotein (N) was significantly lower than in the untreated cultures (Fig. 5B). The concentration of peptide required for an antiviral effect ex vivo was similar to the concentration that inhibited spread and fusion after 6 h in vitro (Fig. 2 and 3). These two peptides penetrated the brain explants and retained their antiviral efficacy.
Inhibition of MV infection in vivo by the cholesterol-conjugated dimer peptide.
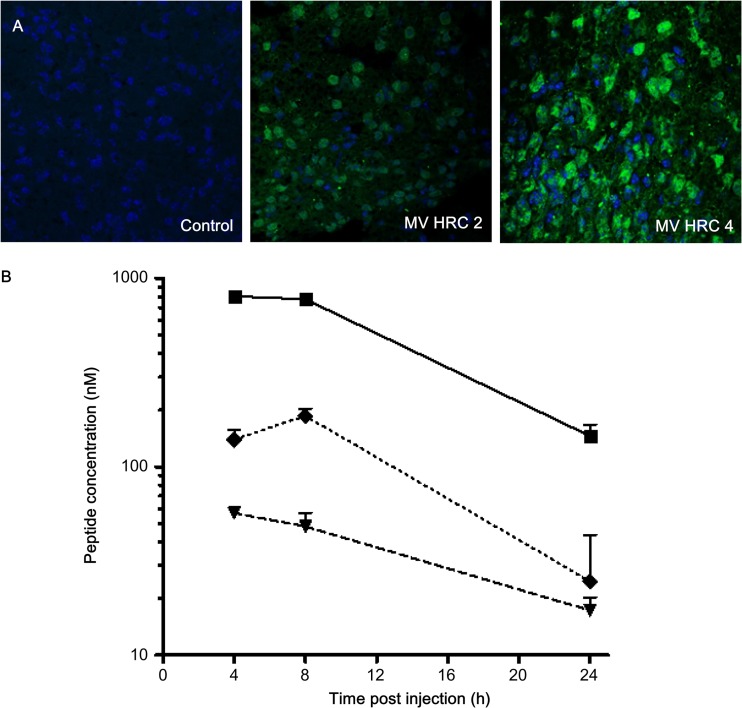
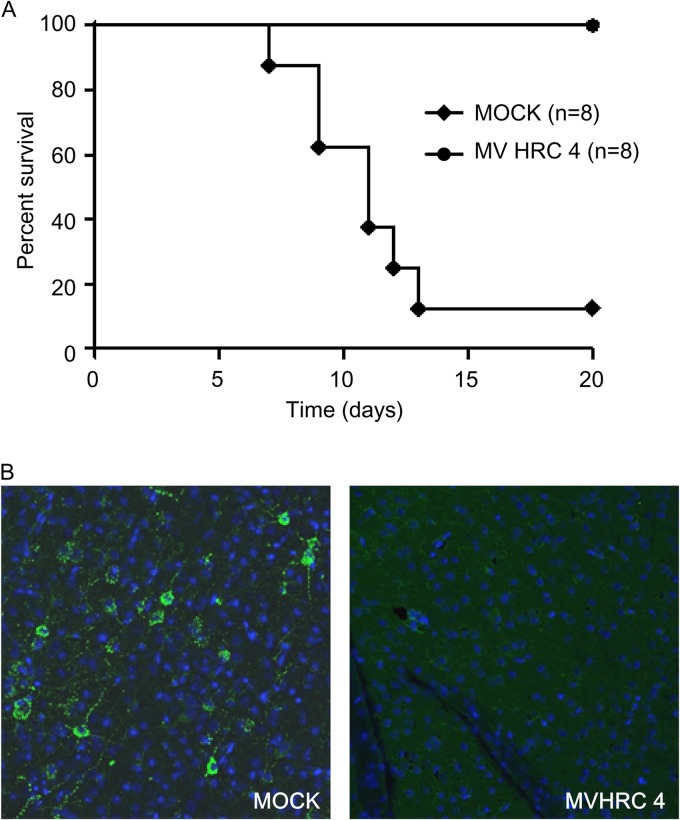
We have previously shown that i.n. MV infection of suckling mice expressing the human SLAM transgene caused a lethal acute neurological syndrome (41, 49). This suckling mouse model was used to analyze whether cholesterol-coupled MV HRC peptides protect mice from fatal MV encephalitis. Initial experiments showed no toxicity in mice after 1 week of treatment with 6 mg/kg of the MV HRC2 and HRC4 peptides. We determined whether the peptides leave the circulation and cross the blood-brain barrier to penetrate the CNS in suckling SLAM TG mice 8 h after s.c. injection (Fig. 6A). Both peptides were found in the CNS. Since the MV HRC4 peptide was the most potent inhibitor in vitro, before testing it in vivo, we performed a pharmacokinetic analysis of MV HRC4 in mice after s.c. injection and detected the peptide in the brains and lungs of mice even 24 h after administration (Fig. 6B). Bioavailability of the peptide after i.n. administration showed restriction to the lungs, where it attained a high concentration (Table 2). Prophylactic administration of the MV HRC4 peptide (i.n. administration starting 24 h before infection and followed by daily s.c. administration) protected 100% of the animals (Fig. 7A). Immunofluorescence analysis of the brains of infected animals (Fig. 7B) revealed the presence of the viral nucleoprotein (N) in the brains of nontreated animals but no detectable viral antigen in surviving treated-animal brains, suggesting that the peptide prevented CNS infection.
Fig 6.
Biodistribution of MV HRC peptides. (A) Mice were injected s.c. with MV HRC2 or HRC4 or the vehicle (control) at 6 mg/kg. Animals were sacrificed 8 h later, and their brains were stained with peptide-specific Abs. (B). Mice were injected s.c. with MV HRC4 at 6 mg/kg. At the indicated times points, the animals were sacrificed and the peptide concentrations in serum (square, smooth line), brain (triangle, dashed line), and lung (diamond, dotted line) samples were quantitated. The value at each point is the mean result obtained from three animals (± the standard deviation).
Table 2.
MV HRC4 bioavailability at 4 h postadministration
| Sample analyzed | Peptide concn (nM) |
|
|---|---|---|
| I.n. administration | S.c. injection | |
| Brain | ND | 57 |
| Lung | 1,760 | 139 |
| Serum | ND | 803 |
ND, not detected.
Fig 7.
MV HRC4 protects suckling mice from lethal MV encephalitis. One-week-old SLAM TG mice received an i.n. challenge of MV G954 at 24 h after MV-HRC peptide treatment and were followed for 5 weeks postinfection. Controls animals (MOCK) were injected with the vehicle alone. (A) Mice received prophylactic i.n. administration of peptides, followed by s.c. injection after infection, daily for 7 days. The statistical significance of the difference between the MV HRC4- and mock-injected groups was analyzed with the Mantel-Cox test (P = 0.0004). (B) Immunofluorescence analysis of MV N expression in the brain of either mock-treated animals on the day that neurological signs appeared or surviving MV HRC4-treated animals at the end of the experiment. Representative images from three mice analyzed in each group are presented (magnification, ×40).
DISCUSSION
Measles is a reemerging disease. Although vaccination programs have significantly reduced the incidence of measles, outbreaks still occur, with resurgence in industrialized countries during last few years. There were approximately 42,000 laboratory-confirmed measles cases worldwide in 2012. (http://www.who.int/immunization_monitoring/diseases/measles_monthlydata/en/). Specific antiviral therapies are urgently needed to complement vaccination and achieve the global elimination of measles (4, 52, 53). MV therapeutics could be used for the rapid control of local outbreaks, protection of immunocompromised people and infants prior to vaccination, and improved management of acute and persistent disease (54–57). Complications of MV infection occur in up to 40% of cases, and those involving the CNS are rare but serious. Primary MV encephalitis occurs in 1 to 3 of 1,000 infected patients, with recovery of infectious virus from the cerebrospinal fluid or brain (5, 6). Acute postinfectious encephalomyelitis also occurs during or shortly after acute measles but seems to be associated with an autoimmune etiology and MV is not isolated. SSPE occurs in 4 to 11 of 100,000 cases of acute measles, causing progressive ataxia, seizures, and dementia (7). Another form of progressive, MV-induced CNS disease, measles inclusion body encephalitis, occurs in immunosuppressed patients 1 to 6 months following MV infection and is characterized by seizures, motor and sensory deficits, and lethargy, with either an acute or a subacute fatal course. Nonrestricted virus replication results in cytolytic viral replication in brain tissue (6, 58, 59). There are no specific therapies for these complications of CNS infection (23, 51, 60, 61), and attempts to treat SSPE with various antiviral drugs, including ribavirin, interferons, and isoprinosin, have been disappointing (2, 4, 21, 23, 51, 60–62). Here, we provide evidence that MV HRC-derived peptides have the potential to fill this void.
Peptides derived from the HRC region of MV F have been shown to have antiviral activity (63), and we have found for other viral fusion-inhibitory peptides that dimerization and cholesterol conjugation increase inhibitory efficacy (30, 32). We show here that MV peptide antiviral efficacy increased with dimerization and cholesterol conjugation. In keeping with the established mechanism of action of HR-derived peptides, the HRC peptides also inhibit fusion mediated by MV H and F; surprisingly, however, the fusion inhibition was reduced over time. Only the dimerized, cholesterol-conjugated peptides retained fusion-inhibitory activity over a 6-h period (despite both cholesterol-tagged peptides being stable in serum for several days; data not shown). The dimerized peptides were efficacious in vivo. These results, like our previous analysis of NiV peptide inhibitors (30, 32), point to fusion-inhibitory efficacy as a predictive tool of in vivo efficacy; future efforts will focus on enhancing fusion inhibition, with the idea that such inhibitors will perform better in vivo.
To evaluate the efficiency of peptides for MV CNS infection, we used a suckling TG mouse model that expresses human SLAM. This model provides a simple readout of antiviral efficacy and an accurate method for testing novel anti-MV agents (41, 45), and the study was designed to establish proof of the concept of the utility of HRC peptides in preventing and treating the neurological complications of measles.
The dimeric, cholesterol-conjugated peptide protected 100% of the mice from MV infection, making the peptide a strong candidate for prophylactic therapy. Most interestingly, this inhibitor blocks the CNS spread of MV. Previous peptide inhibitors have not shown this promising combination of features. For example, the anti-fusion tripeptide FIP inhibited infection by vaccine MV at a very high concentration (200 μM) (20, 23) but had no activity in vivo. The HIV fusion-inhibitory peptide enfuvirtide is used as salvage therapy for multidrug-resistant HIV-1-infected patients, but its negligible distribution in the CNS precludes its use for HIV-1 dementia and encephalitis (64). Thus, the brain-penetrating capacity of cholesterol-conjugated peptides is a key feature that supports their further development as therapeutics.
Several options will be explored to improve the efficacy of the anti-MV peptide treatments described here. For example, on the basis of our finding that the PEG4 spacer used here enhanced the activity of NiV and HPIV3 inhibitors (30), optimization of the length of the linker that joins the monomers may increase antiviral potency (65). Optimization of the interhelical packing interactions of the HRC peptide with the cognate HRN region of MV F is also likely to be beneficial (30). Since it seems likely that MV spread in the CNS requires F (51), peptides optimized versus F may be ideal for the treatment of MV in the CNS. Some SSPE strains have mutations in the F protein that enhance fusion activity (66), and the efficacy of HRC peptides versus those strains remains to be determined.
Already at this stage, the peptides described here show potential for the therapy of MV-induced CNS complications for which no effective therapy exists. Antiviral peptides may provide a combined prophylaxis and therapy for immunocompromised individuals, infants prior to vaccination, and patients who decline vaccination and for rapid control of local outbreaks (2). Finally, the efficacy of the MV peptides for prophylaxis opens a new avenue for rapid protection against highly contagious MV infection during the global outbreaks that will continue to occur until the goal of eradication is attained.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants NS0737781 and NS076385 to M.P. and AI101333-02 to A.M. from NIH (NINDS) and to B.H. from INSERM. We are grateful to Ashton Kutcher and Jonathan Ledecky for their support, to Dan and Nancy Paduano for support of innovative research projects, and to the Friedman Family Foundation for renovation of our laboratories at Weill Cornell Medical College. We are grateful for the Friedman Research Scholar Award to M.P. and for support from the Fondation pour la Recherche Médicale to J.W.
We acknowledge Y. Yanagi (Kyushu University, Fukyoka, Japan), who kindly provided MV-IC323-eGFP. We thank Helene Becq Clot-Faybesse and Jean-Luc Gaiarsa of IMED, Marseille, France, and Nicola Kuczewski and Remi Gervais of CRNL for their help in the development of the brain explant model and Denis Gerlier, Nicolas Baillet, Araf Khaled, and the other members of the Immunobiology of Viral Infection group (CIRI U1111) for their help with this study. We acknowledge PBES-ENS for the help with animal experiments. We thank Jacob Moscona-Skolnik for critically reading and editing the manuscript.
Footnotes
Published ahead of print 9 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02436-13.
REFERENCES
- 1.Simons E, Ferrari M, Fricks J, Wannemuehler K, Anand A, Burton A, Strebel P. 2012. Assessment of the 2010 global measles mortality reduction goal: results from a model of surveillance data. Lancet 379:2173–2178 [DOI] [PubMed] [Google Scholar]
- 2.Moss WJ, Griffin DE. 2012. Measles. Lancet 379:153–164 [DOI] [PubMed] [Google Scholar]
- 3.De Serres G, Markowski F, Toth E, Landry M, Auger D, Mercier M, Belanger P, Turmel B, Arruda H, Boulianne N, Ward BJ, Skowronski DM. 2013. The largest measles epidemic in North America in a decade—Quebec, Canada, 2011: contribution of susceptibility, serendipity and super-spreading events on elimination. J. Infect. Dis. 207:990–998 [DOI] [PubMed] [Google Scholar]
- 4.Plemper RK, Snyder JP. 2009. Measles control—can measles virus inhibitors make a difference? Curr. Opin. Investig. Drugs. 10:811–820 [PMC free article] [PubMed] [Google Scholar]
- 5.Hosoya M. 2006. Measles encephalitis: direct viral invasion or autoimmune-mediated inflammation? Intern. Med. 45:841–842 [DOI] [PubMed] [Google Scholar]
- 6.Buchanan R, Bonthius DJ. 2012. Measles virus and associated central nervous system sequelae. Semin. Pediatr. Neurol. 19:107–114 [DOI] [PubMed] [Google Scholar]
- 7.Allen IV, McQuaid S, McMahon J, Kirk J, McConnell R. 1996. The significance of measles virus antigen and genome distribution in the CNS in SSPE for mechanisms of viral spread and demyelination. J. Neuropathol. Exp. Neurol. 55:471–480 [DOI] [PubMed] [Google Scholar]
- 8.Weissenhorn W, Hinz A, Gaudin Y. 2007. Virus membrane fusion. FEBS Lett. 581:2150–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White JM. 2007. The first family of cell-cell fusion. Dev. Cell 12:667–668 [DOI] [PubMed] [Google Scholar]
- 10.Lamb RA, Paterson RG, Jardetzky TS. 2006. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 344:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swanson K, Wen X, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. 2010. Structure of the Newcastle disease virus F protein in the post-fusion conformation. Virology 402:372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welch BD, Liu Y, Kors CA, Leser GP, Jardetzky TS, Lamb RA. 2012. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc. Natl. Acad. Sci. U. S. A. 109:16672–16677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen X, Krause JC, Leser GP, Cox RG, Lamb RA, Williams JV, Crowe JE, Jr, Jardetzky TS. 2012. Structure of the human metapneumovirus fusion protein with neutralizing antibody identifies a pneumovirus antigenic site. Nat. Struct. Mol. Biol. 19:461–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison SC. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plattet P, Plemper RK. 2013. Envelope protein dynamics in paramyxovirus entry. mBio 4(4):e00413–13. 10.1128/mBio.00413-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mühlebach MD, Mateo M, Sinn PL, Prufer S, Uhlig KM, Leonard VH, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, Ramachandran S, McCray PB, Cichutek K, von Messling V, Lopez M, Cattaneo R. 2011. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 480:530–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noyce RS, Bondre DG, Ha MN, Lin LT, Sisson G, Tsao MS, Richardson CD. 2011. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 7:e1002240. 10.1371/journal.ppat.1002240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehrengruber MU, Ehler E, Billeter MA, Naim HY. 2002. Measles virus spreads in rat hippocampal neurons by cell-to-cell contact and in a polarized fashion. J. Virol. 76:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makhortova NR, Rall GF, Prasolov VS. 2007. Receptor-independent spread of measles virus is unique to neurons, and involves fusion of synaptic membranes. Mol. Biol. (Mosk.) 41:186–188 (In Russian.) [PubMed] [Google Scholar]
- 21.Lin WH, Kouyos RD, Adams RJ, Grenfell BT, Griffin DE. 2012. Prolonged persistence of measles virus RNA is characteristic of primary infection dynamics. Proc. Natl. Acad. Sci. U. S. A. 109:14989–14994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence DM, Patterson CE, Gales TL, D'Orazio JL, Vaughn MM, Rall GF. 2000. Measles virus spread between neurons requires cell contact but not CD46 expression, syncytium formation, or extracellular virus production. J. Virol. 74:1908–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makhortova NR, Askovich P, Patterson CE, Gechman LA, Gerard NP, Rall GF. 2007. Neurokinin-1 enables measles virus trans-synaptic spread in neurons. Virology 362:235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert DM, Barney S, Lambert AL, Guthrie K, Medinas R, Davis DE, Bucy T, Erickson J, Merutka G, Petteway SR., Jr 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. U. S. A. 93:2186–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao Q, Compans RW. 1996. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology 223:103–112 [DOI] [PubMed] [Google Scholar]
- 26.Baker KA, Dutch RE, Lamb RA, Jardetzky TS. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309–319 [DOI] [PubMed] [Google Scholar]
- 27.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U. S. A. 91:9770–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu M, Blacklow SC, Kim PS. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075–1082 [DOI] [PubMed] [Google Scholar]
- 29.Porotto M, Yokoyama C, Orefice G, Kim H-S, Moscona A. 2009. Kinetic dependence of paramyxovirus entry inhibition. J. Virol. 83:6947–6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porotto M, Rockx B, Yokoyama C, Talekar A, DeVito I, Palermo L, Liu J, Cortese R, Lu M, Feldmann H, Pessi A, Moscona A. 2010. Inhibition of Nipah virus infection in vivo: targeting an early stage of paramyxovirus fusion activation during viral entry. PLoS Pathog. 6(10):e1001168. 10.1371/journal.ppat.1001168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingallinella P, Bianchi E, Ladwa NA, Wang YJ, Hrin R, Veneziano M, Bonelli F, Ketas TJ, Moore JP, Miller MD, Pessi A. 2009. Addition of a cholesterol group to an HIV-1 peptide fusion inhibitor dramatically increases its antiviral potency. Proc. Natl. Acad. Sci. U. S. A. 106:5801–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pessi A, Langella A, Capito E, Ghezzi S, Vicenzi E, Poli G, Ketas T, Mathieu C, Cortese R, Horvat B, Moscona A, Porotto M. 2012. A general strategy to endow natural fusion-protein-derived peptides with potent antiviral activity. PLoS One 7:e36833. 10.1371/journal.pone.0036833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kouomou DW, Wild TF. 2002. Adaptation of wild-type measles virus to tissue culture. J. Virol. 76:1505–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashimoto K, Yanagi Y. 2002. Measles virus entry as examined by a recombinant virus expressing green fluorescent protein. Uirusu 52:169–175 (In Japanese.) [PubMed] [Google Scholar]
- 35.Porotto M, Orefice G, Yokoyama C, Mungall B, Realubit R, Sganga M, Aljofan M, Whitt M, Glickman F, Moscona A. 2009. Simulating henipavirus multicycle replication in a screening assay leads to identification of a promising candidate for therapy. J. Virol. 83:5148–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porotto M, Fornabaio M, Kellogg GE, Moscona A. 2007. A second receptor binding site on human parainfluenza virus type 3 hemagglutinin-neuraminidase contributes to activation of the fusion mechanism. J. Virol. 81:3216–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moosmann P, Rusconi S. 1996. Alpha complementation of LacZ in mammalian cells. Nucleic Acids Res. 24:1171–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoppini L, Buchs PA, Muller D. 1991. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods 37:173–182 [DOI] [PubMed] [Google Scholar]
- 39.Grigorov B, Rabilloud J, Lawrence P, Gerlier D. 2011. Rapid titration of measles and other viruses: optimization with determination of replication cycle length. PLoS One 6:e24135. 10.1371/journal.pone.0024135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giraudon P, Jacquier MF, Wild TF. 1988. Antigenic analysis of African measles virus field isolates: identification and localisation of one conserved and two variable epitope sites on the NP protein. Virus Res. 10:137–152 [DOI] [PubMed] [Google Scholar]
- 41.Sellin CI, Davoust N, Guillaume V, Baas D, Belin MF, Buckland R, Wild TF, Horvat B. 2006. High pathogenicity of wild-type measles virus infection in CD150 (SLAM) transgenic mice. J. Virol. 80:6420–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathieu C, Guillaume V, Sabine A, Ong KC, Wong KT, Legras-Lachuer C, Horvat B. 2012. Lethal Nipah virus infection induces rapid overexpression of CXCL10. PLoS One 7:e32157. 10.1371/journal.pone.0032157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plumet S, Duprex WP, Gerlier D. 2005. Dynamics of viral RNA synthesis during measles virus infection. J. Virol. 79:6900–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plumet S, Gerlier D. 2005. Optimized SYBR green real-time PCR assay to quantify the absolute copy number of measles virus RNAs using gene specific primers. J. Virol. Methods 128:79–87 [DOI] [PubMed] [Google Scholar]
- 45.Sellin CI, Horvat B. 2009. Current animal models: transgenic animal models for the study of measles pathogenesis. Curr. Top. Microbiol. Immunol. 330:111–127 [DOI] [PubMed] [Google Scholar]
- 46.Vigant F, Lee B. 2011. Hendra and Nipah infection: pathology, models and potential therapies. Infect. Disord. Drug Targets 11:315–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eckert DM, Kim PS. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777–810 [DOI] [PubMed] [Google Scholar]
- 48.Talekar A, Pessi A, Porotto M. 2011. Infection of primary neurons mediated by Nipah virus envelope proteins: role of host target cells in antiviral action. J. Virol. 85:8422–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sellin CI, Jegou JF, Renneson J, Druelle J, Wild TF, Marie JC, Horvat B. 2009. Interplay between virus-specific effector response and Foxp3 regulatory T cells in measles virus immunopathogenesis. PLoS One 4:e4948. 10.1371/journal.pone.0004948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Druelle J, Sellin CI, Waku-Kouomou D, Horvat B, Wild FT. 2008. Wild type measles virus attenuation independent of type I IFN. Virol. J. 5:22. 10.1186/1743-422X-5-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young VA, Rall GF. 2009. Making it to the synapse: measles virus spread in and among neurons. Curr. Top. Microbiol. Immunol. 330:3–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ndungu JM, Krumm SA, Yan D, Arrendale RF, Reddy GP, Evers T, Howard R, Natchus MG, Saindane MT, Liotta DC, Plemper RK, Snyder JP, Sun A. 2012. Nonnucleoside inhibitors of the measles virus RNA-dependent RNA polymerase: synthesis, structure-activity relationships, and pharmacokinetics. J. Med. Chem. 55:4220–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nilsson A, Chiodi F. 2011. Measles outbreak in Africa—is there a link to the HIV-1 epidemic? PLoS Pathog. 7:e1001241. 10.1371/journal.ppat.1001241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abzug MJ, Qin M, Levin MJ, Fenton T, Beeler JA, Bellini WJ, Audet S, Sowers SB, Borkowsky W, Nachman SA, Pelton SI, Rosenblatt HM. 2012. Immunogenicity, immunologic memory, and safety following measles revaccination in HIV-infected children receiving highly active antiretroviral therapy. J. Infect. Dis. 206:512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stein-Zamir C, Shoob H, Abramson N, Zentner G. 2012. Who are the children at risk? Lessons learned from measles outbreaks. Epidemiol. Infect. 140:1578–1588 [DOI] [PubMed] [Google Scholar]
- 56.Torner N, Anton A, Barrabeig I, Lafuente S, Parron I, Arias C, Camps N, Costa J, Martínez A, Torra R, Godoy P, Minguell S, Ferrús G, Cabezas C, Domínguez A, Measles Elimination Program Surveillance Network of Catalonia, Spain 2013. Epidemiology of two large measles virus outbreaks in Catalonia: what a difference the month of administration of the first dose of vaccine makes. Hum. Vaccin. Immunother. 9:675–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fowlkes A, Witte D, Beeler J, Audet S, Garcia P, Curns A, Yang C, Fudzulani R, Broadhead R, Bellini WJ, Cutts F, Helfand RF. 2011. Persistence of vaccine-induced measles antibody beyond age 12 months: a comparison of response to one and two doses of Edmonston-Zagreb measles vaccine among HIV-infected and uninfected children in Malawi. J. Infect. Dis. 204(Suppl 1):S149–S157 [DOI] [PubMed] [Google Scholar]
- 58.Urbanska EM, Chambers BJ, Ljunggren HG, Norrby E, Kristensson K. 1997. Spread of measles virus through axonal pathways into limbic structures in the brain of TAP1−/− mice. J. Med. Virol. 52:362–369 [DOI] [PubMed] [Google Scholar]
- 59.Norrby E, Kristensson K. 1997. Measles virus in the brain. Brain Res. Bull. 44:213–220 [DOI] [PubMed] [Google Scholar]
- 60.O'Donnell LA, Rall GF. 2010. Blue moon neurovirology: the merits of studying rare CNS diseases of viral origin. J. Neuroimmune Pharmacol. 5:443–455 [DOI] [PubMed] [Google Scholar]
- 61.Reuter D, Schneider-Schaulies J. 2010. Measles virus infection of the CNS: human disease, animal models, and approaches to therapy. Med. Microbiol. Immunol. 199:261–271 [DOI] [PubMed] [Google Scholar]
- 62.Griffin DE, Lin WH, Pan CH. 2012. Measles virus, immune control, and persistence. FEMS Microbiol. Rev. 36:649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wild TF, Buckland R. 1997. Inhibition of measles virus infection and fusion with peptides corresponding to the leucine zipper region of the fusion protein. J. Gen. Virol. 78(Pt 1):107–111 [DOI] [PubMed] [Google Scholar]
- 64.Price RW, Parham R, Kroll JL, Wring SA, Baker B, Sailstad J, Hoh R, Liegler T, Spudich S, Kuritzkes DR, Deeks SG. 2008. Enfuvirtide cerebrospinal fluid (CSF) pharmacokinetics and potential use in defining CSF HIV-1 origin. Antivir. Ther. 13:369–374 [PMC free article] [PubMed] [Google Scholar]
- 65.Francis JN, Redman JS, Eckert DM, Kay MS. 2012. Design of a modular tetrameric scaffold for the synthesis of membrane-localized d-peptide inhibitors of HIV-1 entry. Bioconjug. Chem. 23:1252–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watanabe S, Shirogane Y, Suzuki SO, Ikegame S, Koga R, Yanagi Y. 2013. Mutant fusion proteins with enhanced fusion activity promote measles virus spread in human neuronal cells and brains of suckling hamsters. J. Virol. 87:2648–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.