Abstract
Herpes simplex virus 1 (HSV-1) Us11 protein is a double-stranded RNA-binding protein that suppresses type I interferon production through the inhibition of the cytoplasmic RNA sensor RIG-I. Whether additional cellular mediators are involved in this suppression remains to be determined. In this study, we report on the requirement of cellular double-stranded RNA-binding protein PACT for Us11-mediated perturbation of type I interferon production. Us11 associates with PACT tightly to prevent it from binding with and activating RIG-I. The Us11-deficient HSV-1 was indistinguishable from the Us11-proficient virus in the suppression of interferon production when PACT was compromised. More importantly, HSV-1-induced activation of interferon production was abrogated in PACT knockout murine embryonic fibroblasts. Our findings suggest a new mechanism for viral evasion of innate immunity through which a viral double-stranded RNA-binding protein interacts with PACT to circumvent type I interferon production. This mechanism might also be used by other PACT-binding viral interferon-antagonizing proteins such as Ebola virus VP35 and influenza A virus NS1.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) commonly infects humans, causing cold sores, fever blisters, and genital sores. It also establishes a lifelong persistent infection in trigeminal ganglion during which only the latency-associated transcript, but not other lytic genes, is expressed. Infection with HSV-1 triggers the host innate immune response through the recognition of pathogen-associated molecular patterns by cellular pattern recognition receptors, including both Toll-like receptors and RIG-I-like receptors (1). As a result, antiviral cytokines such as type I interferons (IFNs) are produced. To facilitate its replication and persistence, HSV-1 has developed various countermeasures to suppress the production and action of type I IFNs and IFN-stimulated genes (ISGs) (2).
Immediate-early (IE) protein ICP0, a master regulator of HSV-1 infection and virus-host interaction, is capable of antagonizing innate immunity at multiple levels (2, 3). The delayed-early protein ICP34.5, which is also known as γ34.5 or γ134.5 for its increased expression in later phases of viral infection, is required in certain cell types to prevent an antiviral response where protein synthesis is abruptly inhibited prior to the completion of the virus life cycle (2–5). ICP34.5 also antagonizes the IFN response (2, 4). In addition to ICP0 and ICP34.5, the Us11 gene encodes another protein that counteracts IFN production and signaling (5–9). Us11 is a multifunctional double-stranded RNA (dsRNA)-binding protein expressed in the late stage of infection and incorporated in the virion (10–12). Us11 associates with PKR, PACT, 2′,5′-oligoadenylate synthetase (OAS), RIG-I, and MDA5 in the cell (7, 13–15). It uses its RNA-binding domain to interact with PKR kinase, leading to the prevention of eIF2α phosphorylation and the inhibition of autophagy (7, 13, 16). It also binds with OAS, RIG-I, and MDA5 to impede their function in IFN induction and IFN response (14, 15). PACT is a cellular dsRNA-binding protein originally identified as a binding partner and activator of PKR (17). Although Us11 binds with PACT and inhibits PKR activation, its interaction with PKR but not with PACT is required for the inhibition of PKR activity (13).
RIG-I is a prototypic cytoplasmic sensor of virus-derived RNAs (18, 19). RIG-I-like receptors are critically involved in the sensing of HSV infection (20, 21). Particularly, HSV-1-encoded small noncoding RNAs derived from the latency-associated transcript are sensed by RIG-I (22). Optimal function of RIG-I requires PACT, which interacts with and potently activates RIG-I in a PKR-independent manner (23). Us11 interacts with RIG-I and its homolog MDA5 to suppress their activation of type I IFN production (15). Us11 also interacts with PACT, but the biological function of this interaction is unclear (13). In particular, it remains to be understood whether the interaction between Us11 and PACT might play a role in the perturbation of RIG-I-dependent IFN production.
In the present study, we investigated the interaction of Us11 with PACT and determined the requirement of PACT in Us11-induced suppression of the innate antiviral response. Our findings reveal a new mechanism for viral evasion of innate immunity, by which a viral dsRNA-binding and IFN-antagonizing protein interacts with PACT to impede the association with and activation of RIG-I.
MATERIALS AND METHODS
Cells and viruses.
HEK293 and HEK293T cells were maintained and propagated in Dulbecco's modified Eagle medium. Mouse embryonic fibroblasts (MEFs) were derived from wild-type C57/B6 and PACT−/− mice (24, 25) kindly provided to Kuan-Teh Jeang's laboratory in the National Institutes of Health by Ganes Sen (Cleveland Clinic, OH, USA). Lack of PACT expression in PACT−/− MEFs was previously verified by Western blotting (26). Human telomerase reverse transcriptase (hTERT)-immortalized MEFs were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. Sendai virus was obtained from the American Type Culture Collection (Manassas, VA, USA). Recombinant HSV-1-EGFP-Us11 (where EGFP is enhanced green fluorescent protein), HSV-1-pAUs11, HSV-1-Δ34.5, and HSV-1-Δ34.5-(IE)Us11 viruses were propagated and purified as previously described (27, 28).
Plasmids and siRNAs.
Expression vectors for Flag-RIG-I, Flag-PACT, Myc-PACT, Us11, and EGFP-Us11 were described previously (13, 23, 27). Expression plasmids for truncated Us11 mutants Us11N and Us11C containing N- and C-terminal parts of the protein, respectively, were constructed by PCR. Primers for Us11N are 5′-ATCGAAGCTTTTATGAGCCAGACCCAACCC-3′ (forward) and 5′-ATCGCTGCAGCTATGTCCTGGGGATTGTTGGC-3′ (reverse). Primers for Us11C are 5′-ATCGAAGCTTTTCCGCGTGTTCCCCGGGA-3′ (forward) and 5′-ATCGCTGCAGCTATACAGACCCGCGAGCCG-3′ (reverse). Expression plasmids of domain 1- and domain 2-deleted PACT mutants (PACTΔD1 and PACTΔD2) were constructed by overlap extension PCR (29). Primers for PACTΔD1 are 5′-ATCGGGATCCATGTCCCAGAGCAGGCACGG-3′ (forward), 5′-ATCGAAGCTTTTACTTTCTTTCTGCTAT-3′ (reverse), 5′-CAGCTAAGCCAGGGAAAACAAAAGCCAATGCAAGTATTTGCT-3′ (forward overlap), and 5′-AGCAAATACTTGCATTGGCTTTTGTTTTCCCTGGCTTAGCTG-3′ (reverse overlap). Primers for PACTΔD2 are 5′-ATCGGGATCCATGTCCCAGAGCAGGCACGG-3′ (forward), 5′-AGCAAATACTTGCATTGGCTTTTGTTTTCCCTGGCTTAGCTG-3′ (reverse), 5′-AGCAACCAAAGAACCAGCTTAATAGTAATATTTCTCCAGAGA-3′ (forward overlap), and 5′-AAATGTGGTTCTCTGGAGAAATATTACTATTAAGCTGGTTCT-3′ (reverse overlap). Small interfering RNAs (siRNAs) against PACT, siPACT1 and siPACT2 (siPACT1/2), were described elsewhere (23).
Biochemical fractionation.
HEK293T cells were infected with HSV-1-EGFP-Us11 virus. Cells were harvested at 14 h postinfection and lysed in fractionation buffer (1% NP-40, 150 mM NaCl, 20 mM Tris, pH 8.0). Cell lysates were clarified and fractionated on a Sephadex 200 column as described previously (30). Fractions were run on an SDS-PAGE gel and blotted with anti-green fluorescent protein (GFP) and anti-PACT from Abcam (Cambridge, MA, USA).
Protein and RNA analysis.
Western blotting, coimmunoprecipitation, dual-luciferase assays, and quantitative reverse transcription-PCR (RT-PCR) were performed as described previously (31–33). For immunoprecipitation, mouse anti-Flag (M2) from Sigma-Aldrich (St. Louis, MO, USA) and rabbit anti-green fluorescent protein (GFP) from Abcam were used. For Western blotting, mouse anti-Flag (M5) from Sigma, mouse anti-GFP (B-2) from Santa Cruz (Dallas, TX, USA), mouse anti-ICP0 from Santa Cruz, and mouse anti-β-actin from Sigma were used. For quantitative RT-PCR, the level of target mRNA was calculated from 2−ΔCT by the comparative threshold cycle (CT) method (31). Primers used in quantitative RT-PCRs are 5′-GCACTGGCTGGAATGAGACTA-3′ (human IFN-β forward), 5′-CTCCTTGGCCTTCAGGTAAT-3′ (human IFN-β reverse), 5′-GCATCTGCCTCCCCATATT-3′ (human CCL5 forward), 5′-AGCACTTGCCACTGGTGTAG-3′ (human CCL5 reverse), 5′-GACCTGACGGTGAAGATGCT-3′ (human ISG15 forward), 5′-GAAGGTCAGCCAGAACAGGT-3′ (human ISG15 reverse), 5′-GCAGCCAAGTTTTACCGAAG-3′ (human ISG56 forward), 5′-GCCTTTCTCCGAAGTTTCCT-3′ (human ISG56 reverse), 5′-CACAGCCCTCTCGATCAACT-3′ (mouse IFN-β forward), 5′-GCATCTTCTCCGTCATCTCC-3′ (mouse IFN-β reverse), 5′-TTCTTGGGACTGATGCTGGT-3′ (mouse interleukin-6 [IL-6] forward), and 5′-GCCATTGCACAACTCTTTTCT-3′ (mouse IL-6 reverse). Primers used to amplify glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA were described elsewhere (31).
RESULTS AND DISCUSSION
Us11 targeting of PACT.
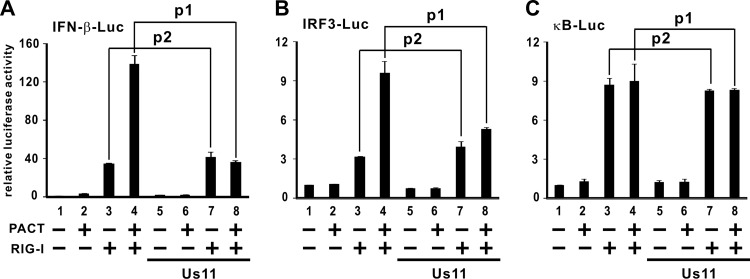
RIG-I can activate IFN production, and PACT potently augments this activity (23). Us11 is thought to directly bind RIG-I and MDA5 to inhibit their functions (15), but it also physically binds PACT (13). Thus, it will be of interest to see whether the inhibitory effect of Us11 on IFN production is PACT dependent or not. To shed light on this, we analyzed the inhibitory activity of Us11 in the presence of RIG-I and PACT. In line with our previous findings (23), PACT potentiated the stimulatory effect of RIG-I on luciferase reporter expression driven by IFN-β promoter (IFN-β-Luc) or by interferon regulatory factor 3 (IRF3)-binding enhancer elements (IRF3-Luc), but not by κB elements (κB-Luc) (Fig. 1A to C). Interestingly, in HEK293 cells, where the level of endogenous PACT is relatively low (23), Us11 did not suppress RIG-I activity on IFN-β-Luc or IRF3-Luc in the absence of ectopically expressed PACT (Fig. 1A and B, compare lanes 3 and 7). In other words, RIG-I was functionally intact in the activation of IFN production in the presence of both RIG-I and Us11. In contrast, Us11 almost completely blunted the PACT-augmented activation of RIG-I when both PACT and RIG-I were overexpressed (Fig. 1A and B, compare lanes 4 and 8). As reported previously (23) and as verified in the results shown in Fig. 1, while RIG-I activates both IRF3 and NF-κB, PACT can stimulate RIG-I-induced activation only of IRF3 and not of NF-κB. If Us11 targets RIG-I directly, it would probably suppress both IRF3 and NF-κB. In contrast, if Us11 counteracts PACT, it should not affect NF-κB activation. Indeed, Us11 had no effect on the activation of κB-Luc reporter by RIG-I (Fig. 1C). Thus, Us11 apparently inhibited IFN production by antagonizing PACT but not RIG-I directly.
Fig 1.
Us11 inhibits PACT-induced activation of RIG-I. (A) Influence on IFN-β promoter. HEK293 cells were transfected with pIFN-β-Luc reporter and expression plasmids for Flag-PACT, Flag-RIG-I, and Us11. Dual-luciferase assays were performed at 33 h posttransfection. Results represent the means ± standard deviations of three independent measurements of firefly luciferase activity normalized to Renilla luciferase activity. Differences between the selected groups were statistically assessed by a two-tailed Student's t test: p1 = 0.002; p2 = 0.087. (B) Influence on IRF3-binding elements (p1 = 0.012; p2 = 0.083). (C) Influence on κB enhancer (p1 = 0.416; p2 = 0.393).
Us11 associates with PACT to impede its interaction with RIG-I.
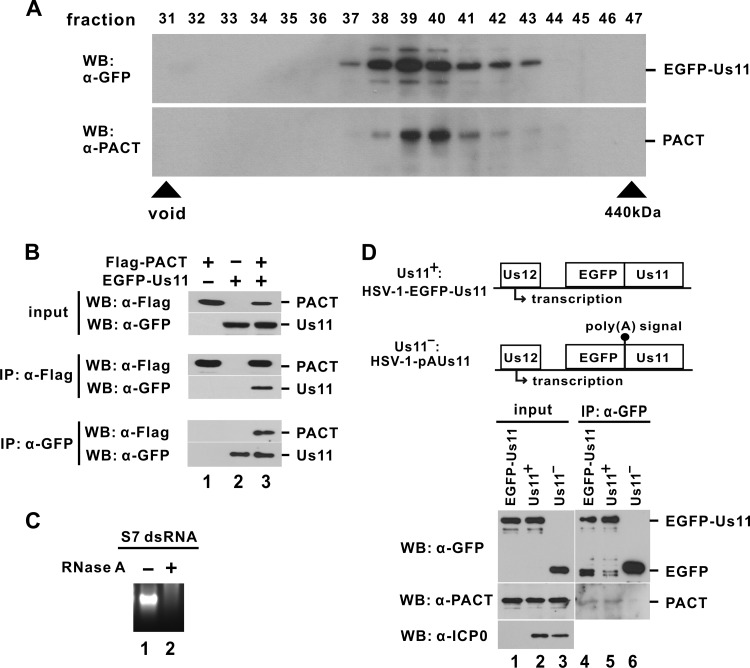
Since both Us11 and PACT are dsRNA-binding proteins (7, 12, 23), their association could be mediated through dsRNA. Although Us11 interacts with PACT in vitro (13), it is not known whether they associate with each other in a dsRNA-independent manner in infected cells. To verify this, we infected HEK293T cells with HSV-1-EGFP-Us11, a recombinant virus expressing an EGFP-Us11 fusion protein (27, 28). Fusion of EGFP with Us11 does not affect biological function of Us11 but provides a useful marker for the tracing of infected cells (27, 28). The expression level of endogenous PACT in HEK293T cells is relatively higher than in HEK293 cells. Biochemical fractionation in the presence of RNase A indicated the presence of PACT and EGFP-Us11 in the same fractions (fractions 38 to 42) of the protein extract of infected cells (Fig. 2A). Reciprocal coimmunoprecipitation and immunoblotting experiments confirmed RNase A-insensitive association of EGFP-Us11 and Flag-PACT in transfected HEK293T cells (Fig. 2B), consistent with the possibility that this interaction is RNA independent. In particular, both proteins were found in the precipitate prepared with anti-Flag or anti-GFP (Fig. 2B, compare lane 3 to lanes 1 and 2). To verify the digestion of dsRNA by RNase A in our experimental setting, we incubated an in vitro-transcribed and annealed dsRNA derived from influenza A virus genomic segment 7 (S7 dsRNA) with RNase A in the same buffer used in our fractionation and immunoprecipitation experiments. Complete degradation of S7 dsRNA indicated the effectiveness of RNase A in dsRNA digestion under our conditions (Fig. 2C, compare lanes 1 and 2). Similar results were also obtained when we repeated our fractionation and immunoprecipitation experiments in the presence of RNase III, which degrades dsRNA more efficiently (data not shown). Hence, the association between Us11 and PACT was not sensitive to RNase A or RNase III digestion and thus was unlikely mediated through dsRNA. Furthermore, immunoprecipitation was also performed with lysates of HEK293T cells infected with Us11-proficient HSV-1-EGFP-Us11 and Us11-deficient HSV-1-pAUs11 viruses (27). Notably, the Us11 locus of HSV-1-pAUs11 is not expressed, as depicted in Fig. 2D. Endogenous PACT protein was found in the EGFP-Us11-containing immune complex obtained from EGFP-Us11-overexpressing and HSV-1-EGFP-Us11-infected cells but not from cells infected with HSV-1-pAUs11 (Fig. 2D, compare lanes 4 and 5 to lane 6). Thus, Us11 associates with endogenous PACT in infected cells.
Fig 2.
Association of Us11 with PACT. (A) Cofractionation of Us11 and PACT in HSV-1-infected cells. HEK293T cells were infected with HSV-1-EGFP-Us11 (multiplicity of infection of 1). Cells were harvested at 14 h postinfection. Protein extract was incubated with 25 μg/ml RNase A for 15 min at 4°C and then fractionated by Superdex 200 gel filtration. Column fractions were analyzed by Western blotting (WB). α, anti. (B) Coimmunoprecipitation of ectopically expressed Us11 and PACT. HEK293T cells were transfected with the indicated expression plasmids. Immunoprecipitations (IP) were carried out at 48 h posttransfection with mouse anti-Flag (α-Flag) or rabbit anti-GFP (α-GFP) in the presence of 25 μg/ml RNase A. (C) dsRNA digestion by RNase A. In vitro-transcribed and annealed dsRNA of about 1 kb derived from influenza A virus genomic segment 7 (S7 dsRNA) was incubated with 25 μg/ml RNase A for 15 min in the same buffer as described above. RNA was then analyzed by agarose gel electrophoresis. (D) Coimmunoprecipitation of Us11 and PACT in HSV-1-infected cells. HEK293T cells were infected with HSV-1-EGFP-Us11 or HSV-1-pAUs11 (multiplicity of infection of 1) or transfected with an EGFP-Us11 expression plasmid. Proteins were analyzed at 14 h after infection or transfection. Diagrams at the top of the panel depict the difference between HSV-1-EGFP-Us11 and HSV-1-pAUs11 viruses. The open reading frame of Us11 is disrupted by the upstream poly(A) signal in the HSV-1-pAUs11 virus. Expression of HSV-1 immediate-early protein ICP0, a marker of productive infection, was probed with mouse monoclonal anti-ICP0 (α-ICP0).
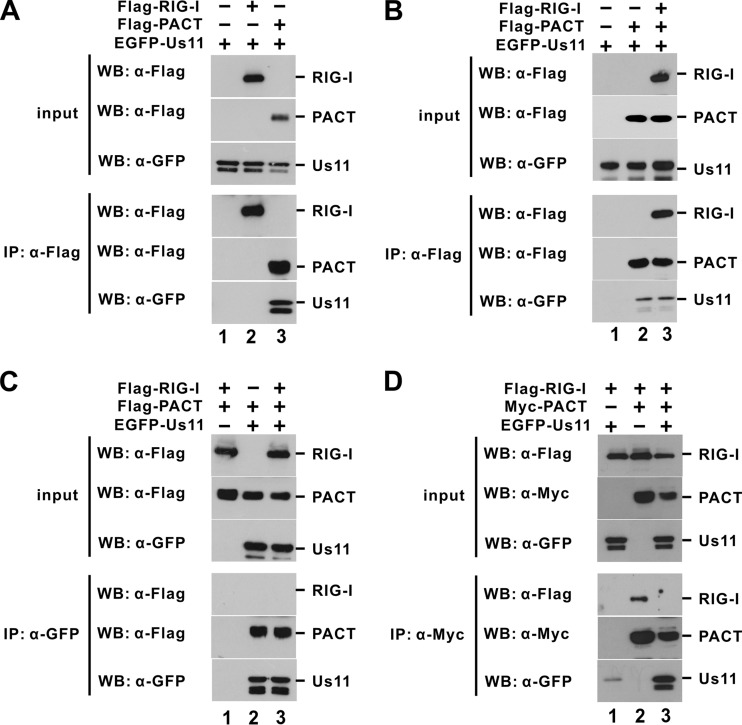
Us11 interacts with PACT (Fig. 2) and RIG-I (15). To perturb the function of RIG-I, Us11 could either impede the interaction between PACT and RIG-I or inhibit the activity of RIG-I in a triple complex containing all three proteins. To explore these two possibilities, we investigated whether PACT and RIG-I are mutually exclusive in the interaction with Us11. Different combinations of Us11, PACT, and RIG-I were expressed in HEK293T cells, and immunoprecipitation was subsequently performed to examine their interactions. When we overexpressed PACT and Us11, Us11 was detected in the protein complex that contains PACT (Fig. 3A, lane 3, B, lane 2, and C, lane 2). However, Us11 was not found in the RIG-I-containing complex when Us11 and RIG-I were overexpressed (Fig. 3A, lane 2). Although an interaction between Us11 and RIG-I could not be totally excluded, this result did suggest that Us11 might interact with PACT more potently or directly. We next performed reciprocal immunoprecipitation and immunoblotting experiments with HEK293T cells overexpressing RIG-I, PACT, and Us11. Only PACT and not RIG-I was found in the Us11-containing complex although Us11 was detected in the precipitate that harbors PACT or RIG-I (Fig. 3B and C, compare lanes 3). In the absence of Us11, PACT and RIG-I formed a complex (Fig. 3D, lane 2). However, when all three proteins were overexpressed, RIG-I disappeared from the complex that contains PACT (Fig. 3D, lane 3). Meanwhile, Us11 was detected in the PACT-containing complex (Fig. 3D, lane 3). These results were consistent with the model in which Us11 interacts directly with PACT to prevent it from interacting with RIG-I.
Fig 3.
Us11 prevents PACT from binding with RIG-I. HEK293T cells were transfected with plasmids expressing the indicated proteins. Immunoprecipitation (IP) and Western blotting (WB) were performed at 48 h posttransfection.
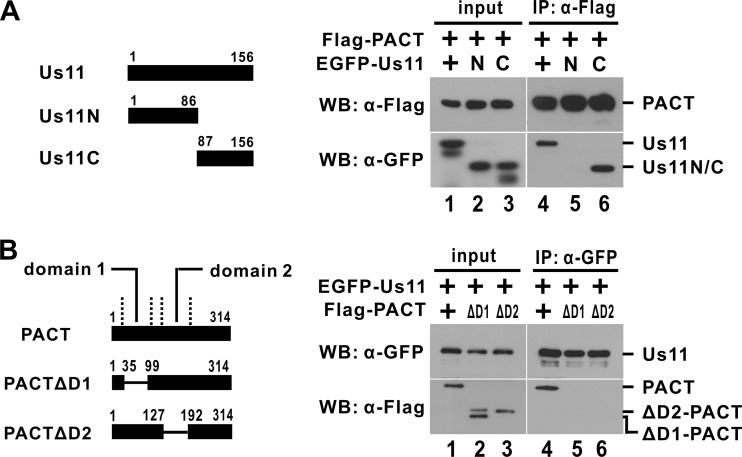
To map the interaction domains in Us11 and PACT, truncated or deletion mutants were constructed and analyzed (Fig. 4). Us11C containing the C-terminal RNA-binding domain, but not Us11N carrying the transactivation domain, was capable of interacting with PACT (Fig. 4A, compare lanes 5 and 6). On the other hand, PACT mutants ΔD1 and ΔD2 with deletions of dsRNA-binding domains 1 and 2, respectively, lost their ability to interact with Us11 (Fig. 4B, compare lane 4 to lanes 5 and 6). Thus, generally consistent with previous findings obtained from a glutathione S-transferase (GST) pulldown assay in vitro (13), RNA-binding domains on both proteins are required for the association between Us11 and PACT in vivo.
Fig 4.
Mapping of binding domains. HEK293T cells were transfected with plasmids expressing the indicated proteins. Immunoprecipitation (IP) and Western blotting (WB) were performed at 48 h posttransfection.
We demonstrated the importance of the C-terminal dsRNA-binding domain of Us11 in its interaction with PACT (Fig. 4). The same domain was also used in the interaction with RIG-I, MDA5, PKR, and OAS (13–15). The mechanism by which Us11 inhibits PACT function remains to be elucidated. Particularly, although we showed that Us11 and RIG-I are mutually exclusive in binding with PACT (Fig. 3), the interplay of Us11, PACT, RIG-I, and MDA5 in the activation of type I IFN production merits further investigations. MDA5 has also been implicated in the IFN antagonism of Us11 (15). Since PACT can also activate MDA5 (23), it will be of interest to see whether the suppression of MDA5 by Us11 is also mediated through a direct effect on PACT. A more detailed analysis of the dynamic interaction of Us11 with PACT, RIG-I, and MDA5 in infected cells by using biochemical cofractionation as in the experiment shown in Fig. 2A will shed significant light on the mechanism by which Us11 suppresses IFN production.
Results from our immunoprecipitation experiments suggested that Us11 associates with PACT more tightly or directly than with RIG-I (Fig. 3). In our setting, we were even unable to detect an interaction between Us11 and RIG-I. We therefore argued that Us11 suppresses RIG-I function through direct interaction with and inhibition of PACT. We did not exclude the possibility that Us11 might still interact with and inhibit RIG-I and MDA5, as previously reported (15). Plausibly, this could be mediated indirectly through RNA or an adaptor protein. Mapping the interaction domains and comparing the binding affinities between purified recombinant Us11 and PACT, between PACT and RIG-I, and between Us11 and RIG-I proteins using in vitro affinity binding assays, fluorescence anisotropy measurement, and surface plasmon resonance technology would clarify whether Us11, indeed, interacts with PACT with high affinity to prevent it from interacting with RIG-I and MDA5.
Requirement of PACT for Us11 suppression of IFN production.
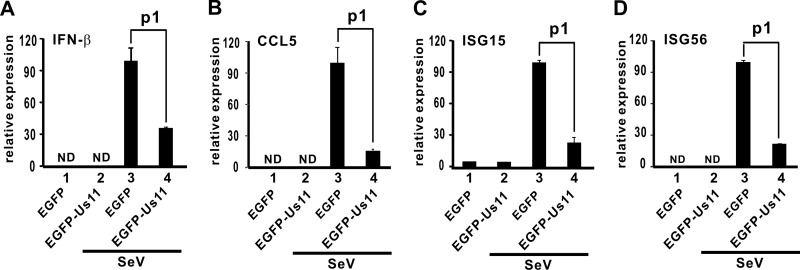
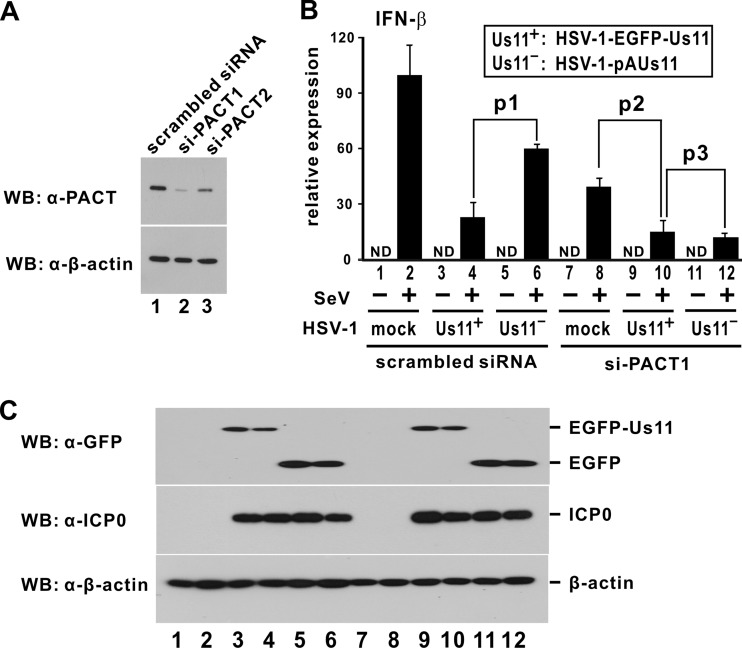
Sendai virus (SeV) is a potent inducer of type I IFNs. To verify the activity of Us11 to antagonize IFN production, we asked how expression of EGFP-Us11 might affect the induction of IFN and IFN-stimulated genes by SeV. We observed a significant reduction in the levels of IFN-β, CCL5, ISG15, and ISG56 transcripts in HEK293T cells stably expressing EGFP-Us11 (Fig. 5A to D, compare lanes 3 and 4). Thus, EGFP-Us11 can sufficiently suppress IFN production induced by SeV. With this in mind, we compared Us11-proficient HSV-1-EGFP-Us11 and Us11-deficient HSV-1-pAUs11 viruses in the suppression of SeV-induced IFN production in PACT-compromised cells. The knockdown effect of two siRNAs against PACT mRNA, siPACT1 and siPACT2, in HEK293T cells was verified by Western blotting (Fig. 6A). In the absence of siPACT1/2, infection with HSV-1-EGFP-Us11 dampened IFN-β induction by SeV (Fig. 6B, compare lanes 2 and 4). This effect was less pronounced when cells were infected with HSV-1-pAUs11 (Fig. 6B, compare lanes 4 and 6). However, in PACT knockdown cells transfected with siPACT1, HSV-1-EGFP-Us11 and HSV-1-pAUs11 were equally competent in the suppression of SeV-induced IFN-β production (Fig. 6B, compare lanes 10 and 12). In this setting, both viruses circumvented IFN production in a PACT-independent manner. The expression of EGFP-Us11 and ICP0, a marker for HSV-1 infection, in infected cells was verified (Fig. 6C). Similar results were also obtained with siPACT2 (data not shown). Thus, the inhibitory role of Us11 on SeV-induced IFN production requires PACT.
Fig 5.
Suppression of IFN-β production by EGFP-Us11. HEK293T cells stably expressing EGFP or EGFP-Us11 were infected with SeV (100 hemagglutinating units/ml). Relative expression of IFN-β mRNA (A) and IFN-stimulated transcripts CCL5, ISG15, and ISG56 (B to D) was analyzed by quantitative RT-PCR at 14 h postinfection and normalized to the level of GAPDH mRNA. Statistical analysis was performed with a two-tailed Student's t test. P values (p1) are as follows: 0.006 (A), 0.008 (B), 0.0002 (C), and 0.00003 (D). ND, not detected.
Fig 6.
Suppression of IFN-β induction by Us11-proficient and Us11-deficient HSV-1 viruses in PACT-compromised cells. (A) Knockdown of PACT expression by siRNAs. HEK293T cells were transfected with siPACT1/2 or scrambled siRNA. Proteins were analyzed at 72 h posttransfection. (B) Comparison of Us11-proficient and Us11-deficient viruses in the suppression of IFN-β induction in PACT-depleted cells. HEK293T cells were transfected with siPACT1 or scrambled siRNA. Cells were infected first with SeV (100 hemagglutinating units/ml) at 72 h posttransfection and then with HSV-1-EGFP-Us11 or HSV-1-pAUs11 (multiplicity of infection of 1) after another 8 h. Levels of IFN-β mRNA were determined by quantitative RT-PCR at 14 h after HSV-1 infection. Results represent the means ± standard deviations of three measurements of expression levels normalized to the level of GAPDH mRNA as calculated from 2−ΔCT. The differences between lanes 4 and 6 as well as between lanes 8 and 10 are statistically significant (p1 = 0.008 and p2 = 0.004) by a two-tailed Student's t test, whereas the difference between lanes 10 and 12 is statistically insignificant (p3 = 0.287). ND, not detected. (C) Verification of protein expression.
Neither HSV-1-EGFP-Us11 nor HSV-1-pAUs11 induced IFN-β in infected cells (Fig. 6B, lanes 3 and 5). This was generally consistent with previous findings on wild-type HSV-1 (2). HSV-1 encodes multiple viral proteins that antagonize IFN production (2–4). The induction of IFNs by either Us11-proficient or Us11-deficient HSV-1 would be efficiently suppressed by virus-encoded IFN-antagonizing proteins such as ICP0 and ICP34.5 (3, 4). In view of this, in the next part of our study we employed ICP34.5-deficient viruses to investigate IFN induction by HSV-1 and the influence of Us11 on this induction.
Both HSV-1-EGFP-Us11 and HSV-1-pAUs11 viruses were capable of suppressing SeV-induced IFN production in PACT-compromised cells (Fig. 6B, compare lane 8 to lanes 10 and 12). This suppression might be mediated through other IFN-antagonizing proteins such as ICP0 and ICP34.5 in a PACT- and Us11-independent manner. More experiments are required to clarify whether PACT is dispensable for ICP0- and ICP34.5-mediated suppression of innate antiviral response.
PACT acts upstream of RIG-I and MDA5 in the activation of IFN production (23). That is to say, RIG-I and MDA5 functions are severely impaired when PACT is compromised. Although our results did not support a direct interaction between Us11 and RIG-I (Fig. 3), Us11 could still inhibit the activity of RIG-I and MDA5 through PACT or other proteins. Thus, the requirement of RIG-I and MDA5 for Us11-dependent suppression of IFN production should be further investigated in RIG-I−/− and MDA5−/− MEFs.
Requirement of PACT for HSV-1 induction of IFN production.
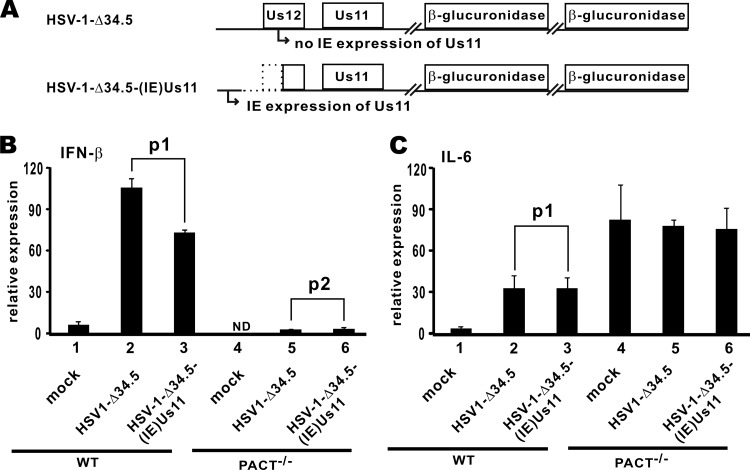
As described above, we used SeV to induce IFN production in HSV-1-infected cells. Since the induction and action of IFNs were inhibited in part by HSV-1 ICP34.5 (2, 4), here we employed an ICP34.5-deleted virus named HSV-1-Δ34.5 to induce IFNs. Us11 was not expressed in the early phase of infection when the cells were collected for analysis. Another virus, HSV-1-Δ34.5-(IE)Us11, from which Us11 is expressed in the immediate-early phase of infection (Fig. 7A), was also included in our analysis. The latter virus would allow us to analyze the effect of Us11 expression in the immediate-early phase on IFN production (27). These two viruses induced IFN-β expression swiftly after infection of wild-type MEFs that were PACT proficient. As expected, the induction was less pronounced in HSV-1-Δ34.5-(IE)Us11-infected cells due to immediate-early expression of Us11 that antagonizes IFN production (Fig. 7B, compare lanes 2 and 3). To shed light on the role of PACT in HSV-1 induction of type I IFNs, we employed PACT−/− MEFs in which the PACT locus was genetically disrupted. As previously characterized in the literature and by us, no functional PACT protein is expressed in these MEFs (24–26). In sharp contrast to the results obtained in PACT-proficient cells, no IFN-β was induced when these PACT−/− MEFs were infected with either virus (Fig. 7B, lanes 5 and 6). That is to say, PACT is required for IFN induction by HSV-1.
Fig 7.
IFN-β induction by HSV-1 was abrogated in PACT−/− cells. (A) Diagrams of the genomic structure of recombinant viruses. In HSV-1-Δ34.5 virus, both ICP34.5 alleles have been replaced with the β-glucuronidase gene. No Us11 is expressed in the immediate-early (IE) phase. In HSV-1-Δ34.5-(IE)Us11 virus, a spontaneous deletion results in the expression of Us11 from the IE promoter of Us12. (B and C) Viral induction of IFN-β and IL-6 in PACT−/− cells. Wild-type (WT) and PACT−/− MEFs were infected with HSV-1-Δ34.5 or HSV-1-Δ34.5-(IE)Us11 (multiplicity of infection of 2.5). Cells were harvested at 4 h postinfection, and total RNA was extracted. Expression of IFN-β (B) and IL-6 (C) mRNAs relative to GAPDH transcript was analyzed by quantitative RT-PCR. RNA levels were derived from 2−ΔCT. The difference between lanes 2 and 3 in panel B is statistically significant (p1 = 0.009) by a two-tailed Student's t test, whereas the differences between lanes 5 and 6 in panel B as well as between lanes 2 and 3 in panel C are statistically insignificant (p2 = 0.324 in panel B and p1 = 0.957 in panel C).
We also examined the induction of IL-6, the expression of which is controlled primarily by NF-κB, in MEFs infected with the two mutant HSV-1 viruses. IL-6 expression was activated in PACT-proficient wild-type MEFs infected with either virus (Fig. 7C, lanes 2 and 3). Consistent with earlier results on the effect of Us11 on NF-κB activation obtained in transfected cells overexpressing Us11 (Fig. 1C), immediate-early expression of Us11 did not affect IL-6 induction (Fig. 7C, compare lanes 2 and 3). In the absence of PACT, the basal expression of IL-6 was elevated for unknown reasons (Fig. 7C, lanes 4 to 6). In this setting, the two viruses were unable to further activate IL-6 expression. Nevertheless, Us11 was not influential in the activation of IL-6 production.
Exactly how host cells sense HSV-1 to induce type I IFNs remains mysterious (1, 2). RIG-I has been implicated in this process (1, 20, 21). More recent evidence points to the importance of DNA sensors such as IFI16, cGAS, and STING (34–36). Our findings that neither HSV-1-Δ34.5 nor HSV-1-Δ34.5-(IE)Us11 virus induced IFN-β in PACT−/− MEFs suggested the requirement of PACT for the sensing of HSV-1. It remains to be elucidated as to whether and how PACT cooperates with RIG-I or other sensors to mediate the activation of IFN production by HSV-1. PACT is also known to affect PKR and Dicer function (13, 30). Involvement of PKR and/or Dicer in the HSV-1-induced innate immune response could not be excluded and merits further analysis. Theoretically, Us11 and other HSV-1 proteins might also counteract the action of PACT in the sensing of HSV-1. To clarify this, the influence of PACT and Us11 on the induction of IFN by HSV-1 DNA should be assessed.
We obtained two lines of evidence concerning the influence of Us11 on NF-κB activation. First, expression of Us11 did not influence RIG-I-induced activation of NF-κB (Fig. 1C). Second, HSV-1-Δ34.5 virus did not induce more IL-6 than HSV-1-Δ34.5-(IE)Us11 (Fig. 7C). Because PACT does not affect RIG-I-dependent activation of NF-κB (23), our results that Us11 did not inhibit NF-κB activation are in keeping with the notion that Us11 directly suppresses PACT but not RIG-I. Us11 was previously found to inhibit NF-κB activation induced by SeV (15). More experiments are needed to clarify whether this might be mediated through an RIG-I-independent mechanism. The induction of IL-6 in mock-infected PACT−/− MEFs was indicative of NF-κB activation (Fig. 7C). PACT is known to affect NF-κB activation through PKR (37). The activation of NF-κB in PACT−/− MEFs might be independent of RIG-I and would be assessed in detail in the next phase of our study.
In summary, we demonstrated suppression of PACT-induced type I IFN production by HSV-1 Us11 protein. PACT-mediated enhancement of RIG-I activation was largely ablated by Us11 (Fig. 1), which interacts with PACT to impede its interaction with RIG-I (Fig. 2 to 4). The elevated induction of IFN-β during the infection of PACT-competent cells with Us11-deficient HSV-1-pAUs11 was not seen in PACT-compromised cells (Fig. 5 and 6). Finally, PACT was indispensable for the activation of type I IFN production by HSV-1 (Fig. 7). Our findings revealed PACT as an essential factor in viral induction of IFNs and a new target of a viral IFN-antagonizing protein.
Other viral IFN-antagonizing proteins such as influenza A virus NS1 (38, 39) and Ebola virus VP35 (40, 41) can also interact with PACT and perturb RIG-I function. Suppression of PACT-augmented activation of RIG-I might therefore represent a viral countermeasure to combat the host antiviral response commonly used by other viruses. One recent report on mutual antagonism between VP35 and PACT lent further support to this emerging new concept (42). It remains to be seen whether NS1 and PACT might also antagonize each other (43). In this regard, the influence of PACT on the function of Us11 in the life cycle of HSV-1 also warrants further analysis.
ACKNOWLEDGMENTS
We thank Takashi Fujita and Ganes Sen for reagents and Vincent Tang and Sam Yuen for critical readings of the manuscript.
This work was supported by the Hong Kong Health and Medical Research Fund (10091202 and 12111312), Hong Kong Research Grants Council (HKU7677/10 M and HKU1/CRF/11G), and SK Yee Medical Research Fund (2011).
Footnotes
Published ahead of print 25 September 2013
REFERENCES
- 1.Melchjorsen J. 2012. Sensing herpes: more than Toll. Rev. Med. Virol. 22:106–121 [DOI] [PubMed] [Google Scholar]
- 2.Paladino P, Mossman KL. 2009. Mechanisms employed by herpes simplex virus 1 to inhibit the interferon response. J. Interferon Cytokine Res. 29:599–607 [DOI] [PubMed] [Google Scholar]
- 3.Boutell C, Everett RD. 2013. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J. Gen. Virol. 94:465–481 [DOI] [PubMed] [Google Scholar]
- 4.Broberg EK, Hukkanen V. 2005. Immune response to herpes simplex virus and γ134.5 deleted HSV vectors. Curr. Gene Ther. 5:523–530 [DOI] [PubMed] [Google Scholar]
- 5.Cassady KA, Gross M, Roizman B. 1998. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic translation initiation factor 2. J. Virol. 72:8620–8626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulvey M, Poppers J, Ladd A, Mohr I. 1999. A herpesvirus ribosome-associated, RNA-binding protein confers a growth advantage upon mutants deficient in a GADD34-related function. J. Virol. 73:3375–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poppers J, Mulvey M, Khoo D, Mohr I. 2000. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J. Virol. 74:11215–11221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulvey M, Camarena V, Mohr I. 2004. Full resistance of herpes simplex virus type 1-infected primary human cells to alpha interferon requires both the Us11 and γ134.5 gene products. J. Virol. 78:10193–10196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishioka K, Ikuta K, Sato Y, Kaneko H, Sorimachi K, Fukushima E, Saijo M, Suzutani T. 2013. Herpes simplex virus type 1 virion-derived US11 inhibits type 1 interferon-induced protein kinase R phosphorylation. Microbiol. Immunol. 57:426–436 [DOI] [PubMed] [Google Scholar]
- 10.Roller RJ, Roizman B. 1990. The herpes simplex virus Us11 open reading frame encodes a sequence-specific RNA-binding protein. J. Virol. 64:3463–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoo D, Perez C, Mohr I. 2002. Characterization of RNA determinants recognized by the arginine- and proline-rich region of Us11, a herpes simplex virus type 1-encoded double-stranded RNA binding protein that prevents PKR activation. J. Virol. 76:11971–11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant KF, Cox JC, Wang H, Hogle JM, Ellington AD, Coen DM. 2005. Binding of herpes simplex virus-1 US11 to specific RNA sequences. Nucleic Acids Res. 33:6090–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters GA, Khoo D, Mohr I, Sen GC. 2002. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J. Virol. 76:11054–11064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez R, Mohr I. 2007. Inhibition of cellular 2′-5′ oligoadenylate synthetase by the herpes simplex virus type 1 Us11 protein. J. Virol. 81:3455–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing J, Wang S, Lin R, Mossman KL, Zheng C. 2012. Herpes simplex virus 1 tegument protein Us11 downmodulates the RLR signaling pathway via direct interaction with RIG-I and MDA-5. J. Virol. 86:3528–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lussignol M, Queval C, Bernet-Camard MF, Cotte-Laffitte J, Beau I, Codogno P, Esclatine A. 2013. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J. Virol. 87:859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel RC, Sen GC. 1998. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 17:4379–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]
- 19.Goubau D, Deddouche S, Reis E Sousa C. 2013. Cytosolic sensing of viruses. Immunity 38:855–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasmussen SB, Jensen SB, Nielsen C, Quartin E, Kato H, Chen ZJ, Silverman RH, Akira S, Paludan SR. 2009. Herpes simplex virus infection is sensed by both Toll-like receptors and retinoic acid-inducible gene-like receptors, which synergize to induce type I interferon production. J. Gen. Virol. 90:74–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melchjorsen J, Rintahaka J, Soby S, Horan KA, Poltajainen A, Ostergaard L, Paludan SR, Matikainen S. 2010. Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS-dependent and MDA5/MAVS/RNA polymerase III-independent pathways. J. Virol. 84:11350–11358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da Silva LF, Jones C. 2013. Small non-coding RNAs encoded within the herpes simplex virus type 1 latency associated transcript (LAT) cooperate with the retinoic acid inducible gene I (RIG-I) to induce beta-interferon promoter activity and promote cell survival. Virus Res. 175:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kok KH, Lui PY, Ng MH, Siu KL, Au SW, Jin DY. 2011. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe 9:299–309 [DOI] [PubMed] [Google Scholar]
- 24.Rowe TM, Rizzi M, Hirose K, Peters GA, Sen GC. 2006. A role of the double-stranded RNA-binding protein PACT in mouse ear development and hearing. Proc. Natl. Acad. Sci. U. S. A. 103:5823–5828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters GA, Seachrist DD, Keri RA, Sen GC. 2009. The double-stranded RNA-binding protein, PACT, is required for postnatal anterior pituitary proliferation. Proc. Natl. Acad. Sci. U. S. A. 106:10696–10701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Houzet L, Jeang KT. 2012. Tombusvirus P19 RNA silencing suppressor (RSS) activity in mammalian cells correlates with charged amino acids that contribute to direct RNA-binding. Cell Biosci. 2:41. 10.1186/2045-3701-2-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulvey M, Poppers J, Sternberg D, Mohr I. 2003. Regulation of eIF2α phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J. Virol. 77:10917–10928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benboudjema L, Mulvey M, Gao Y, Pimplikar SW, Mohr I. 2003. Association of the herpes simplex virus type 1 Us11 gene product with the cellular kinesin light-chain-related protein PAT1 results in the redistribution of both polypeptides. J. Virol. 77:9192–9203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 30.Kok KH, Ng MH, Ching YP, Jin DY. 2007. Human TRBP and PACT interact with each other and associate with Dicer to facilitate the production of small interfering RNA. J. Biol. Chem. 282:17649–17657 [DOI] [PubMed] [Google Scholar]
- 31.Tang HM, Gao WW, Chan CP, Siu YT, Wong CM, Kok KH, Ching YP, Takemori H, Jin DY. 2013. LKB1 tumor suppressor and salt-inducible kinases negatively regulate human T-cell leukemia virus type 1 transcription. Retrovirology 10:40. 10.1186/1742-4690-10-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siu KL, Kok KH, Ng MH, Poon VK, Yuen KY, Zheng BJ, Jin DY. 2009. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3 · TANK · TBK1/IKKε complex. J. Biol. Chem. 284:16202–16209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng MH, Ho TH, Kok KH, Siu KL, Li J, Jin DY. 2011. MIP-T3 is a negative regulator of innate type I interferon response. J. Immunol. 187:6473–6482 [DOI] [PubMed] [Google Scholar]
- 34.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paludan SR, Bowie AG. 2013. Immune sensing of DNA. Immunity 38:870–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao TS, Fitzgerald KA. 2013. The cGAS-STING pathway for DNA sensing. Mol. Cell 51:135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D'Acquisto F, Ghosh S. 2001. PACT and PKR: turning on NF-κB in the absence of virus. Sci. STKE 2001:re1. 10.1126/stke.2001.89.re1 [DOI] [PubMed] [Google Scholar]
- 38.Li S, Min JY, Krug RM, Sen GC. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13–21 [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Li M, Zheng H, Muster T, Palese P, Beg AA, Garcia-Sastre A. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of α/β interferon. J. Virol. 74:11566–11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardenas WB, Loo YM, Gale M, Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits α/β interferon production induced by RIG-I signaling. J. Virol. 80:5168–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabozzi G, Nabel CS, Dolan MA, Sullivan NJ. 2011. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J. Virol. 85:2512–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luthra P, Ramanan P, Mire CE, Weisend C, Tsuda Y, Yen B, Liu G, Leung DW, Geisbert TW, Ebihara H, Amarasinghe GK, Basler CF. 2013. Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe 14:74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kok KH, Jin DY. 2013. Balance of power in host-virus arms races. Cell Host Microbe 14:5–6 [DOI] [PubMed] [Google Scholar]