Abstract
The herpes simplex virus 1 (HSV-1) immediate-early protein, infected cell protein 22 (ICP22), is required for efficient replication in restrictive cells, for virus-induced chaperone-enriched (VICE) domain formation, and for normal expression of a subset of viral late proteins. Additionally, ICP22 is important for optimal acute viral replication in vivo. Previous studies have shown that the US1 gene that encodes ICP22, produces an in-frame, N-terminally truncated form of ICP22, known as US1.5. To date, studies conducted to characterize the functions of ICP22 have not separated its functions from those of US1.5. To determine the individual roles of ICP22 and US1.5, we made viral mutants that express either ICP22 with an M90A mutation in the US1.5 initiation codon (M90A) or US1.5 with three stop codons introduced upstream of the US1.5 start codon (3×stop). Our studies showed that, in contrast to M90A, 3×stop was unable to replicate efficiently in the eyes and trigeminal ganglia of mice during acute infection, to efficiently establish a latent infection, or to induce VICE domain formation and was only mildly reduced in its replication in restrictive HEL-299 cells and murine embryonic fibroblasts (MEFs). Both mutants enhanced the expression of the late viral proteins virion host shutoff (vhs) and glycoprotein C (gC) and inhibited viral gene expression mediated by HSV-1 infected cell protein 0 (ICP0). When we tested our mutants' sensitivity to type I interferon (beta interferon [IFN-β]) in restrictive cells, we noticed that the plating of the ICP22 null (d22) and 3×stop mutants was reduced by the addition of IFN-β. Overall, our data suggest that US1.5 partially complements the functions of ICP22.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) infects a majority of the world's population, causing diseases that range from vesicular eruptions around the mouth and keratitis to life-threatening encephalitis (1). During its life cycle, the virus switches between a lytic replication cycle and a latent form of infection (2). The lytic replication cycle is characterized by an ordered gene cascade that begins with immediate-early (IE) genes, followed by the early (E) and then late (L) genes (3). The latent form of infection, established when the virus reaches the sensory neurons innervating the primary site of infection, is characterized by an overall lack of lytic gene expression, with the exception of the latency-associated transcripts (LATs) (4). This latent infection is lifelong and can be reactivated via different stress stimuli.
Of the five IE proteins, infected cell protein 22 (ICP22) is the least studied, and its functions during viral replication are not fully understood. This is partially due to early reports that showed that ICP22 is dispensable for HSV-1 replication in Vero cells (5). Further studies demonstrated that ICP22 is required for efficient replication in a subset of cell lines, including primary human cell lines and rodent cells, which are designated restrictive cells (6, 7). In addition, ICP22 was shown to be necessary for optimal acute ocular and neuronal replication and the establishment of latency (7, 8). Another well-characterized function of ICP22 is the modification of the phosphorylation status of the C terminus of the large subunit of the host RNA polymerase II (9–11). Although the significance of this function in viral replication is not clear, it has been proposed that the altered modification of RNA polymerase II either enhances HSV-1 genome transcription or helps suppress cellular transcription (12). Recently, ICP22 was also shown to be required for the formation of virus-induced chaperone-enriched (VICE) domains (13). These domains were initially described to form in response to viral infection (14, 15) and contain many chaperone and polyubiquitinated proteins. Consequently, VICE domains are thought to have a role in nuclear protein quality control in viral infection (16).
ICP22 is encoded by the US1 gene and is composed of 420 amino acids. This protein has been shown to be extensively modified posttranslationally and appears to be phosphorylated, at least in part, by the virus-encoded kinase UL13 (17). Examining the functions of ICP22 has been complicated by the coexpression of an in-frame N-terminally truncated form, termed US1.5 (18). The US1.5 protein was initially proposed to be the product of a transcript which is distinct from that of ICP22 and whose translation initiates at the codon corresponding to methionine (M)147 of the ICP22 open reading frame (ORF) (18). When the expression of US1.5 was examined by Bowman and Schaffer, the authors reported that they were unable to detect a second transcript encoded by the US1 gene (19). Furthermore, mutagenesis studies established that expression or translation of the US1.5 protein originated from an alternate codon (i.e., M90) in the ICP22 ORF (19).
None of the studies conducted to characterize ICP22 has separated its functions from those of US1.5. Although viral mutants have been made to express only the US1.5 protein, these mutants were engineered based on the previously reported initiation codon (i.e., M147) of US1.5 (20, 21). The goal of the current study was to separate the functions of ICP22 from those of US1.5. Consequently, we generated mutant viruses from plasmids by mutating the mapped US1.5 initiation codon M90 to alanine (M90A) so that only ICP22 is expressed or by introducing three stop codons upstream of the initiation codon of US1.5 (3×stop) (19) so that only US1.5 is expressed. We then tested these mutant viruses for acute replication and establishment of latency in the mouse ocular model and for virus replication and VICE domain formation in cell culture-based assays. Moreover, we examined the inhibitory effect of both proteins on the transactivated gene expression of the HSV-1 IE protein, ICP0, and whether they contributed to the plating efficiency of HSV-1 in the presence and absence of type I interferon (beta interferon [IFN-β]). Our data indicate that although both ICP22 and US1.5 are able to enhance the maximum expression of the late viral proteins virion host shutoff (vhs) and glycoprotein C (gC) and inhibit ICP0-mediated gene expression in cell culture, only ICP22 is required for efficient acute viral replication and the establishment of latency in vivo and for the formation of VICE domains. Additionally, our data demonstrate a new function for ICP22 in counteracting the antiviral effects of IFN-β.
MATERIALS AND METHODS
Cells and viruses.
Vero cells and HEL-299 cells were obtained from the American Type Culture Collection (ATCC). Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum (FBS), 2 mM l-glutamine, 100 μg/ml penicillin, and 100 U/ml streptomycin. HEL-299 cells were grown in minimum essential medium (MEM) alpha supplemented with 10% FBS, l-glutamine, and antibiotics as described for Vero cells. CD-1 mouse embryonic fibroblasts (MEFs) (kindly provided by Kristi Neufeld) and 129 MEFs (prepared in our laboratory under a protocol approved by the University of Kansas Institutional Animal Care and Use Committee) were grown in the same medium as Vero cells except that the DMEM was supplemented with 15% FBS. The wild-type HSV-1 strain KOS (passage 11) was used in our study. All the viruses were propagated in and titers were determined on Vero cells.
Construction of mutant and marker rescue (MR) viruses.
To make our mutants in a KOS background, we initially made the ICP22 null mutant, d22, by marker transfer. For the construction of this virus, Vero cells were seeded on 60-mm dishes at 5 × 105 cells/plate. Twenty-four hours later, cells were cotransfected with 1 μg of KOS viral DNA and 2.5 μg of AgeI-linearized pUCNS:lacZ plasmid (22) (kindly provided by Stephen Rice). Cells were transfected with Fugene HD (Roche) at a ratio of 3:1 (μl of transfection reagent to μg of DNA) according to the manufacturer's recommendations. Mutants were identified by blue/white selection in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Blue plaques were picked and plaque purified at least three times. Correct insertion of the mutation was confirmed by BamHI digestion of viral DNA and Southern blot analyses (Fig. 1B). To create the M90A and 3×stop mutant viruses, pAlter-M90A and pAlter-3×stop plasmids (kindly provided by Priscilla Schaffer's lab) (19) were digested with EcoRI and KpnI and cotransfected with d22 viral DNA. White plaques were picked and purified at least three times. Correct insertion of the M90A mutation was confirmed with BamHI and PciI digests and Southern blot analyses (Fig. 1B). Correct insertion of the 3×stop mutation was confirmed with SacII digests and Southern blot analyses (Fig. 1B). M90AMR and 3×stopMR viruses were made by cotransfecting the corresponding mutant viral DNA with the wild-type pAlter-ICP22 plasmid cut with EcoRI and KpnI. Plaques were picked randomly, and the rescuants were initially identified by PCR and restriction enzyme digestion analyses; rescue by the wild-type ICP22 sequences was confirmed by Southern blot analyses (H. H. Mostafa and D. J. Davido, unpublished data).
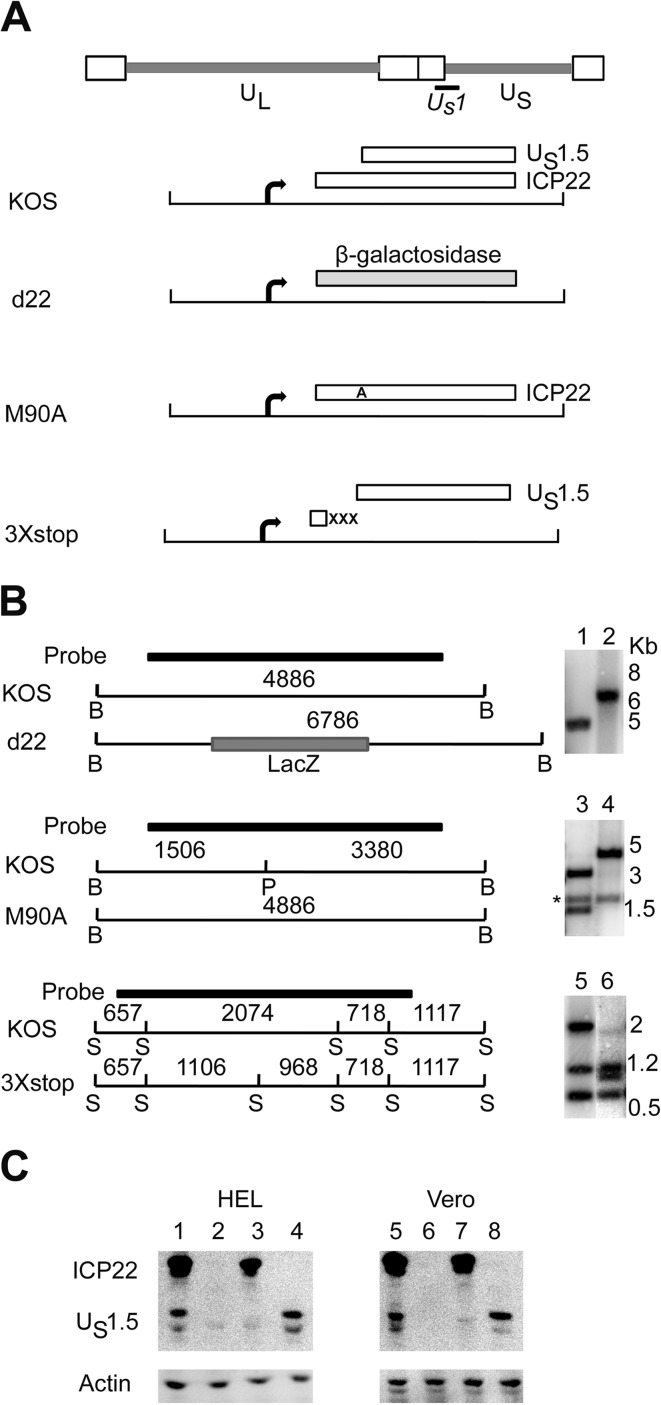
Fig 1.
Construction of ICP22 and US1.5 mutant viruses. (A) Diagram of the HSV-1 genome showing the US1 gene, which encodes ICP22 and US1.5 protein. The open boxes denote the repeated sequences flanking the unique long (UL) and unique short (US) segments. The ICP22 null mutant, d22, has the lacZ gene (which codes for β-galactosidase) inserted in place of the ICP22 ORF. M90A is mutated such that only ICP22, but not US1.5, is produced as the initiation codon of US1.5 is mutated to alanine. 3×stop produces only US1.5 and not ICP22 as this mutant has three stop codons inserted upstream of the initiation codon of US1.5. (B) The restriction enzyme digestion patterns with the fragment lengths (indicated in base pairs) on the left (B, BamHI; P, PciI; S, SacII) and Southern blots shown on the right. The M90A and 3×stop mutations were designed to eliminate a PciI site and add an SacII site, respectively. Lanes 1 and 2, KOS and d22, respectively, cut with BamHI; lanes 3 and 4, KOS and M90A, respectively, cut with PciI; lanes 5 and 6, KOS and 3×stop, respectively, cut with SacII. The star in the second blot denotes a 1.9-kb BamHI fragment of the opposite US/RS junction (21). (C) ICP22 and US1.5 protein expression of KOS (lanes 1 and 5), d22 (lanes 2 and 6), M90A (lanes 3 and 7), and 3×stop (lanes 4 and 8) mutant viruses at 8 h postinfection in HEL-299 cells or Vero cells, as determined by Western blot analyses.
Acute viral replication in mice.
Infections were performed as previously described (23). Briefly, CD-1 outbred female mice (6 to 7 weeks old) were purchased from Charles Rivers Laboratories (Shrewsbury, MA) and cared for according to Guide for the Care and Use of Laboratory Animals (24). The protocol for using these mice was approved by the University of Kansas Institutional Animal Care and Use Committee. Mice were anesthetized by intraperitoneal injection of ketamine (75 to 100 mg/kg of body weight) and xylazine (10 mg/kg of body weight). Corneas of mice were scarified with a 26-gauge needle and infected with KOS, d22, M90A, 3×stop, or each MR virus at 2 × 105 PFU of virus per eye in 5 μl of medium. To determine acute ocular titers, the eyes of infected mice were swabbed at 4 h and on days 1, 3, and 5 postinfection using moistened cotton-tipped swabs, and samples were placed in 500 μl of Vero cell growth medium. To determine acute replication in the trigeminal ganglia (TG), mice were euthanized by CO2 asphyxiation at days 3 and 5 postinfection, and TG were removed and placed in 500 μl of 1% FBS growth medium and 100 μl of 1-mm glass beads. Samples were homogenized with a Mini-Beadbeater 8 (BioSpec). Virus titers for all samples were determined on Vero cells, and differences in virus titers were evaluated using a Student's t test.
Viral pathogenicity scoring.
At day 8 postinfection, mice were scored for the appearance of gross lesions based on the following scoring system: 0, no signs of infection; 1, swollen eyelids; 2, less than 20% removal of hair around the eyes; 3, 20 to 70% removal of hair between the eyes; and 4, more than 70% removal of hair between the eyes. Statistical analyses were performed using a Student's t test.
Latent viral genome loads in TG.
At day 28 postinfection, TG were collected, and DNA was isolated from each TG as previously described (25). PCR primers specific for the HSV-1 UL50 gene were used to measure viral DNA loads (26), and primers for the mouse adipsin gene were used as a loading control (27). Real-time PCR was performed in a total volume of 25 μl per sample of FastStart SYBR green Master (Roche, Indianapolis, IN) and primers in a StepOnePlus Real-Time PCR System (Applied Biosystems). For UL50 PCR samples, 125 ng of DNA was used per reaction mix, and for adipsin PCR samples, 10 ng of DNA was used per reaction mix. All samples were analyzed in duplicate. Standard curves for each PCR condition were carried out as described previously (27) to quantify the amount of viral DNA present in each sample relative to the adipsin gene using the 2−ΔΔCT (where CT is threshold cycle) method (28). Statistical analyses were performed using a one-way analysis of variance (ANOVA) test.
Viral yield assays.
HEL-299 cells or MEFs were plated at 1 × 105 cells per well in 12-well plate. Twenty-four hours later, cells were infected at a multiplicity of infection (MOI) of 0.1 with each virus. One hour after addition of the virus, cells were washed with acid wash buffer for 1 min to inactivate any unabsorbed virus, then washed with phosphate-buffered saline (PBS), and supplemented with HEL-299 or MEF growth medium. Twenty-four hours postinfection, cells were harvested, and viral yields were determined by standard plaque assays. For viral yield assays in the presence of IFN-β, the same protocol was followed as above, with the exception that HEL-299 cells and MEFs were pretreated with human IFN-β (catalog number 11410-2; PBL InterferonSource) or mouse IFN-β (catalog number 12400-1; PBL InterferonSource), respectively, at 1,000 units per ml for 16 h before the addition of virus. The same concentration of IFN-β was added to PBS washes and media during the course of infection.
Western blot analysis.
HEL-299 cells and MEFs were plated on 12-well plates at 1 × 105 cells per well. Twenty-four hours postplating, cells were infected at an MOI of 2 for each virus. Samples were harvested at 8 or 24 h postinfection in 50 μl of 1× Laemmli buffer (100°C) supplemented with 1× protease inhibitors (1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride [PMSF]). Samples were heated at 95°C for 5 min, vortexed, and centrifuged. Ten microliters per sample was resolved on a 6% SDS-PAGE gel. Proteins were transferred to nitrocellulose membranes using a semidry transfer unit (catalog number TE77; GE Healthcare). Each membrane was blocked in 5% bovine serum albumin (BSA) in 0.1% Tween 20 diluted in Tris-buffered saline (TBS-T) for 1 h at room temperature. The following primary antibodies were used: for ICP22, rabbit polyclonal antibody 413 produced by Bethyl Laboratories (19), diluted 1:5,000; for β-actin, a rabbit polyclonal antibody (catalog number sc-1616; Santa Cruz Biotechnology), diluted 1:1,000; for vhs, a rabbit polyclonal antibody described in Read et al. (29), diluted 1:100; and for gC, a mouse monoclonal antibody (catalog number sc-56982; Santa Cruz Biotechnology), diluted 1:250. The membranes were washed three times in TBS-T, and probed with peroxidase-conjugated goat antirabbit (Jackson ImmunoResearch) diluted 1:1,000 and peroxidase-conjugated goat antimouse (Jackson ImmunoResearch) diluted 1:1,000 at room temperature for 1 h. Membranes were washed three times in TBS-T and developed using SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific). Images were captured with a Kodak 4000R image station, and the band intensities were measured by densitometry using ImageJ.
Immunofluorescence studies.
HEL-299 cells or Vero cells were plated on glass coverslips in 24-well plates for 24 h (5 × 104 per well). Cells were then infected at an MOI of 0.1. After 24 h of infection, cells were fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature, washed with PBS, and blocked for 1 h in 5% normal goat serum and 0.2% Triton X-100 diluted in PBS at 37°C. The primary antibodies used were ICP22 rabbit polyclonal 413, described above in our Western blot studies, diluted 1:500, and Hsc70 rat monoclonal (catalog number SPA-815; Stressgen), diluted 1:500. Cells were incubated in primary antibodies for 1 h at 37°C and then washed at least three times. Each secondary antibody was added for 1 h at 37°C. The secondary antibodies used were goat anti-rabbit dy488 (Jackson ImmunoResearch), diluted 1:500, and goat anti-rat Alexa-Fluor 568 (Invitrogen),diluted 1:1,000. Cells were then washed three times with PBS, mounted with a Prolong Antifade Kit (Invitrogen), and examined by fluorescence microscopy (Nikon Eclipse TE-2000-U4).
Luciferase assays.
Vero cells were plated at 5 × 104 cells per well in 24-well plates for 24 h. Cells were cotransfected with 50 ng of the reporter plasmid (VP16 promoter-luciferase construct) (30) alone or with 100 ng of pAlter-1+ICP0 (pICP0), or pAlter-1 that expresses both ICP22 and US1.5 (pICP22), only ICP22 (pM90A) (19), or only US1.5 (p3×stop) (19), all driven by their native promoters, or with a combination of 100 ng of pICP0 and 100 ng of pICP22, pM90A, or p3×stop. In all cases, salmon sperm DNA was added for a total of 1 μg per well. Transfections were performed according to the manufacturer's protocol using Fugene HD at a ratio of 3:1. Cells were transfected for 48 h, wells were washed with PBS, and each well was then harvested in 100 μl of 1× passive lysis buffer (PLB) (catalog number E1941; Promega) by rocking at room temperature for 15 min. Samples were harvested in microcentrifuge tubes, vortexed for 30 s, and briefly centrifuged at room temperature. Fifty microliters of each sample was then added to 50 μl of luciferase assay buffer (125 mM sodium MES [2-(N-morpholino)ethanesulfonic acid] pH 7.8, 25 mM magnesium acetate, 2 mg of ATP per ml) as previously described (31). Luciferase assays were conducted using the Promega Luciferase Assay System 1000; 50 μl of 1 mM d-luciferin was added per sample, and there was a 2-s delay before a 10-s read (HT Multicode Microplate Reader; Biotec Synergy). Light units are displayed as percentages normalized to the pICP0 values, and statistical analyses were performed using a Mann-Whitney U test.
Plaque reduction assays.
HEL-299 cells or MEFs were plated in 24-well plates at 5 × 104 cells per well for 24 h. Cells were then mock treated or pretreated with human IFN-β as described above for viral yield assays. Sixteen hours later, cells were infected with serially diluted KOS, d22, M90A, or 3×stop virus. At 1 h postinfection, cells were overlaid with 0.5% methylcellulose in the appropriate cell culture medium. Three days later, cells were fixed in 4% formaldehyde in PBS for 15 min at room temperature, washed with PBS, and stained for 1 h at room temperature with an anti-HSV-1 antibody (catalog number B0114; Dako), diluted 1:350, at room temperature with shaking. Plates were washed once with PBS and incubated with horseradish peroxidase-conjugated antibody (Jackson ImmunoResearch), diluted 1:500, for 1 h at room temperature. Plates were washed once with PBS, and plaques were visualized using an aminoethylcarbazole (AEC) peroxidase substrate kit (Vector Laboratories) according to the manufacturer's recommendations. Pictures were taken with a scanner, and the number of pixels within a given plaque (representing its area) was measured by Adobe Photoshop. Twenty plaques were examined for each virus.
RESULTS
Construction of ICP22 mutant viruses.
In order to separate the functions of ICP22 and the US1.5 protein during viral infection, we constructed two mutant viruses. The M90A mutant (Fig. 1A and B) was generated by homologous recombination as previously described (19), except that we used an ICP22 deletion virus (d22) that we generated in a KOS background (Fig. 1A, B, and C). M90A contains a methionine-to-alanine mutation in codon 90 of ICP22, which is the initiating codon for US1.5 (Fig. 1A and B). Consequently, this mutant expresses ICP22 and not US1.5 (Fig. 1C) (19). The 3×stop mutant virus was generated in a manner similar to that used for the M90A mutant virus. This mutant introduces three stop codons upstream of the initiation codon of US1.5 (Fig. 1A and B) and expresses the US1.5 protein and not ICP22 (Fig. 1C).
ICP22 but not US1.5 is required for efficient acute viral replication in vivo.
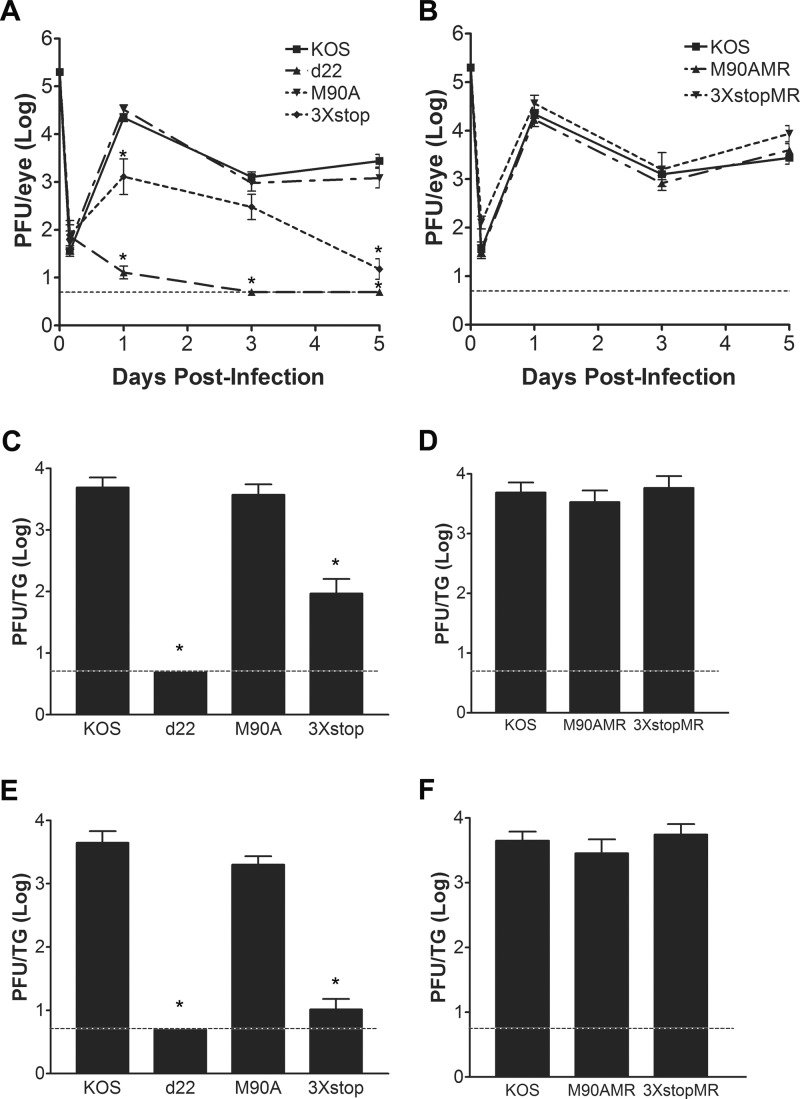
Genetic studies have demonstrated the importance of ICP22 for acute replication in vivo when the productive replication of various ICP22 mutant viruses was examined in different animals models (7, 8, 32). In order to test the individual roles of ICP22 and the US1.5 protein during acute viral replication in vivo, we used a mouse ocular model of HSV-1 infection. The eyes of mice were infected with KOS, d22, M90A, or 3×stop and, in the case of M90A and 3×stop, with their corresponding marker rescue (MR) viruses. Eye swabs were taken at 4 h and at days 1, 3, and 5 postinfection to monitor ocular replication, or trigeminal ganglion homogenates were prepared on days 3 and 5 postinfection to monitor acute replication in neurons. As shown in Fig. 2A, the M90A mutant virus replicated to levels comparable to the level of KOS in the eyes of mice for all time points tested. In contrast, the replication of the 3×stop mutant in the eyes was significantly reduced relative to that of KOS on day 1 (17-fold; t test, P = 0.01) and day 5 (184-fold; t test, P = 6.8 × 10−7) postinfection. The replication of d22 in the eyes of mice was significantly reduced at days 1, 3, and 5 postinfection by 1722-, 255-, and 554-fold, respectively (t test, P values of 5.9 × 10−11, 1.7 × 10−19, and 6.5 × 10−18, respectively). The replication of M90AMR and 3×stopMR viruses in the eyes was comparable to that of KOS (Fig. 2B). When acute replication of the M90A and 3×stop mutants in TG was examined, only the 3×stop mutant showed decreased growth at day 3 (59-fold; t test, P = 3.6 × 10−5) (Fig. 2C) and day 5 (429-fold; t test, P = 2.3 × 10−10) (Fig. 2E). The M90A mutant replicated as efficiently as KOS on both days (Fig. 2C and E). We were unable to detect progeny virus in the TG of d22-infected mice on days 3 and 5 postinfection (t test, P = 6.7 × 10−16 and P = 3.3 × 10−18, respectively) (Fig. 2C and E). The M90AMR and 3×stopMR viruses replicated at levels similar to the level of KOS in TG on both days (Fig. 2D and F). Thus, ICP22, not US1.5, is the major contributor to acute ocular and, consequently, neuronal replication.
Fig 2.
Acute replication of M90A and 3×stop mutants and MR viruses in vivo. (A and B) Acute ocular replication of M90A and 3×stop mutants and MR viruses in mice. CD-1 mice were infected with 2 × 105 PFU per eye. At the indicated time points, the eyes of mice were swabbed, and viral titers were determined by standard plaque assays. (C to F) Acute TG replication of M90A and 3×stop mutants and MR viruses in mice. CD-1 mice were infected with 2 × 105 PFU per eye. On day 3 (C and D) or day 5 (E and F) postinfection, mice were sacrificed, TG were removed and homogenized, and viral titers were determined by standard plaque assays. *, P < 0.05, compared to KOS (Student's t test). Error bars represent the standard errors of the means (n = 8 samples/virus/time point). In all cases, the dashed line is the limit of detection.
ICP22 but not US1.5 is required for external pathological lesions in mice.
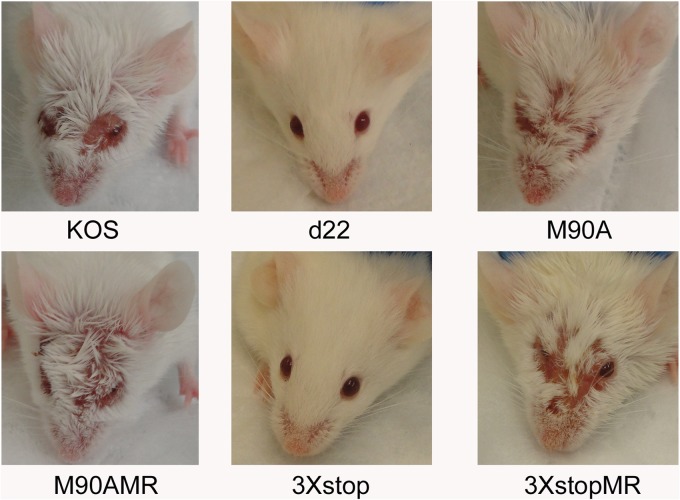
We scored the pathological lesions caused by the ocular infection of mice with KOS, d22, M90A, 3×stop, M90AMR, and 3×stopMR at day 8 postinfection. The severity of infection was ranked on a scale of 0 to 4 for each group, where 0 indicates no signs of infection and 4 indicates more than 70% loss of hair from between the eyes, which is the result of scratching or secondary inflammation. As shown in Fig. 3, the infection with the 3×stop mutant was similar to that with d22 in that it did not produce any signs of disease, with both groups of mice having pathology scores of 0 (t test, P = 9 × 10−5). Infection with the M90A mutant, on the other hand, was similar to that with KOS and the MR viruses, where inoculated mice showed obvious signs of infection (scores of 2.5 ± 0.5 for the M90A, KOS, and 3×stopMR viruses and 2.33 ± 0.47 for the M90AMR virus [means ± standard deviations; n = 5 to 6 mice per virus]) (Fig. 3). Our data indicate that, in contrast to ICP22, the expression of US1.5 is insufficient to induce signs of viral pathogenesis in mice.
Fig 3.
Gross pathological effects of M90A and 3×stop mutants and MR viruses. CD-1 mice were infected with 2 × 105 PFU per eye. A representative picture from each group of infected mice at 8 days postinfection is shown.
ICP22 is required for efficient establishment of latency.
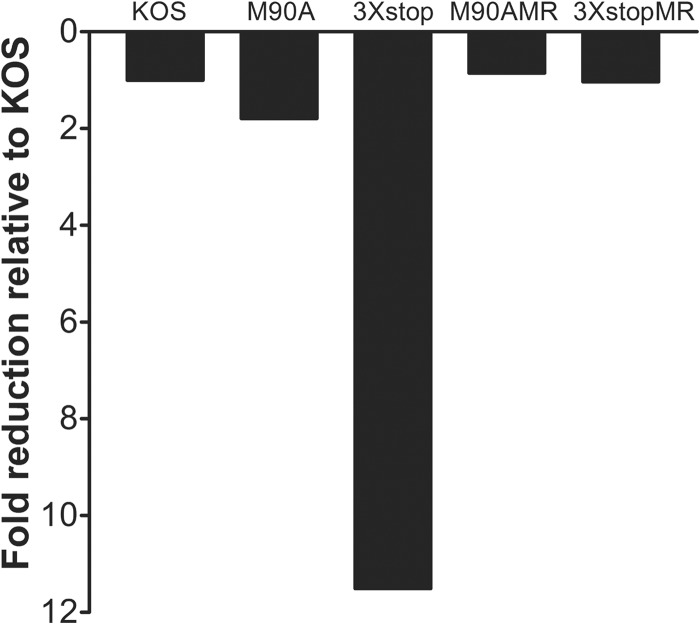
To examine the ability of our mutants to establish a latent infection, TG from infected mice were collected at 28 days postinfection, and viral DNA loads were determined using real-time PCR. As shown in Fig. 4, relative to KOS, the 3×stop mutant was significantly reduced for viral DNA levels by 11.5-fold (one way ANOVA, P < 0.05). The M90A mutant, on the other hand, was able to establish latency comparable to the levels of the KOS and MR viruses (Fig. 4). The fold reduction of the d22 mutant was comparable to that of the mock-infected mice (≥568-fold; one way ANOVA, P < 0.05) (H. H. Mostafa and D. J. Davido, unpublished data). The defect in establishing an efficient latent infection with both d22 and 3×stop is likely attributed to their impaired acute replication phenotypes in the eyes and TG, as has been observed with other HSV-1 mutants (33, 34).
Fig 4.
Establishment of latency of M90A and 3×stop mutants and MR viruses. CD-1 mice were infected with 2 × 105 PFU per eye. At 28 days postinfection, mice were sacrificed, TG were removed, and genomic DNA was isolated from samples. The amount of HSV-1 DNA present in each sample was quantified by real-time PCR (n = 10 to 12 TG per group). Results shown are the fold reductions compared to KOS levels. The fold reduction for d22 was comparable to that of the mock-infected TG (≥568-fold).
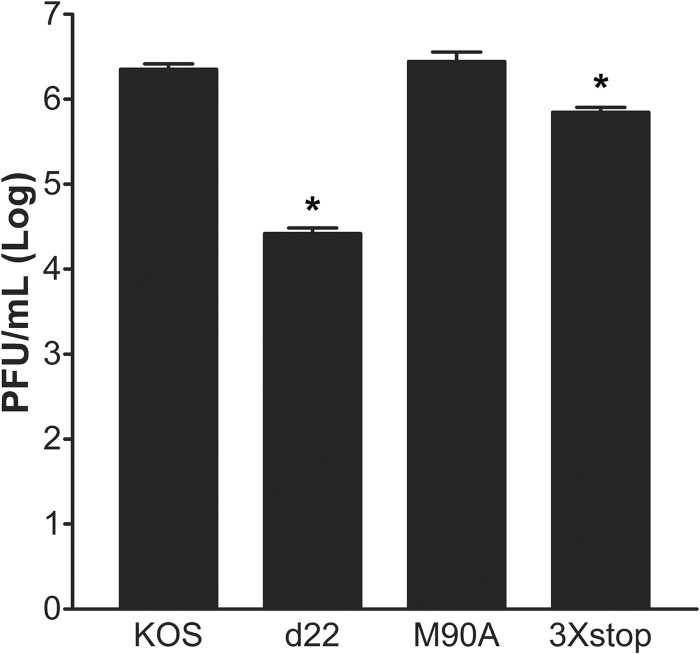
ICP22 or US1.5 is sufficient for HSV-1 replication in restrictive cells.
It has been previously established that ICP22 is required for efficient viral growth in certain cell lines, which have been termed restrictive cell lines (7). In order to test the roles of ICP22 and US1.5 in viral replication in restrictive cells, we performed a viral yield assay in HEL-299 cells (7). HEL-299 cells were infected at an MOI of 0.1 with KOS, d22, M90A, or the 3×stop mutant. At 24 h postinfection, cells and progeny virus were collected, and viral yields were determined with a standard plaque assay. As shown in Fig. 5, the M90A mutant replicated to levels that were comparable to the level of KOS; however, d22 replication was significantly reduced (83-fold; t test, P = 2.5 × 10−7). Replication of the 3×stop mutant was only modestly impaired relative to that of KOS (4.5-fold; t test, P = 0.003) (Fig. 5). Furthermore, the replication of the 3×stop mutant was significantly reduced relative to that of the d22 mutant (18.4-fold; t test, P = 5.5 × 10−8). Similar results were seen in mouse embryo fibroblasts (MEFs) of CD-1 mice (Mostafa and Davido, unpublished). These data indicate that ICP22 is sufficient to enhance viral growth in HEL-299 and MEF cells; however, the US1.5 protein can, to a great extent, complement the function of full-length ICP22.
Fig 5.
Growth of M90A and 3×stop mutant viruses on HEL-299 cells. HEL-299 cells were infected at an MOI of 0.1 with the indicated viruses. At 24 h postinfection, cells were harvested, and viral titers were determined by a standard plaque assay. *, P < 0.05 compared to KOS (Student's t test). Error bars represent the standard errors of the means.
Both ICP22 and US1.5 enhance the expression of late viral proteins in cell culture.
ICP22 has been demonstrated to enhance the expression of a subset of late viral proteins, including viral glycoprotein C (gC) and virion host shutoff (vhs) (6, 7, 9, 35). In order to test the individual roles of ICP22 and US1.5 in the expression of late viral proteins, we infected HEL-299 cells with KOS, d22, M90A, and 3×stop for 24 h and examined vhs and gC protein levels by Western blotting. Compared to levels in KOS-infected samples, the expression levels of gC and vhs were reduced in d22-infected cells but not in M90A- or 3×stop-infected cells (Fig. 6). Similar results were also observed at 12 h postinfection (Mostafa and Davido, unpublished). Our data show that both ICP22 and US1.5 are capable of stimulating late viral protein expression. Analogous results were observed in MEFs (CD-1 strain) and Vero cells (Mostafa and Davido, unpublished).
Fig 6.
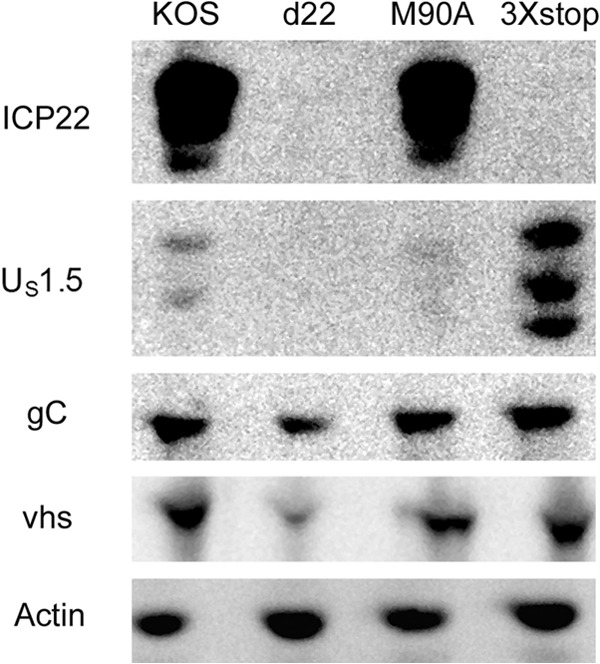
Examination of late viral gene expression for M90A and 3×stop mutant viruses. HEL-299 cells were infected at an MOI of 2 with the indicated viruses. At 24 h postinfection, cells were harvested, and protein levels from cell extracts were determined by Western blot analysis.
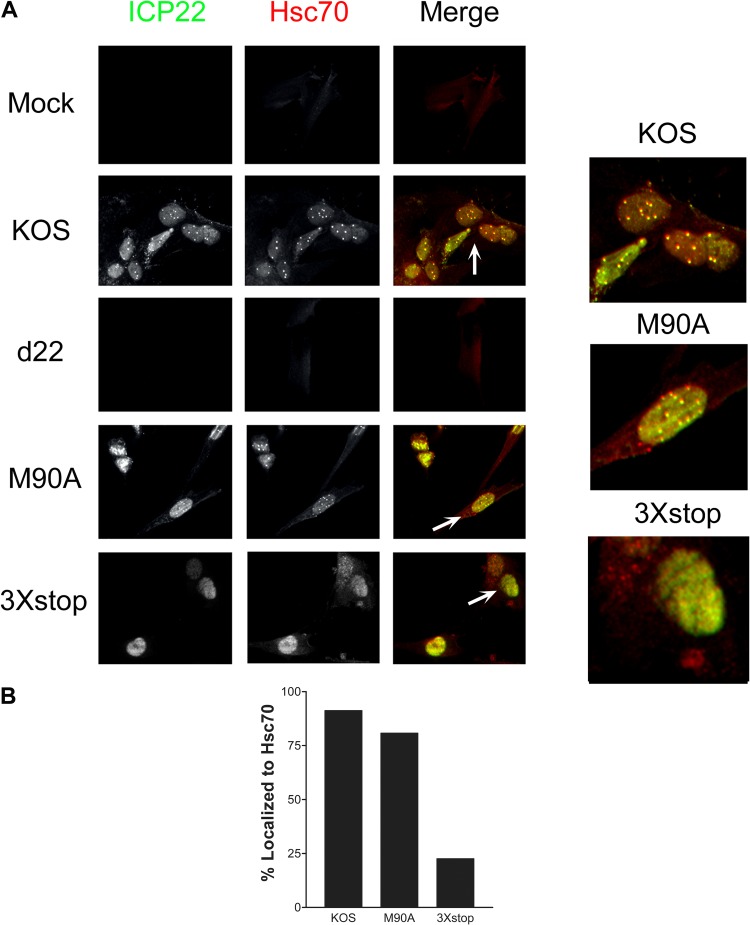
US1.5 does not induce VICE domains.
ICP22 has been shown to be required for the formation of VICE domains (13) during productive HSV-1 infection. To test the individual contributions of ICP22 and US1.5 in the formation of VICE domains, HEL-299 cells were mock infected or infected with KOS, d22, M90A, or 3×stop for 24 h, and the localization of ICP22 and the VICE domain marker, Hsc70, was examined by immunofluorescence. Our experiments showed that the M90A mutant was able to form VICE domains that appeared identical to those of KOS (Fig. 7A and B), in which ICP22 colocalized with Hsc70 in the nucleus. The 3×stop mutant, however, was unable to induce Hsc70 puncta (Fig. 7A and B). Although the US1.5 protein localized to the nucleus, it failed to form the speckled pattern of ICP22 staining observed with KOS- and M90A-infected nuclei (Fig. 7A). Notably, the staining pattern of Hsc70 in the presence of US1.5 localized to the nucleus, which was different from that of mock- and d22-infected cells (Fig. 7A). This indicates that although US1.5 fails to induce the formation of VICE domains, it is still capable of relocalizing Hsc70 to the nucleus. Similar results were observed in Vero cells (Mostafa and Davido, unpublished).
Fig 7.
ICP22 but not US1.5 induces VICE domain formation. (A) HEL-299 cells were mock infected or infected with the indicated viruses at an MOI of 0.1. At 24 h postinfection, cells were fixed and stained for ICP22 and Hsc70 and examined by fluorescence microscopy. (B) At least 100 cells that expressed ICP22 and/or US1.5 were examined for each virus, and the percentages of these cells that colocalized with Hsc70 are shown.
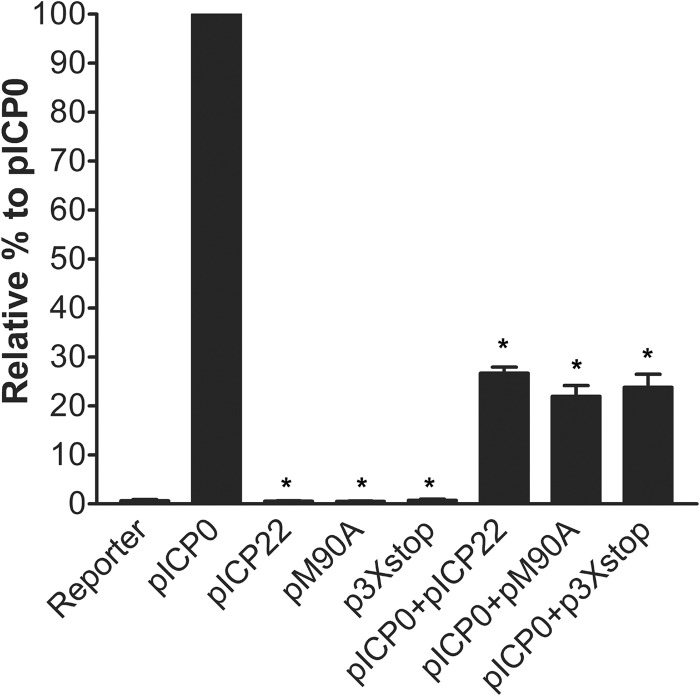
M90A and 3×stop inhibit ICP0-directed transactivation of an HSV promoter-luciferase reporter.
The IE protein ICP0 is a promiscuous transactivator of all classes of HSV-1 genes (36). One published report demonstrated that ICP22 represses ICP0-transactivated expression of reporter genes in transient transfections (37), which could be either directly through interfering with ICP0's transactivation activity or indirectly by reducing ICP0's expression (38, 39). To investigate if this inhibitory function is carried out by ICP22, US1.5, or both, we performed a luciferase assay to test the effect of both proteins on ICP0-mediated gene expression. ICP0 expression led to the maximum activity of the VP16 promoter and was given the 100% value (Fig. 8). Neither the pICP22 plasmid (which expresses both ICP22 and US1.5), pM90A, or p3×stop was able to activate VP16 promoter. Combining pICP22, pM90A, or p3×stop with pICP0 led to a significant inhibitory effect on ICP0's transactivation of the VP16 promoter (Mann Whitney U-test, P = 0.002). Our results demonstrate that both ICP22 and US1.5 have significant and comparable inhibitory effects on the ICP0-directed transactivation in reporter gene experiments.
Fig 8.
Inhibition of ICP0's transactivated gene expression by vectors that express either ICP22 or US1.5. Vero cells were transfected with an HSV-1 reporter plasmid (50 ng; pGL3-VP16) and a plasmid expressing wild-type ICP0 (pICP0), both ICP22 and US1.5 (pICP22), ICP22 alone (pM90A), or US1.5 alone (p3×stop) or a combination of pICP0 and pICP22, pM90A, or p3×stop for 48 h. Cell extracts were analyzed in luciferase assays to monitor ICP0's transactivating activity. *, P < 0.05 compared to pICP0 (Mann-Whitney U test). The error bars indicate the standard errors of the means.
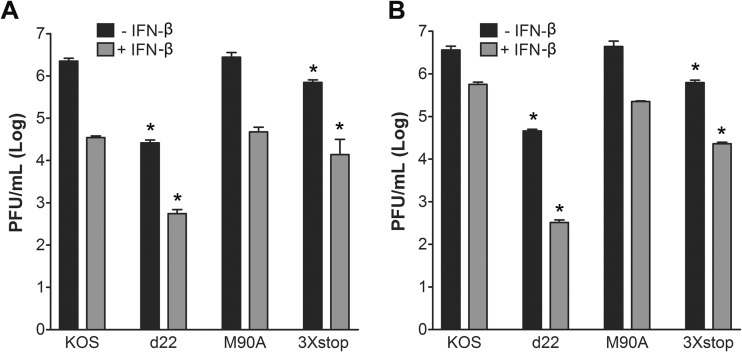
ICP22 but not US1.5 counteracts the effects of beta interferon (IFN-β) in restrictive cell lines.
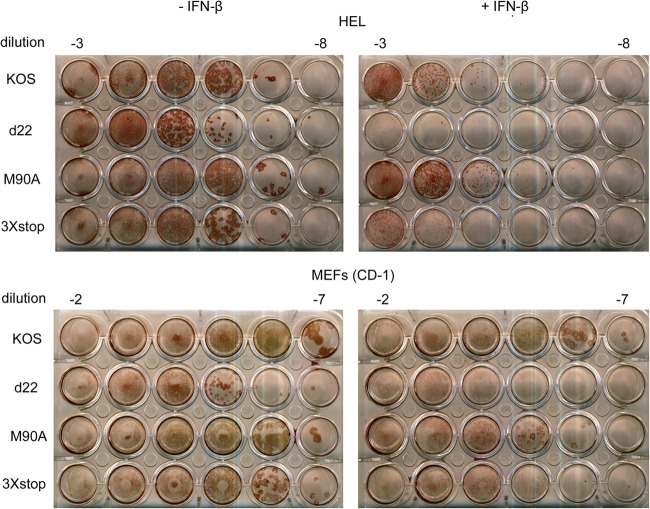
In order to understand the reduction in the ability of the 3×stop mutant to replicate acutely in vivo, we wanted to test if there is a relationship between ICP22 or US1.5 and the type I IFN response. An earlier publication examined the plaquing of an ICP22 null mutant in IFN-treated Vero cells, a cell line permissive for ICP22 mutant replication (5), and concluded that ICP22 is not required to counteract an established type I IFN response (40). Consequently, we decided to perform plaque reduction assays in a restrictive cell line, HEL-299 cells. Interestingly, we found a noticeable reduction in the number of plaques formed by d22 in the presence of IFN-β relative to KOS (Table 1). The overall plaque size for d22 was smaller than that of KOS. Furthermore, the reduction in plaque size for d22 was further enhanced by IFN-β compared to that for KOS (46-fold versus 10-fold reduction) (Fig. 9). For M90A, the reduction in the number of plaques in the presence of IFN-β was comparable to that for KOS (Table 1), and the plaque sizes with (9-fold reduction) and without IFN-β were similar for the two viruses. Interestingly, although the reduction in the number of plaques formed by 3×stop after the addition of IFN-β was comparable to that for KOS (Table 1), the plaques appeared smaller than those of KOS only after the addition of IFN-β (37-fold versus 10-fold reduction) (Fig. 9). In order to correlate this observation to the in vivo acute replication data, we decided to repeat the same experiment in CD-1 MEFs. Our results showed that the size of d22 and 3×stop plaques was markedly smaller than that of the wild-type plaques, a phenotype that was enhanced in IFN-β-treated cells (34-fold and 21-fold reductions, respectively, versus a 2-fold reduction with KOS) (Fig. 9). Consistent with these experiments, there was a reduction in the number of plaques for d22 and 3×stop that exceeded the decreases observed for KOS and M90A (Table 1). When we repeated the same experiments on MEFs of strain 129, we observed a further reduction in the number of plaques for d22 and 3×stop (Table 1). To determine if the plaque reduction phenotype correlated with reductions in viral replication, we performed yield assays in the absence or presence of IFN-β. As shown in Fig. 10A, reductions in viral replication after the addition of IFN-β were similar for KOS, d22, M90A, and 3×stop mutants (∼50-fold decrease) on HEL-299 cells, and similar results were noticed on 129 MEFs (Mostafa and Davido, unpublished). In CD-1 MEFs (Fig. 10B), IFN-β further impaired d22 replication (140-fold compared to 6-fold reduction of KOS), while IFN-β only slightly reduced the growth of M90A and 3×stop compared to that of KOS (19- and 27-fold reductions, respectively). These data indicate that ICP22 but not US1.5 is required for efficient plaquing in the restrictive cell lines, HEL-299 and MEFs, in the presence of a preestablished interferon response and that the degree of this requirement is cell type dependent.
Table 1.
Plaque reduction assays of KOS and ICP22 mutants in the absence and presence of IFN-β
| Virus | No. of plaques without IFN-β/no. of plaques with IFN-βin:a |
|||||
|---|---|---|---|---|---|---|
| HEL-299 cells |
CD-1 MEFs |
129 MEFs |
||||
| Expt 1 | Expt 2 | Expt 1 | Expt 2 | Expt 1 | Expt 2 | |
| KOS | 6.7 | 15 | 6.7 | 2 | 37.5 | 20 |
| d22 | 50 | 133 | 45.5 | 20 | 1,100 | 1,400 |
| M90A | 6.7 | 6.2 | 4 | 6.7 | 27 | 35 |
| 3×stop | 20 | 13 | 14 | 20 | 125 | 175 |
HEL-299 cells or MEF cells were infected with serially diluted viruses in the absence or presence of 1,000 U/ml of IFN-β. At 3 days postinfection, cells were fixed and immunostained for plaque formation. Plaques were counted for all samples, and the ratios of plaque numbers between untreated and treated plates were determined. The data shown are results from two independent experiments for each cell line.
Fig 9.
Plaque reduction assays. HEL-299 cells or CD-1 MEFs cells were infected with serial dilutions of the indicated viruses in the absence or the presence of 1,000 U/ml IFN-β. At 3 days postinfection, cells were fixed and immunostained for plaque formation.
Fig 10.
Growth of M90A and 3×stop with and without IFN-β. HEL-299 cells (A) or CD-1 MEFs (B) were infected at an MOI of 0.1 in the presence or in the absence of 1,000 U/ml IFN-β with the indicated viruses. At 24 h postinfection, cells were collected, and the viral titers were determined by a standard plaque assay. * P < 0.05, compared to KOS (Student's t test). Error bars represent the standard errors of the means.
DISCUSSION
The HSV-1 genome contains ∼90 genes, and, with some exceptions, each gene encodes a single protein. The US1 gene is one such exception as it codes for two proteins: ICP22 and its N-terminally truncated form, US1.5 (18). To date, reports that have examined the functions of ICP22 have largely not separated its biological activities from those of US1.5. The goal of this study was to characterize the individual contributions of ICP22 and US1.5 in the HSV-1 life cycle by generating mutant viruses (i.e., M90A and 3×stop) that can express only one of the two proteins. Our data show that the US1.5 protein is dispensable for acute replication in mice and for the induction of VICE domains. Also, our data indicate that either protein can functionally enhance the expression of late viral proteins in cell culture and inhibit ICP0-mediated gene expression in transient-transfection assays. Additionally, we identified a new role for ICP22 in enhancing viral plaquing in the presence of IFN-β, a function that was not fully compensated by US1.5 alone. Lastly, the M90A mutant behaved like KOS in our assays, indicating that the methionine-to-alanine amino acid substitution at residue 90 did not impact the functional activities of ICP22 tested in our study.
We observed that the expression of US1.5 alone (expressed from 3×stop) was marginally reduced for viral replication in restrictive HEL-299 and MEF cells; however, its acute replication in the eyes and TG of mice was remarkably reduced relative to KOS. The diminished replication of 3×stop in TG may be attributed to its reduced ocular replication (34), given that the acute TG titers of 3×stop at days 3 and 5 followed a similar trend for replication in the eyes at days 1 and 5 postinfection (Fig. 2A, C, and E). This impaired acute replication phenotype both in the eyes and TG likely explains 3×stop's defect in establishing an efficient latent infection as similar results have been observed with other mutants of HSV-1 (33, 34). This decrease in the establishment of latency by 3×stop would be expected to negatively impact reactivation. For this reason, we did not perform ex vivo reactivation studies with this mutant.
The deficiency of the 3×stop mutant in acute replication in vivo suggests that the US1.5 protein lacks a critical function of ICP22. What could this function be? A potential answer is that it is unable to induce VICE domains (Fig. 7A and B). Notably, our results provide a finer map of the region of ICP22 required for VICE domain formation, reducing it from the first 146 N-terminal residues as previously described (13) to the first 89 N-terminal amino acids. Because the functional significance of VICE domains in the HSV-1 life cycle is not yet fully understood, it is possible that the requirement of VICE domains in viral growth could be cell type dependent and specifically required for in vivo replication. Thus, this defect in the VICE domains observed with the 3×stop virus may contribute to its impaired acute replication in vivo. Additional studies will be needed to determine the role VICE domains play in HSV-1 replication and how ICP22 facilitates their formation. While VICE domain formation is affected in 3×stop-infected cells, US1.5 is expected to modify the C terminus of the large subunit of the host RNA polymerase II based on ICP22 mapping studies by Bastian and Rice as residues 147 to 420 of ICP22 are sufficient to modify RNA polymerase II (21). Although the mechanism by which ICP22 alters RNA polymerase II to enhance viral gene expression is unknown, the expected presence of this activity in US1.5 may explain the intermediate levels of acute ocular and neuronal replication observed with 3×stop in mice compared to levels with KOS and d22.
Although the 3×stop mutant's expression level of US1.5 protein might be lower than that of ICP22 expressed from either KOS or M90A (Fig. 1C and 6), the protein is still able to fully carry out some functions of ICP22; these functions include enhancing late viral protein expression, inhibiting ICP0 transactivated gene expression, and largely enabling efficient viral replication in cell culture. Additionally, we also tested if overexpression of US1.5 using the human cytomegalovirus (HCMV) promoter (19) might lead to VICE domain formation in transiently transfected HEL-299 cells. In contrast to results for ICP22, we did not detect VICE domains in these experiments (Mostafa and Davido, unpublished). Thus, while it may be argued that the expression levels of US1.5 dictate its ability to complement ICP22's functions, these experiments do not support this possibility.
Another plausible explanation for the impaired replication of the 3×stop mutant is related to the host's antiviral IFN response. Specifically, our results demonstrate that the 3×stop and d22 mutants are hypersensitive to IFN-β in cell culture. Thus, we have identified a new function of ICP22 in that it counteracts the effects of this type I IFN. Interestingly, this phenotype correlated very well with the impaired replication phenotype in the restrictive cell line HEL-299 and MEFs of the CD-1 mouse strain (Fig. 5 and 10) and in MEFs of strain 129 and the neuronal cell line SK-N-SH (Mostafa and Davido, unpublished). On the other hand, the hypersensitivity of both mutants to IFN-β was not detected in the permissive cell line, Vero cells (40; also Mostafa and Davido, unpublished), which are known to have a defect in IFN production (41). This could explain, in part, the replication phenotypes of 3×stop and d22 in mice. Because the decrease in the plaque sizes observed in plaque reduction assays (Fig. 9) was not accompanied by a similar reduction in viral growth (Fig. 10) after addition of IFN-β, there is the possibility that the observed phenotype is due to a defect in cell-to-cell spread rather than a defect in viral replication. Whether this function of ICP22 is mediated directly or indirectly through other viral factors will require more studies to determine. Related to this possibility, the reduction in the levels of the late viral protein vhs might partially contribute to this phenotype in the case of the d22 mutant. vhs is an endoribonuclease that degrades viral and cellular mRNA and is capable of interfering with an interferon-induced antiviral state (42). This mechanism is not applicable to the 3×stop phenotype, which is able to enhance wild-type levels of vhs protein (Fig. 6). Additionally, there could be a potential association between the formation of VICE domains and the interferon response based on published reports that have linked the cellular chaperone machinery to type I IFN (43–45).
It is notable that the acute replication of the ICP22 null mutant, d22, was greatly reduced in the eyes relative to the level of the 3×stop mutant both at day 1 and at day 3. Consequently, d22 was unable to acutely replicate in TG or establish a detectable latent infection in our experiments. The absence of both ICP22 and US1.5 in d22 significantly attenuated lytic and latent infections in mice, a result which mirrors the in vivo phenotypes of other ICP22 mutants (8, 20, 32). Many of these mutants do not express optimal levels of late viral gene products, including vhs, US11, and/or gC. As previously mentioned, it has been hypothesized that the expression of these late genes is dependent on VICE domain formation and/or the altered modification of the C terminus of the host RNA polymerase II (46). Ultimately, this defect in late viral gene expression likely reduces d22 growth in mice. Additionally, a reduction in gC levels could thwart the ability of HSV-1 to counteract complement-mediated neutralization (47–50), which might further contribute to the defective acute replication phenotype of d22 in vivo. Lastly, the composition of virion proteins in the ICP22 mutant 22/n199 is altered, and this mutant replicates poorly in mice (8). It was hypothesized that 22/n199 virions are less stable than wild-type virions and that this contributes to its in vivo phenotype. Consequently, it is plausible that the composition of d22 virions is similarly altered and that this negatively impacts its replication.
Our results did not define a unique function for US1.5 or a defect in HSV-1 replication in its absence. This raises the question of why HSV-1 has evolved to express the US1.5 protein. One possibility might be that the viral requirement for US1.5 is conditional and is enhanced only under certain circumstances during the HSV-1 life cycle. Moreover, while US1.5 is not required for HSV-1 replication in our assays, a role for US1.5 in the HSV-1 life cycle may be apparent in other model systems or in its natural host, humans. Finally, since we find that many, if not most, of the known functions of ICP22 can be carried out by US1.5, it may be that US1.5 can serve as a biological backup for ICP22 should ribosome readthrough be blocked at the translation initiation codon of ICP22.
ACKNOWLEDGMENTS
This work was supported by the University of Kansas.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the University of Kansas.
We thank members of the Davido lab and Stephen Rice for discussions related to this project and critical reading of the manuscript. We also thank Kristi Neufeld for cells and reagents.
Footnotes
Published ahead of print 2 October 2013
REFERENCES
- 1.Roizman R, Knipe DM, Whitley RJ. 2007. Herpes simplex viruses, p 2501–2601 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2.Knipe DM, Cliffe A. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6:211–221 [DOI] [PubMed] [Google Scholar]
- 3.Roizman B, Kozak M, Honess RW, Hayward G. 1975. Regulation of herpesvirus macromolecular synthesis: evidence for multilevel regulation of herpes simplex 1 RNA and protein synthesis. Cold Spring Harbor Symp. Quant. Biol. 39:687–701 [DOI] [PubMed] [Google Scholar]
- 4.Stevens JG. 1987. Defining herpes simplex genes involved in neurovirulence and neuroinvasiveness. Curr. Eye Res. 6:63–67 [DOI] [PubMed] [Google Scholar]
- 5.Post LE, Roizman B. 1981. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell 25:227–232 [DOI] [PubMed] [Google Scholar]
- 6.Poffenberger KL, Raichlen PE, Herman RC. 1993. In vitro characterization of a herpes simplex virus type 1 ICP22 deletion mutant. Virus Genes 7:171–186 [DOI] [PubMed] [Google Scholar]
- 7.Sears AE, Halliburton IW, Meignier B, Silver S, Roizman B. 1985. Herpes simplex virus 1 mutant deleted in the α22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J. Virol. 55:338–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orlando JS, Balliet JW, Kushnir AS, Astor TL, Kosz-Vnenchak M, Rice SA, Knipe DM, Schaffer PA. 2006. ICP22 is required for wild-type composition and infectivity of herpes simplex virus type 1 virions. J. Virol. 80:9381–9390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice SA, Long MC, Lam V, Schaffer PA, Spencer CA. 1995. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J. Virol. 69:5550–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durand LO, Advani SJ, Poon AP, Roizman B. 2005. The carboxyl-terminal domain of RNA polymerase II is phosphorylated by a complex containing cdk9 and infected-cell protein 22 of herpes simplex virus 1. J. Virol. 79:6757–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser KA, Rice SA. 2007. Herpes simplex virus immediate-early protein ICP22 triggers loss of serine 2-phosphorylated RNA polymerase II. J. Virol. 81:5091–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice SA. 2011. Multiple roles of immediate-early protein ICP22 in HSV-1 replication. In Weller SK. (ed), Alphaherpesviruses: molecular virology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 13.Bastian TW, Livingston CM, Weller SK, Rice SA. 2010. Herpes simplex virus type 1 immediate-early protein ICP22 is required for VICE domain formation during productive viral infection. J. Virol. 84:2384–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burch AD, Weller SK. 2004. Nuclear sequestration of cellular chaperone and proteasomal machinery during herpes simplex virus type 1 infection. J. Virol. 78:7175–7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burch AD, Weller SK. 2005. Herpes simplex virus type 1 DNA polymerase requires the mammalian chaperone hsp90 for proper localization to the nucleus. J. Virol. 79:10740–10749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livingston CM, Ifrim MF, Cowan AE, Weller SK. 2009. Virus-induced chaperone-enriched (VICE) domains function as nuclear protein quality control centers during HSV-1 infection. PLoS Pathog. 5:e1000619. 10.1371/journal.ppat.1000619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purves FC, Roizman B. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein α22. Proc. Natl. Acad. Sci. U. S. A. 89:7310–7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter KL, Roizman B. 1996. The promoter and transcriptional unit of a novel herpes simplex virus 1 α gene are contained in, and encode a protein in frame with, the open reading frame of the α22 gene. J. Virol. 70:172–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowman JJ, Schaffer PA. 2009. Origin of expression of the herpes simplex virus type 1 protein U(S)1.5. J. Virol. 83:9183–9194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogle WO, Roizman B. 1999. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J. Virol. 73:4305–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bastian TW, Rice SA. 2009. Identification of sequences in herpes simplex virus type 1 ICP22 that influence RNA polymerase II modification and viral late gene expression. J. Virol. 83:128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long MC, Leong V, Schaffer PA, Spencer CA, Rice SA. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 73:5593–5604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mostafa HH, Thompson TW, Kushnir AS, Haenchen SD, Bayless AM, Hilliard JG, Link MA, Pitcher LA, Loveday E, Schaffer PA, Davido DJ. 2011. Herpes simplex virus 1 ICP0 phosphorylation site mutants are attenuated for viral replication and impaired for explant-induced reactivation. J. Virol. 85:12631–12637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Research Council 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC [Google Scholar]
- 25.Halford WP, Schaffer PA. 2000. Optimized viral dose and transient immunosuppression enable herpes simplex virus ICP0-null mutants to establish wild-type levels of latency in vivo. J. Virol. 74:5957–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schrimpf JE, Tu EM, Wang H, Wong YM, Morrison LA. 2011. B7 costimulation molecules encoded by replication-defective, vhs-deficient HSV-1 improve vaccine-induced protection against corneal disease PLoS One 6:e22772. 10.1371/journal.pone.0022772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strand SS, Leib DA. 2004. Role of the VP16-binding domain of vhs in viral growth, host shutoff activity, and pathogenesis. J. Virol. 78:13562–13572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 29.Read GS, Karr BM, Knight K. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 67:7149–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kushnir AS, Davido DJ, Schaffer PA. 2010. Role of nuclear factor Y in stress-induced activation of the herpes simplex virus type 1 ICP0 promoter. J. Virol. 84:188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flory E, Weber CK, Chen P, Hoffmeyer A, Jassoy C, Rapp UR. 1998. Plasma membrane-targeted Raf kinase activates NF-κB and human immunodeficiency virus type 1 replication in T lymphocytes. J. Virol. 72:2788–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poffenberger KL, Idowu AD, Fraser-Smith EB, Raichlen PE, Herman RC. 1994. A herpes simplex virus type 1 ICP22 deletion mutant is altered for virulence and latency in vivo. Arch. Virol. 139:111–119 [DOI] [PubMed] [Google Scholar]
- 33.Katz JP, Bodin ET, Coen DM. 1990. Quantitative polymerase chain reaction analysis of herpes simplex virus DNA in ganglia of mice infected with replication-incompetent mutants. J. Virol. 64:4288–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leib DA, Coen DM, Bogard CL, Hicks KA, Yager DR, Knipe DM, Tyler KL, Schaffer PA. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63:759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purves FC, Ogle WO, Roizman B. 1993. Processing of the herpes simplex virus regulatory protein α22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. U. S. A. 90:6701–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Everett RD. 1984. Transactivation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 3:3135–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowman JJ, Orlando JS, Davido DJ, Kushnir AS, Schaffer PA. 2009. Transient expression of herpes simplex virus type 1 ICP22 represses viral promoter activity and complements the replication of an ICP22 null virus. J. Virol. 83:8733–8743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo L, Wu WJ, Liu LD, Wang LC, Zhang Y, Wu LQ, Guan Y, Li QH. 2012. Herpes simplex virus 1 ICP22 inhibits the transcription of viral gene promoters by binding to and blocking the recruitment of P-TEFb. PLoS One 7:e45749. 10.1371/journal.pone.0045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cun W, Guo L, Zhang Y, Liu L, Wang L, Li J, Dong C, Wang J, Li Q. 2009. Transcriptional regulation of the herpes simplex virus 1α-gene by the viral immediate-early protein ICP22 in association with VP16. Sci. China C Life Sci. 52:344–351 [DOI] [PubMed] [Google Scholar]
- 40.Mossman KL, Saffran HA, Smiley JR. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emeny JM, Morgan MJ. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43:247–252 [DOI] [PubMed] [Google Scholar]
- 42.Chee AV, Roizman B. 2004. Herpes simplex virus 1 gene products occlude the interferon signaling pathway at multiple sites. J. Virol. 78:4185–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang M, Wang C, Zhu X, Tang S, Shi L, Cao X, Chen T. 2011. E3 ubiquitin ligase CHIP facilitates Toll-like receptor signaling by recruiting and polyubiquitinating Src and atypical PKCζ. J. Exp. Med. 208:2099–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shang L, Tomasi TB. 2006. The heat shock protein 90-CDC37 chaperone complex is required for signaling by types I and II interferons. J. Biol. Chem. 281:1876–1884 [DOI] [PubMed] [Google Scholar]
- 45.Yang K, Shi H, Qi R, Sun S, Tang Y, Zhang B, Wang C. 2006. Hsp90 regulates activation of interferon regulatory factor 3 and TBK-1 stabilization in Sendai virus-infected cells. Mol. Biol. Cell 17:1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rice SA, Davido DJ. 2013. HSV-1 ICP22: hijacking host nuclear functions to enhance viral infection. Future Microbiol. 8:311–321 [DOI] [PubMed] [Google Scholar]
- 47.Hook LM, Lubinski JM, Jiang M, Pangburn MK, Friedman HM. 2006. Herpes simplex virus type 1 and 2 glycoprotein C prevents complement-mediated neutralization induced by natural immunoglobulin M antibody. J. Virol. 80:4038–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedman HM, Cohen GH, Eisenberg RJ, Seidel CA, Cines DB. 1984. Glycoprotein C of herpes simplex virus 1 acts as a receptor for the C3b complement component on infected cells. Nature 309:633–635 [DOI] [PubMed] [Google Scholar]
- 49.Hung SL, Peng C, Kostavasili I, Friedman HM, Lambris JD, Eisenberg RJ, Cohen GH. 1994. The interaction of glycoprotein C of herpes simplex virus types 1 and 2 with the alternative complement pathway. Virology 203:299–312 [DOI] [PubMed] [Google Scholar]
- 50.Kostavasili I, Sahu A, Friedman HM, Eisenberg RJ, Cohen GH, Lambris JD. 1997. Mechanism of complement inactivation by glycoprotein C of herpes simplex virus. J. Immunol. 158:1763–1771 [PubMed] [Google Scholar]