Abstract
Optogenetics has emerged as an alternative method for electrical control of the heart, where illumination is used to elicit a bioelectric response in tissue modified to express photosensitive proteins (opsins). This technology promises to enable evocation of spatiotemporally precise responses in targeted cells or tissues, thus creating new possibilities for safe and effective therapeutic approaches to ameliorate cardiac function. Here, we present a comprehensive framework for multi-scale modelling of cardiac optogenetics, allowing both mechanistic examination of optical control and exploration of potential therapeutic applications. The framework incorporates accurate representations of opsin channel kinetics and delivery modes, spatial distribution of photosensitive cells, and tissue illumination constraints, making possible the prediction of emergent behaviour resulting from interactions at sub-organ scales. We apply this framework to explore how optogenetic delivery characteristics determine energy requirements for optical stimulation and to identify cardiac structures that are potential pacemaking targets with low optical excitation threshold.
Electrical stimulation of the heart is a mainstay of modern cardiology, widely used for treating a range of heart rhythm disorders and for improving pump function. Direct current delivery from a lead in contact with the heart is used to evoke local excitation for pacemaking, cardiac resynchronization therapy, and anti-tachycardia pacing; delivery of large currents by imposing strong electric fields (shocks) across the heart is used to elicit a global electrical response that terminates lethal arrhythmias. Despite this widespread and successful usage, the electrical stimulation paradigm is associated with numerous drawbacks. Pacing devices have limited autonomic responsiveness,1 electrochemical reactions at the electrode-tissue interface limit stimulus amplitude and duration,2 and complications can arise during device implantation (infection)3 or extraction (lead fracture).4 Moreover, paced ventricular excitation often results in an asynchronous activation sequence,5 which is a poor substitute for normal sinus rhythm, leading to increased incidence of AF, heart failure, and death.6 High-voltage cardioversion and defibrillation shocks have significant adverse consequences, including cellular electroporation;7 disruption of normal cardiac electrical rhythm8 and mechanical function;9 psychological trauma,10 leading to discontinuation of treatment11 and increased procedure cost;12 and increased mortality.13,14
Optogenetics is an emerging field in which optical illumination is used to elicit a bioelectric response in excitable tissue that has been modified to express light-sensitive ion channel proteins called opsins.15 Photon absorption triggers conformational changes in these proteins, allowing a transient flow of transmembrane current; photoevoked current is potentiated by the energy content of incident light. Depending on which opsin is expressed, photoevoked current can be depolarising (inward) or hyperpolarising (outward). Optical stimulation is envisioned to result in a broad range of applications because optogenetic schemes can selectively deliver light-sensitivity to specific cell types using genetic promoters16 or to specific functional regions,17 as demonstrated in neuroscience. Moreover, since the energy transduction mechanism is fundamentally different from conventional electrical stimulation (transmembrane current as opposed to Faradaic charge transfer), the range of safe pulse widths and amplitudes is dramatically expanded for optical stimulation.18 Once delivery techniques have been adapted and perfected to work effectively in the heart, optogenetics is expected to enable the evocation of precise spatiotemporal patterns of transmembrane current in any genetically targetable subpopulation of cardiac cells for which direct illumination is physically possible. As such, it is clear that optogenetics has the potential to transform the field of cardiac electrophysiology, rewriting the rules that govern the design of safe, effective, and energy-efficient stimulation applications.
Initial experiments attempting light-based stimulation of cardiac tissue have yielded exciting results.19 Optical sensitivity has been inscribed in working heart cells in vitro using the photosensitive cation channel Chlamydomonas channelrhodopsin-2 (ChR2), either by direct gene delivery (GD) using viral vectors20 or by cell delivery (CD), where cardiomyocytes are co-cultured with (and couple to) ChR2-rich inexcitable cells.21 Experiments in transgenic mice have demonstrated the feasibility of optically pacing the mammalian heart in vitro and in vivo.22
Cardiac electrophysiology and electromechanics modelling is recognised as a vital tool for mechanistic enquiry, allowing researchers to test clinically-relevant hypotheses at various scales of structural hierarchy, from the molecular to that of the entire organ.23 Developing the ability to simulate cardiac optogenetics in a realistic manner within a biophysically-detailed whole-heart modelling infrastructure is an essential step in the quest to obtain novel solutions for optical pacing, defibrillation, and other types of electrophysiological control of cardiac behaviour. Tools for modelling cardiac optogenetics will accelerate the translation of this exciting technology to the clinic by providing an accurate and efficient platform for assessing the feasibility of potential optical control strategies.
The first objective of this study is to develop a comprehensive but flexible framework for simulating cardiac optogenetics. To achieve this goal, we identify key properties of light sensitivity, as well as its inscription in tissue and actuation by light, that need to be incorporated in the simulation framework to ensure its broad usefulness and the accuracy of its solutions. We then describe how each of these properties are integrated within a multiscale simulation hierarchy for modelling cardiac function. The second objective of this study is to demonstrate the utility of the framework in simulating a set of illustrative examples of optogenetic applications. We investigate optical stimulation efficiencies in models with different opsin delivery modes and spatial distributions of light-sensitive cells and we explore differential stimulation by cell-specific targeting. Finally, we showcase the modularity of the optogenetics simulation framework by demonstrating the ease with which it can be integrated with models of arbitrary complexity.
Results
Overview of key optogenetic framework components
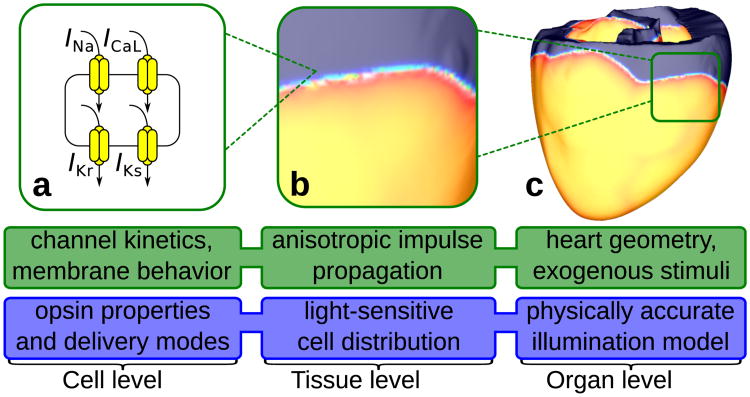
Fig. 1 presents a multiscale model hierarchy for cardiac electrophysiology simulations (description of modules' function is in green). This type of hierarchy has been described and successfully applied in mechanistic studies of cardiac rhythm dysfunction and therapy by our group24, 25 and others.26–28 In order to develop cardiac optogenetic simulation capabilities on the basis of such model hierarchy, we develop and introduce new model features at each spatial scale (cell, tissue, and organ levels, see blue boxes), as described in detail in the following sections. The framework we have developed is capable of representing opsin delivery using gene or cell therapy approaches and incorporates realistic representations of spatial distribution of light-sensitive cells and in vivo illumination constraints.
Figure 1. New features for simulating optogenetics.
Optogenetics simulation components (blue) are developed at each level of the existing multiscale cardiac model hierarchy (green). (a) Fundamental building blocks of our optogenetic modelling at the cell level include photoevoked current in opsins and opsin delivery modes. We represent opsin currents as complex photokinetic processes that depend nonlinearly on light, membrane voltage, and time.29 Opsins are either directly delivered to normal myocytes using viral vectors20 (GD mode) or expressed in inexcitable donor cells (CD mode) which can form gap junctions with host myocytes.21(b) At the tissue level, our optogenetic simulation framework incorporates heterogeneous spatial distribution of light-sensitive cells; this is important because diffuse, patchy patterns have been observed for both transgene30 and donor cell31 distribution in the heart. (c) At the organ level our framework accounts for practical limitations associated with illumination, namely the limited ability of light to penetrate tissue without significant attenuation due to energy absorption and photon scattering.32
Modelling ChR2 properties and delivery modes
Current flow in cardiac tissue is driven by active processes of ionic exchange across myocyte membranes. In a mathematical framework, these processes are represented by the ionic model of myocyte membrane behaviour, where current flow through ion channels, pumps and exchangers and current resulting from sub-cellular calcium cycling are governed by a set of ordinary differential and algebraic equations. The sum of all these currents is denoted by Iion; in our framework, cell-level optogenetic properties were represented by incorporating a light-sensitive component in Iion.
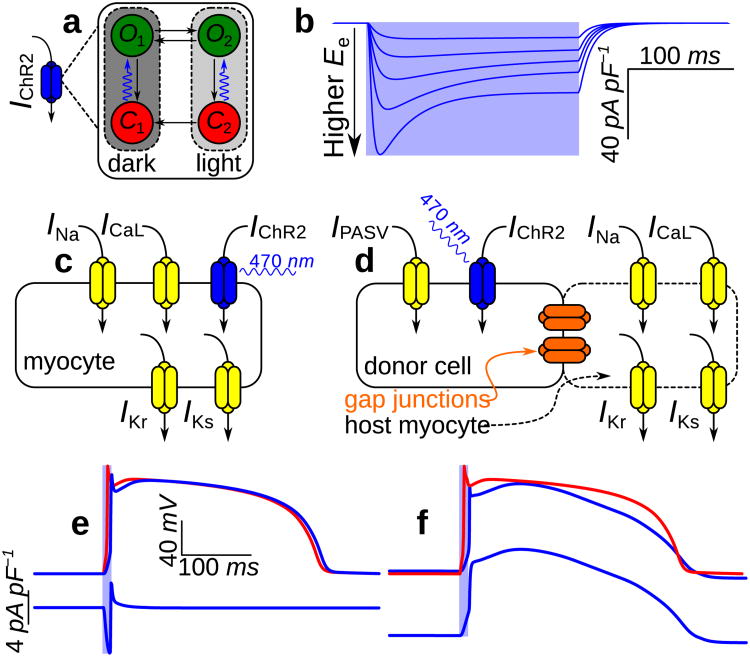
Our model of ChR2-mediated current (IChR2) is built upon a Markov model proposed by Nikolic et al.,29 derived from a channelrhodopsin-1 description by Hegemann et al.33 (Fig. 2a). We adjusted model parameters to better match experimental recordings of IChR2 collected from a stable ChR2-expressing human embryonic kidney (HEK) cell line;21 full details of our IChR2 model can be found in Methods. Fig. 2b shows the irradiance-potentiated response of our IChR2 model to illumination in a cell clamped to resting ventricular myocyte membrane voltage (Vm).
Figure 2. Modelling ChR2 properties and delivery modes at the cell level.
(a) Four-state Markov model for IChR2 with dark- and light-adapted photocycle branches associated with different peak conductances; each branch comprises closed- (red) and open-channel (green) states. Channel opening rates (blue arrows) are directly proportional to the irradiance (Ee) of light absorbed by ChR2 and open channels approach an Ee-dependent equilibrium between open states O1 and O2; all other transitions are purely time-dependent. (b) IChR2 in response to illumination (pale blue background; top to bottom: Ee = 0.4, 1, 2, 4, and 10mW mm−2) with Vm clamped to —85.6mV. Photoevoked ChR2 current has well-defined transient and steady-state phases resulting from the transition from full dark adaptation to an equilibrium between the two operating modes. The IChR2 model qualitatively reproduces experimental records from whole-cell patch-clamp recordings in a stable HEK-ChR2 cell line21, as shown in Supplementary Fig. S1. (c&d) Schematics for modelling gene delivery (GD) and cell delivery (CD) of ChR2; generic ionic currents are shown in myocytes. (e) Optically- (blue) and electrically-evoked (red) action potentials (APs) and underlying IChR2 in a myocyte with gene-delivered ChR2. Illumination at 2× threshold (Ee = 0.468mW mm−2 over 10ms) elicited depolarising IChR2 (bottom), which triggered an optically-evoked AP. (f) Response to illumination in an inexcitable ChR2-rich donor cell (CD; dashed blue) coupled to a normal myocyte by a 500 MΩ resistance (solid blue); a photoevoked AP in the latter cell is compared to an electrically-evoked AP (red). Light delivered to the donor cell was at 2× threshold for optically eliciting an AP in the myocyte (Ee = 0.652mW mm_2 over 10ms). During the AP plateau phase, myocyte Vm (≈ 2.77mV) was between the plateau Vm of the electrically-induced AP (≈ 15.3mV) and the donor cell resting level (−40mV).
When light-sensitivity is inscribed via GD, photoevoked current directly affects excitable cells.20,22 To represent this delivery mode in our framework, the IChR2 model was added to Iion for species- and cell type-specific membrane models of ion kinetics (Fig. 2c), allowing simulation of species- and cell type-specific photoevoked action potentials (APs). When light-sensitivity is inscribed via CD, IChR2 increases Vm in ChR2-rich donor cells, establishing a potential gradient with coupled host myocytes; this gives rise to junctional currents, which depolarise host cells and evoke APs.21 To represent membrane behaviour in light-sensitive donor cells Iion was replaced by IPASV, a model of electrically passive current with properties based on experimental measurements from HEK cells (used here as a model for donor cells; see Methods), combined with the IChR2 model. The resulting model of ChR2-expressing donor cell behaviour was either resistively coupled to a host myocyte model to simulate an optically-excitable tandem pair (Fig. 2d) or integrated in a tissue-scale model to represent a syncytium of donor and host cells, as described in the next section.
Fig. 2e shows a simulated optically-evoked AP (top) in a human ventricular myocyte model34 with ChR2 inscribed by GD; the AP in response to electrical stimulation in the same cell is superimposed for comparison. Differences between the optically and electrically evoked APs in the notch and plateau phases were due to non-instantaneous ChR2 channel closing kinetics. Fig. 2f compares the same electrically-induced AP to the response of a normal myocyte (top) resistively coupled (R = 500 MΩ) to an illuminated ChR2-rich donor cell (CD; bottom). Initial donor cell depolarisation elicited myocyte depolarisation via junctional current, leading to an AP. Following the myocyte upstroke, electrotonic coupling resulted in a plateau phase in both cells. The characteristics observed here qualitatively resembled the experimentally-observed response to optical stimulation in tandem preparations of ChR2-rich HEK cells and myocytes.21
Modelling spatial distribution of light-sensitive cells
At the tissue level, AP propagation due to current flow between cells coupled by gap junctions is modelled by solving one or more partial differential equations.35,36 Preferential conduction in the direction of myocardial fibres is incorporated via spatially-varying anisotropic conductivity tensors. In vivo ChR2 delivery in whole hearts has not yet been reported, but observations from injection of non-optogenetic viral vectors30 or donor cells31 to the heart suggest that ChR2 delivery sites will comprise interdigitated clusters of light-sensitive cells and normal myocytes. This type of distribution has also been observed in cardiac cell monolayers with cell-delivered ChR2.21 Our framework modelled this heterogeneous distribution of light-sensitive cells within target sites of ChR2 delivery.
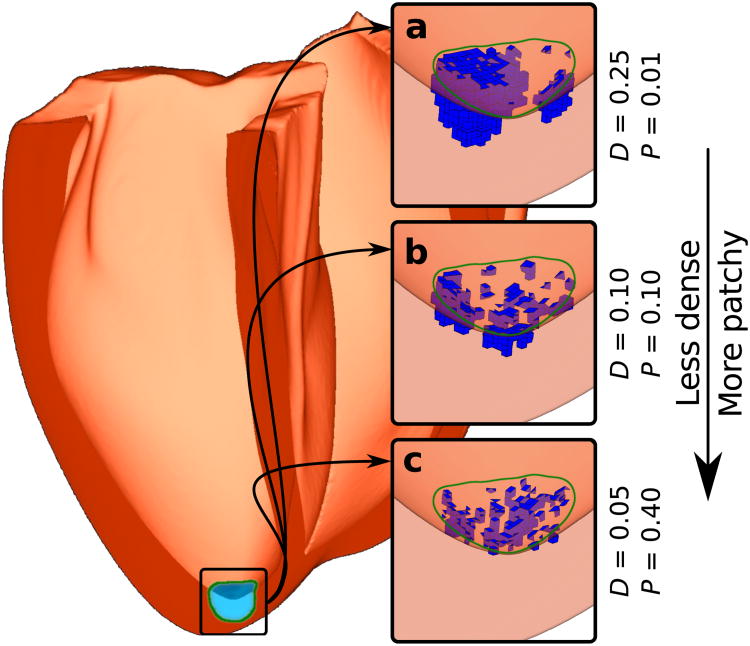
To generate realistic spatial distributions of ChR2-expressing cells, we adapted a stochastic algorithm originally used to model fibrosis patterns in cardiac tissue.37 For each ChR2 delivery target region, the parameters D and P controlled the density (by volume) and level of patchiness of light-sensitive cell clusters, respectively. Different parameter values resulted in a variety of stochastically-generated distributions (Fig. 3a-c) consistent with experimentally-observed patterns of transgene expression in mouse hearts30 and aggregation of injected donor cells in dog hearts.31 Full details of our implementation of this algorithm as well as a description of our approach to modelling electrical propagation in donor cell clusters can be found in Methods.
Figure 3. Modelling spatial distribution of light-sensitive cells at the tissue level.
Human ventricular model with a photosensitisation target (green boundary; hemispherical, 1 cm diameter) near the LV apex. (a-c) Results of applying the light-sensitive cell distribution algorithm to populate the target region with framework-generated ChR2-expressing clusters (blue) for three combinations of the parameters D (density) and P (patchiness).
Modelling light attenuation
State-of-the-art cardiac models typically incorporate 3D heart geometry and fibrous structure from various imaging modalities. For optogenetic applications, issues of light distribution and attenuation in the ventricular walls need to be considered at this scale.32 Photoevoked IChR2 has an insignificant depolarising effect unless the illumination source delivers blue light with a wavelength near 480 nm and with Ee ≥ 0.1 mW mm−2. 29 We modelled optical stimulation with direct endocardial surface illumination with blue light, which is currently the only viable option for depolarising photostimulation — efforts to red-shift excitatory opsins have had limited success17 and two-photon excitation of ChR2, while possible,38 is impractical outside laboratory settings.
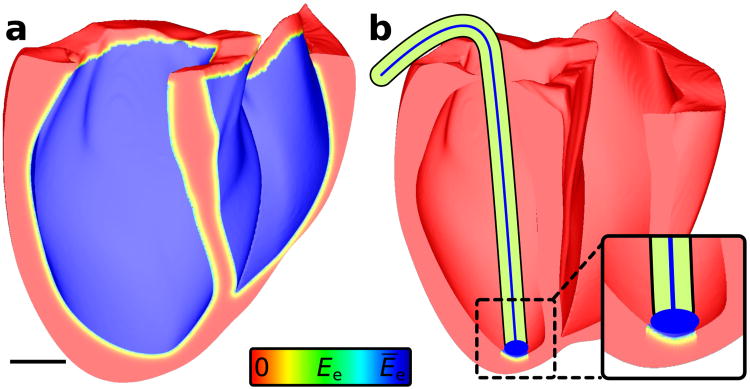
We assumed fixed irradiance values (Ēe) at endocardial surface points being illuminated and modelled attenuation in cells beneath the surface by Ee(r) = Ēee−r/δ, where r is the distance to the nearest site of direct illumination and δ is a wavelength-dependent decay coefficient. Previous simulations of cardiac optical mapping have proven that this formulation (with δ = 570 μm) is an excellent model of depth-dependent attenuation in heart tissue uniformly illuminated by blue light near the range for ChR2 stimulation (488 nm).39 As shown by the spatial distributions of Ee (Fig. 4) resulting from uniform (a) or focal (b) illumination of the endocardium, only a thin (≈ 1.5 mm) layer of cells near the surface received an appreciable proportion of the energy in applied light.
Figure 4. Modelling light attenuation at the organ level.
(a) Spatial profile of effective Ee (normalised to surface irradiance Ēe) in human ventricular model with uniform illumination of the entire endocardium; scale bar: 20 mm. (b) The limited extent of illuminated tissue is emphasised for a more realistic illumination configuration, in which the tip of an optrode — an ensheathed bundle of optical fibres — was pressed against the endocardium and delivered unattenuated light to tissue directly below.
Optogenetic pacing of the human heart
Since experimental studies in cardiac optogenetics are presently in a gestational stage, it is unknown what opsin delivery schemes and illumination strategies are technologically feasible; our newly-developed framework is a platform for simulating speculative scenarios that provide insight and guidance in future optogenetic developments. To demonstrate how insights can be gained by using our framework, we present illustrative examples of simulated cardiac optogenetics in two validated, biophysically-detailed ventricular models;24,25 comprehensive model descriptions are provided in Methods.
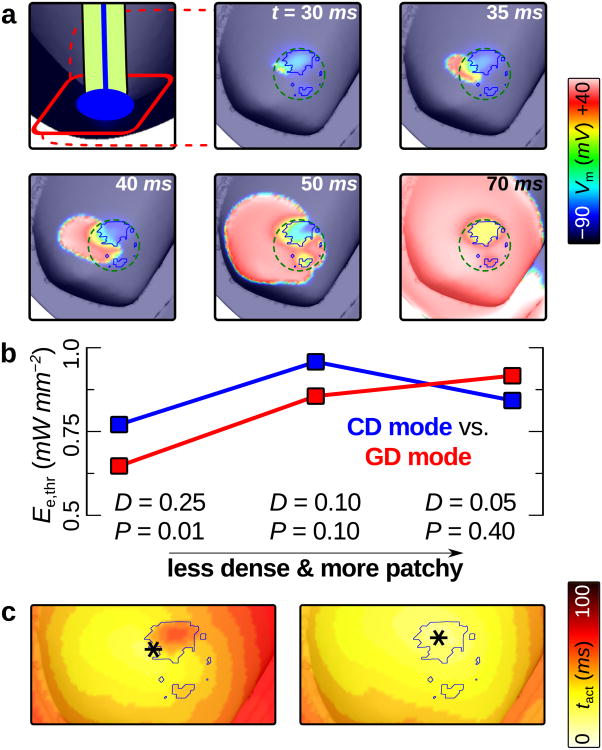
In the first illustrative example, we sought to examine optical stimulation efficiency for different optogenetic configurations (i.e., combinations of ChR2 delivery mode and light-sensitive cell spatial distribution) in an MRI-based model of the human ventricles;24 it is important to understand how these factors affect the energy required to stimulate optically because optogenetic techniques will need to be very efficient in order to be clinically viable and attractive as alternatives to conventional strategies. For the 3 light-sensitive cell spatial distributions shown in Fig. 3a-c for both CD and GD, optical stimulation was applied to the endocardial surface under the simulated optrode and efficiency was quantified by calculating the threshold irradiance required to elicit a propagating response (Ee,thr). These 3 spatial distributions were chosen to represent different stages in the evolution of a ChR2 delivery site over time as transfected or donor cells die off and migrate away from the injection site; the progression was from an initially dense, consolidated mass of light-sensitive cells (high D, low P) to a sparse, patchy distribution (low D, high P), as observed experimentally for GD30 and CD.31
Fig. 5a shows the ventricular response to optical stimulation in a configuration with large, consolidated donor cell clusters. AP propagation initiated on the left-hand side of the delivery site, in a region of extensive interdigitation between ChR2-rich donor cells and myocytes; subsequently, activation spread towards the base of the heart. As summarised in Fig. 5b, the spatial distribution of light-sensitive cells affected Ee,thr differently depending on ChR2 delivery mode. For GD of ChR2 in whole-heart models, as light-sensitive myocyte clusters became smaller and more sparsely distributed, optical stimulation efficiency decreased (Ee,thr rose monotonically); for CD mode, the least dense, patchiest ChR2-expressing donor cell patterns were associated with lower Ee,thr values compared to distributions with intermediate D and P values.
Figure 5. Determinants of optical stimulation efficiency in the human ventricles.
(a) Membrane voltage (Vm) response to 10 ms blue light pulse at t = 0. Cell-delivered ChR2 was distributed as in Fig. 3a (D = 0.25, P = 0.01); blue outlines in zoomed-in panels indicate boundaries of ChR2-rich donor cell clusters. Optical stimulation just above threshold (Ee,thr = 0.769 mW mm−2) was applied to the endocardial surface local to the ChR2 delivery site (dashed green line is contour of illuminated area). Supplementary movie 1 shows an animation of the full sequence. (b)Ee,thr values for all optogenetic delivery/distribution configurations. (c) Activation maps for CD mode (top) and GD mode (bottom) ChR2 in the same pattern as (a). *: sites of earliest activation.
Delivery mode-dependent differences in tissue-level excitation mechanisms are explored in Fig. 5c. Activation maps for the response to illumination given CD (left) and GD (right) of ChR2 in the same consolidated spatial pattern (blue outline) reveal different sites of earliest propagation. For CD, initial excitation was on the boundary of the donor cell cluster, in an area of extensive interdigitation between ChR2-rich inexcitable cells and myocytes; the site of peak donor cell density was associated with delayed activation. For GD, the same site was colocalised with the earliest activation. Similar differences between the responses to illumination for CD and GD of ChR2 were observed in configurations with different spatial distributions. This explained why Ee,thr was lower for CD compared to GD when D was low and P was high (Fig. 5b, right side) — compared to other parameter combinations, these spatial distributions had the lowest peak local density of ChR2-expressing cells and the highest peak local contact area between light-sensitive cell clusters and normal myocardium; in other words, these distributions maximised the amount of interface between electrical sources and sinks during illumination. Thus, when donor cell or viral vector delivery of ChR2 to the intact heart results in light-sensitive cells distributed in small, patchy clusters — as observed for non-optogenetic cardiac cell and gene therapy injections30, 31 — our model predicts that optical stimulation efficiency will be higher for CD compared to GD. This demonstrates how our framework will help guide the development of efficient ChR2 delivery strategies as experiments in cardiac optogenetics move from light-sensitive tissue culture and transgenic animal models to in vivo applications.
Targeted optogenetic stimulation of specialised tissue
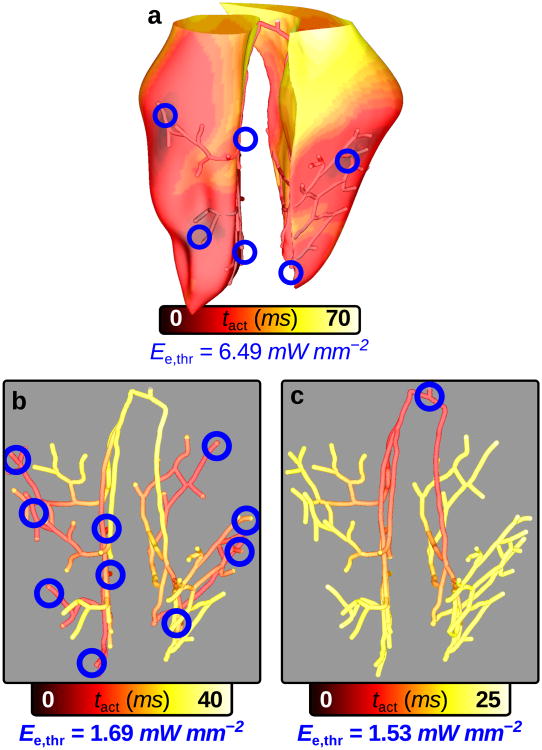
In the second illustrative example, we sought to compare optical stimulation efficiency in models with GD of ChR2 in either the Purkinje system (PS) or ventricular tissue; we used a geometrically-detailed rabbit ventricular model with a representation of the PS.25 Due to increased excitability in Purkinje fibers compared to ventricular cells40 and the cable-like geometric structure of the PS,41 we hypothesised that optical stimulation would elicit propagating responses at lower Ee,thr values in the PS than in the ventricles. Selective ChR2 expression in distinct cell types has already been demonstrated in neuroscience.16, 17 Our simulation results would thus provide insight into whether the pursuit of optogenetic targeting of ventricular or PS cells would be worthwhile, which is important because cell-specific viral targeting remains a challenging process.42
Ventricular models with GD of ChR2 in either the myocardium or PS were optically stimulated with illumination patterns designed to produce sinus-like activation sequences. In one model (Fig. 6a), patchy clusters of light-sensitive myocytes were located in 10 delivery sites corresponding to experimentally-observed early activation sites25 in regions of dense PS arborisation. Each site contained both ChR2-expressing myocytes and non-photosensitive PS cells, since early activations emanate from Purkinje-myocardial junctions; the latter detail was important because it allowed us to use the same 10-site illumination setup to optically stimulate in the second model, which had gene-delivered ChR2 throughout the PS (Fig. 6b). Direct His bundle pacing by optical stimulation from a single optrode was also simulated in this second model (Fig. 6c).
Figure 6. Cell-specific optogenetic targeting in rabbit ventricles with Purkinje system.
Each response shown here was elicited by applying 1.1×Ee,thr to the endocardial surface under each optrode for 2ms at t = 0. In multi-optrode models (A&B), all 10 delivery sites were illuminated simultaneously to achieve synchronous ventricular depolarisation; Ee,thr was the smallest value that initiated a propagating response when applied to each of the 10 sites independently. (a) Response to illumination of ChR2 delivery sites (blue) in regions of dense Purkinje system (PS) arborisation (see text). Sites were hemispherical (2 mm diameter) with patchy GD of ChR2 in ventricular cells only (D = 0.25, P = 0.1). (b) Response to the same illumination pattern as in (a) but with GD optogenetic targeting of the PS only; Ee,thr was 4.24× lower compared to the model in (a). (c) Response to His bundle illumination for the same model as in (b); Ee,thr was 3.84× lower compared to the model in (a) and optical stimulation was only applied by 1 optrode compared to 10.
As shown by activation maps for the ventricles and PS (Fig. 6a) or the PS only (Fig. 6b&c), the response to illumination was sinus-like in all cases; however, optical stimulation efficiency was dramatically higher for the model with GD of ChR2 to PS only, regardless of how it was illuminated. These differences could not be attributed to increased intrinsic PS excitability alone, since our simulations in single cells showed that Ee,thr was only 1.60× lower in individual Purkinje cells compared to ventricular myocytes (0.835mW mm−2 and Ee,thr = 1.34mW mm−2, respectively, for 2 ms of illumination). Even when all ventricular target sites had consolidated instead of patchy spatial distribution, the Ee,thr value (5.78 mW mm−2) was dramatically higher than for the photosensitive PS. Thus, in terms of stimulation efficiency, we conclude that the PS is a very attractive candidate for targeted, cell-specific optogenetic pacing.
Optogenetic Framework Generalisation
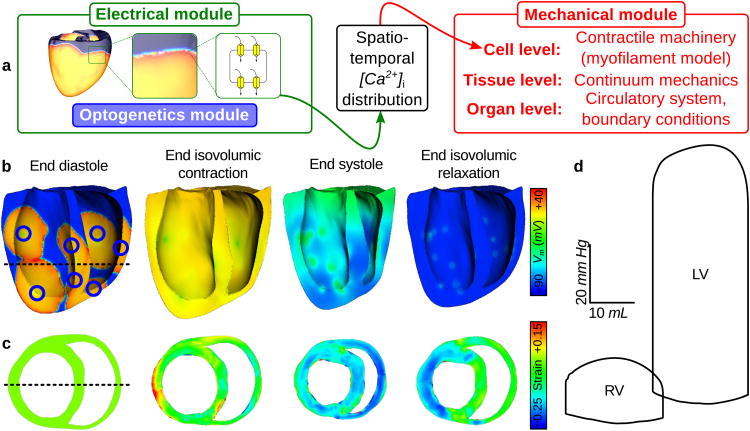
Our newly-developed framework is modular and can easily be combined with components that model other aspects of cardiac physiology. To illustrate this flexibility, we applied the framework to develop a light-sensitive electrophysiological model of the canine ventricles and PS, to which a cardiac mechanics component was added, as in previous studies.43 Integration of electrical, mechanical, and optogenetic modules is illustrated in Fig. 7a. Electromechanical simulation sinus rhythm and multi-site optical stimulation (as in Fig. 6a) was performed using tools developed previously.44
Figure 7. Electromechanical response to optical stimulation in canine ventricles.
(a) The electrical component (green), which encapsulates optogenetic framework model features (blue), is coupled with the mechanical component (red) by passing 3D intracellular [Ca2+] distribution from the ionic model to a cell-level myofilament model at each time step. (b&c) Long-axis membrane voltage (Vm) and short-axis strain profiles (unitless) during the cardiac cycle. Illumination delivered 12.8 mW mm−2 to 10 ventricular ChR2 delivery sites (blue circles) for 10 ms at t = 0. Delivery sites were hemispherical (3mm diameter) with consolidated CD expression. Dashed line in (b) shows position of slice in (c) and vice-versa. Strain was measured with respect to the end diastolic state. PS fibres were simulated but were not rendered graphically; regions of delayed repolarisation due to long intrinsic PS action potential duration are visible in (b). (d) LV and RV pressure-volume (PV) loops for the photoevoked response. These PV loops matched those for sinus rhythm very closely (cross correlation coefficient γ > 0.9). The 10-site illumination pattern shown here resulted in a more vigorous contraction compared to optical pacing from the endocardial apex only (7.34% increase in stroke volume), due to increased depolarisation synchrony (see Supplementary Fig. S2).
Figs. 7b and c show the spatiotemporal sequence of Vm propagation and strain development during a heartbeat elicited by sinus activation-like optical pacing. Initial excitations for optical stimulation differed from those during sinus rhythm because delivery sites were illuminated simultaneously, so the detailed spatiotemporal order of PS excitation was not replicated. The hemodynamic responses in the two models were near identical (see pressure-volume loops in Fig. 7d). These simulations demonstrate the utility of the combined optogenetic-electromechanics model, which could be used to design and optimise novel, light-based therapies for treating contractile dyssynchrony in the heart.
Discussion
In this study, we developed a comprehensive framework for incorporating optogenetic phenomena in detailed multiscale simulations of cardiac electrophysiology. Optogenetic capabilities were developed at each model scale: at the cell level, we modelled photokinetics of the light-sensitive ChR2 channel and represented ChR2 delivery either directly to myocytes (GD) or via donor cells (CD); at the tissue level, we modelled realistic heterogeneous spatial distribution of ChR2-expressing cells; and, at the organ level, we incorporated a physically accurate model for illuminating light-sensitive tissue. We then applied the newly-developed framework in two case studies that aimed to illustrate its capability: first, we showed that spatial distribution of light-sensitive cells affected optical stimulation efficiency differently depending on ChR2 delivery mode; second, we showed that optical stimulation was more efficient when cell-specific optogenetic targeting was used to express ChR2 in Purkinje cells compared to ventricular myocytes. Finally, we combined the optogenetic electrophysiological model with a cardiac mechanics component to demonstrate the modularity of the framework and highlight the ease of integrating optogenetics in multiscale, multi-physics heart models of arbitrary complexity. Here, we discuss results presented in our study and potential future applications of our framework; in the Supplementary discussion section, we elaborate upon the significance of multiscale optogenetic modelling capabilities and the versatility of the framework, we discuss the feasibility and energy efficiency of optical stimulation, and we propose strategies for validating the results of this study in vitro.
Our findings indicate that ChR2 delivery mode and light-sensitive cell spatial distribution play important roles in determining threshold irradiance for optical stimulation. In the context of simulated ChR2 delivery by injection in the intact heart, ChR2-rich donor cells (CD) with low density and high patchiness are associated with lower Ee,thr values than the same distributions of ChR2-expressing myocytes (GD), indicating higher efficiency. This is due to the fact that patchy distributions of cell-delivered ChR2 maximise the interface between of electrical sources (donor cells) and sinks (myocytes at rest) during the excitation phase.
This result is relevant to practical optogenetic therapy because ChR2-expressing cells can die off or migrate away from the original delivery site over time, leading to decreased density and increased patchiness; our model indicates that optical stimulation is more likely to remain viable in spite of such changes if ChR2 is delivered via donor cells (CD) as opposed to viral transfection (GD), which is a hypothesis that could be validated experimentally. Our framework provides an ideal platform for further analysis of this important problem because it accounts for all major factors that might affect optical stimulation efficiency.
We also show that if a PS-specific promoter can be developed to achieve targeted gene delivery with high expression density, optical stimulation can be used to elicit sinus-like paced activations at low irradiances. Purkinje fibres are more easily excited than well-coupled ventricular myocytes due to differences in membrane kinetics40 (cell-level) and dramatically reduced source-sink mismatch due to network geometry45 (tissue-level). Moreover, since PS fibres tend to run along or just beneath the endocardial surface, light attenuation does not have such a dramatic effect. Direct His bundle pacing by conventional electrical stimulation can overcome bundle branch block and restore synchrony in certain cardiac resynchronization therapy candidates but pure His capture is difficult to attain and energy requirements are very high;46 our findings suggest that both of these shortcomings — lack of selectivity and low energy efficiency — could be mitigated by applying optogenetics to facilitate direct optical His bundle pacing.
Selective inscription of light-sensitivity in specific cell types has already been demonstrated in neuroscience16 and should be possible in the PS. That said, the process of engineering a viral vector to accurately and efficiently target a particular cell type without toxic side effects is both costly and difficult.42 Our simulations of optogenetic targeting of Purkinje cells show how the framework can be leveraged to rapidly and accurately assess such high-risk, high-reward therapeutic strategies prior to the investment of considerable resources and effort in the wet lab.
Although our framework integrates all presently-available information regarding cardiac opto-genetics, there are still numerous unknown physiological factors. As such, we are cognisant of the fact that the above conclusions must be subjected to validation in vitro and we have indicated future directions accordingly (see: Supplementary discussion). This inherent uncertainty also reaffirms the importance of our framework's modularity — as new details become available, they can be integrated at the appropriate levels of model hierarchy and new, more accurate simulations can be run.
As the framework continues to be expanded, it can be used to explore optogenetics-based solutions to many open problems in cardiac electrophysiology. Early investigation of multi-pulse pacing for arrhythmia termination suggested that PS stimulation was an important determinant of therapy success47 but subsequent studies have not explored this connection in depth. Direct optical stimulation of the PS only (and not surrounding myocardium) could help elucidate this complex matter and defibrillation by illumination may prove to be a plausible option.
Sustained low-amplitude illumination of depolarising or hyperpolarising opsins18 could be used to dynamically modulate the firing rate of autorhythmic myocytes22 or to add pacemaker functionality to quiescent myocytes.21 Weak hyperpolarising optical stimuli could also be used to suppress ectopic activity near the endocardium, where the safety factor for propagation is ele-vated48 and attenuation does not limit the ability to optically modulate Vm. A similar approach using an excitatory opsin could be used to increase the heart's propensity for triggered activity, providing a valuable diagnostic tool for identifying sites with the highest likelihood of producing ectopic beats.
Another exciting possibility is the use of ChR2 and other opsins to shape AP repolarisation.17 Optogenetics could be used as a novel therapeutic approach to shorten or lengthen AP duration in a regionally heterogeneous manner. This could be especially useful in the treatment of long and short QT syndromes, heart failure, and abnormal ventricular wall motion, since changes in transmural dispersion of repolarisation are thought to catalyse arrhythmogenesis in all of these disorders.49–52
Our computational framework for simulating cardiac optogenetics is a predictive tool that will drive hypothesis development and provide a platform for detailed mechanistic enquiry. The modelling methodologies specified here can be implemented in any computational cardiology software and used to accurately simulate opsin delivery and optical stimulation in cardiac models of arbitrary complexity and for a variety of species. As the field of cardiac optogenetics matures, in silico tools will provide insight and guidance that could help anticipate pitfalls, minimise expensive and time-consuming wet lab experiments, and fully characterise the underlying mechanisms of therapeutic techniques.
Methods
Light-sensitive current model
For this study, we developed a model of ChR2 current (IChR2) based on the 4-state photocycle.29 This model incorporated closed and open states for light- and dark-adapted operating modes. A ChR2 parameter set was generated from previously published values to produce currents qualitatively consistent with those obtained from whole-cell patch-clamp recordings. Some modifications to the original four-state model29 were included; e.g., the ChR2 channel activation rate was captured using a time- and light-dependent state variable p as in Talathi et al.,53 and some parameters were adopted from Grossman et al.54 (see Supplementary Table S1). In addition to light sensitivity, our model incorporated a simplified linear current-voltage relationship and was scaled via the maximum conductance (gChR2 = 2mS cm−2) so that peak current qualitatively matched experimental data from whole-cell patch clamp experiments in a stable HEK cell line expressing ChR2-H134R (a ChR2 mutant with enhanced current), which has been described previously.21
The governing IChR2 equation was:
| (1) |
where γ = 0.1 was the conductance ratio between open states (O2:O1), gChR2 = 2mS cm−2 was the maximal channel conductance, and Echr2 = 0mV was the reversal potential. While open, ChR2 channels approached an Ee-dependent equilibrium between dark- and light-adapted modes; closed channels recovered very slowly from light adaptation (τ = 250 ms) compared to other transition kinetics. A complete listing of IchR2 model equations and parameters is provided in Supplementary Table S1.
Donor cell model for CD mode
Instead of explicitly representing HEK-ChR2 cells, we modelled light-sensitive donor cells using an electrically passive membrane model with IchR2. HEK-ChR2 cells do have other low-conductance channels, but these have not been characterised in detail and contribute negligibly to optical excitability;21 thus, our model includes the key characteristics of donor cells (compared to normal myocytes) — light-sensitivity and the lack of an active response.
The electrically passive membrane model used for donor cells in CD mode was given by:
| (2) |
where gPASV = 14μS cm−2 was the specific membrane conductivity and Vrest = −40 mV was the resting potential. These properties were based on measurements from HEK cells.21,55 Like all excitable cells modelled, specific membrane capacitance was assumed to be 1μF cm−2. In donor cell-myocyte tandem pairs, the two membrane models were coupled by a 500 MΩ resistance. In organ-scale simulations, donor cell clusters (CD) were modelled with reduced, isotropic conductivity: σdonor = 0.2 × σiL, where σiL is the intracellular conductivity along the myocyte axis. We made this choice based on reported properties of HEK cell clusters, which lack the fibre-like arrangement of heart tissue and have diminished cell-cell coupling.21
Light-sensitive cell distribution algorithm
Each simulated ChR2 delivery site was described by a sphere of radius (r) surrounding a node (n) on the endocardial surface of the finite element mesh. We began by building a list of N mesh elements completely contained within the distance r from node n. Then, elements were tagged as light-sensitive one by one until the number of tagged elements was ≥ D × N, where D is the density parameter. The process of choosing which elements to tag was governed by a stochastic distribution algorithm developed for use in 2D cardiac cell monolayers.37 The process of extending this algorithm into 3D was straightforward: rather than examining shared edges to determine the neighbourhood around each cluster, we identified shared faces and applied the distribution process described in the original paper. Briefly, for each new light-sensitive element, the probability of starting a new cluster instead of adding to an existing one was 1 — P, where P is the patchiness parameter. One important constraint was necessitated by the 2D to 3D extension – seed elements for new clusters were always chosen from the subset of elements touching the endocardial boundary; we chose this constraint because transfected and/or donor cells tend to congregate near the injection site.30,31 Supplementary Fig. S3 shows an example of simulated ChR2 delivery in a representative 2D schematic and summarises how node- and element-level optogenetic properties are assigned for GD and CD of ChR2.
Simulation details
We ran monodomain simulations of electrical activity using the CARP software package;36,56 simulation results were visualised using Dr. Edward Vigmond's meshalyzer tool. Ventricular cell membrane kinetics in the human model were simulated according to the formulation developed by ten Tusscher and Panfilov34 as modified by Moreno et al.24 In the rabbit model, we used the Mahajan-Shiferaw AP model in the ventricles57 and the Aslanidi-Sleiman AP model in Purkinje fibres;58 PS-myocardial junction dynamics were simulated using a previously-validated model.59 Other relevant model parameters for the human and rabbit electrophysiological models are provided in Supplementary Tables S2 and S3, respectively.
Supplementary Material
Acknowledgments
Dr. Boyle is supported by a fellowship from the Natural Sciences and Engineering Research Council of Canada. This research was also supported by the U.S. National Institutes of Health, grants R01 HL111649 and R01 HL103428, and by the U.S. National Science Foundation, grants NSF-CDI-1124804 and NSF-OCI-108849.
Footnotes
Author contributions: P.M.B., E.E., and N.A.T. conceived the idea. P.M.B. developed the framework, performed the simulations, interpreted the data, and wrote the manuscript. C.M.A., E.E. and N.A.T. assisted with data interpretation and manuscript preparation. J.C.W. derived IChR2 model parameters from experimental observations of ChR2.
Additional information: Supplementary information accompanies this paper at http://www.nature.com/ncomms
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Binggeli C, et al. Autonomic nervous system-controlled cardiac pacing: a comparison between intracardiac impedance signal and muscle sympathetic nerve activity. Pacing Clin Electrophysiol. 2000;23:1632–1637. doi: 10.1046/j.1460-9592.2000.01632.x. [DOI] [PubMed] [Google Scholar]
- 2.Merrill DR, Bikson M, Jefferys JGR. Electrical stimulation of excitable tissue: design of efficacious and safe protocols. J Neurosci Methods. 2005;141:171–198. doi: 10.1016/j.jneumeth.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 3.Raad D, et al. Implantable electrophysiologic cardiac device infections: a risk factor analysis. Eur J Clin Microbiol Infect Dis. 2012;31:3015–3021. doi: 10.1007/s10096-012-1655-1. [DOI] [PubMed] [Google Scholar]
- 4.Kutarski A, Pietura R, Czajkowski M. Difficult dual stage transcutaneous lead extraction complicated by fracture of both leads. Cardiol J. 2012;19:412–417. doi: 10.5603/cj.2012.0074. [DOI] [PubMed] [Google Scholar]
- 5.Brenyo A, Goldenberg I, Barsheshet A. The downside of right ventricular apical pacing. Indian Pacing Electrophysiol J. 2012;12:102–113. doi: 10.1016/s0972-6292(16)30502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen HR, et al. Long-term follow-up of patients from a randomised trial of atrial versus ventricular pacing for sick-sinus syndrome. Lancet. 1997;350:1210–1216. doi: 10.1016/S0140-6736(97)03425-9. [DOI] [PubMed] [Google Scholar]
- 7.Al-Khadra A, Nikolski V, Efimov IR. The role of electroporation in defibrillation. Circ Res. 2000;87:797–804. doi: 10.1161/01.res.87.9.797. [DOI] [PubMed] [Google Scholar]
- 8.Eysmann SB, Marchlinski FE, Buxton AE, Josephson ME. Electrocardiographic changes after cardioversion of ventricular arrhythmias. Circulation. 1986;73:73–81. doi: 10.1161/01.cir.73.1.73. [DOI] [PubMed] [Google Scholar]
- 9.Mollerus M, Naslund L. Myocardial stunning following defibrillation threshold testing. J Interv Card Electrophysiol. 2007;19:213–216. doi: 10.1007/s10840-007-9150-9. [DOI] [PubMed] [Google Scholar]
- 10.Maisel WH. Pacemaker and icd generator reliability: meta-analysis of device registries. JAMA. 2006;295:1929–1934. doi: 10.1001/jama.295.16.1929. [DOI] [PubMed] [Google Scholar]
- 11.Marcus GM, Chan DW, Redberg RF. Recollection of pain due to inappropriate versus appropriate implantable cardioverter-defibrillator shocks. Pacing Clin Electrophysiol. 2011;34:348–353. doi: 10.1111/j.1540-8159.2010.02971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boodhoo L, et al. The safety and effectiveness of a nurse led cardioversion service under sedation. Heart. 2004;90:1443–1446. doi: 10.1136/hrt.2004.034900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moss AJ, et al. Prophylactic implantation of a defibrillator in patients with myocardial infarction and reduced ejection fraction. N Engl J Med. 2002;346:877–883. doi: 10.1056/NEJMoa013474. [DOI] [PubMed] [Google Scholar]
- 14.Larsen GK, Evans J, Lambert WE, Chen Y, Raitt MH. Shocks burden and increased mortality in implantable cardioverter-defibrillator patients. Heart Rhythm. 2011;8:1881–1886. doi: 10.1016/j.hrthm.2011.07.036. [DOI] [PubMed] [Google Scholar]
- 15.Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsubota T, Ohashi Y, Tamura K, Sato A, Miyashita Y. Optogenetic manipulation of cerebellar Purkinje cell activity in vivo. PLoS One. 2011;6:e22400. doi: 10.1371/journal.pone.0022400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arrenberg AB, Stainier DYR, Baier H, Huisken J. Optogenetic control of cardiac function. Science. 2010;330:971–974. doi: 10.1126/science.1195929. [DOI] [PubMed] [Google Scholar]
- 19.Entcheva E. Cardiac optogenetics. Am J Physiol Heart Circ Physiol. 2013 doi: 10.1152/ajpheart.00432.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abilez OJ, et al. Multiscale computational models for optogenetic control of cardiac function. Biophys J. 2011;101:1326–1334. doi: 10.1016/j.bpj.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia Z, et al. Stimulating cardiac muscle by light: cardiac optogenetics by cell delivery. Circ Arrhythm Electrophysiol. 2011;4:753–760. doi: 10.1161/CIRCEP.111.964247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruegmann T, et al. Optogenetic control of heart muscle in vitro and in vivo. Nat Methods. 2010;7:897–900. doi: 10.1038/nmeth.1512. [DOI] [PubMed] [Google Scholar]
- 23.Trayanova NA. Whole-heart modeling: applications to cardiac electrophysiology and electromechanics. Circ Res. 2011;108:113–128. doi: 10.1161/CIRCRESAHA.110.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno JD, et al. A computational model to predict the effects of class I anti-arrhythmic drugs on ventricular rhythms. Sci Transl Med. 2011;3:98ra83. doi: 10.1126/scitranslmed.3002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle PM, Deo M, Plank G, Vigmond EJ. Purkinje-mediated effects in the response of quiescent ventricles to defibrillation shocks. Ann Biomed Eng. 2010;38:456–468. doi: 10.1007/s10439-009-9829-4. [DOI] [PubMed] [Google Scholar]
- 26.Niederer SA, et al. Biophysical modeling to simulate the response to multisite left ventricular stimulation using a quadripolar pacing lead. Pacing Clin Electrophysiol. 2012;35:204–214. doi: 10.1111/j.1540-8159.2011.03243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop MJ, et al. Development of an anatomically detailed MRI-derived rabbit ventricular model and assessment of its impact on simulations of electrophysiological function. Am J Physiol Heart Circ Physiol. 2010;298:H699–H718. doi: 10.1152/ajpheart.00606.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ten Tusscher KHWJ, Hren R, Panfilov AV. Organization of ventricular fibrillation in the human heart. Circ Res. 2007;100:e87–101. doi: 10.1161/CIRCRESAHA.107.150730. [DOI] [PubMed] [Google Scholar]
- 29.Nikolic K, et al. Photocycles of channelrhodopsin-2. Photochem Photobiol. 2009;85:400–411. doi: 10.1111/j.1751-1097.2008.00460.x. [DOI] [PubMed] [Google Scholar]
- 30.Prasad KMR, Smith RS, Xu Y, French BA. A single direct injection into the left ventricular wall of an adeno-associated virus 9 (AAV9) vector expressing extracellular superoxide dismutase from the cardiac troponin-t promoter protects mice against myocardial infarction. J Gene Med. 2011;13:333–341. doi: 10.1002/jgm.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosen AB, et al. Finding fluorescent needles in the cardiac haystack: tracking human mesenchymal stem cells labeled with quantum dots for quantitative in vivo three-dimensional fluorescence analysis. Stem Cells. 2007;25:2128–2138. doi: 10.1634/stemcells.2006-0722. [DOI] [PubMed] [Google Scholar]
- 32.Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nat Med. 2003;9:123–128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 33.Hegemann P, Ehlenbeck S, Gradmann D. Multiple photocycles of channelrhodopsin. Biophys J. 2005;89:3911–3918. doi: 10.1529/biophysj.105.069716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ten Tusscher KHWJ, Panfilov AV. Alternans and spiral breakup in a human ventricular tissue model. Am J Physiol Heart Circ Physiol. 2006;291:H1088–H1100. doi: 10.1152/ajpheart.00109.2006. [DOI] [PubMed] [Google Scholar]
- 35.Vigmond EJ, Aguel F, Trayanova NA. Computational techniques for solving the bidomain equations in three dimensions. IEEE Trans Biomed Eng. 2002;49:1260–1269. doi: 10.1109/TBME.2002.804597. [DOI] [PubMed] [Google Scholar]
- 36.Vigmond EJ, dos Santos RW, Prassl AJ, Deo M, Plank G. Solvers for the cardiac bidomain equations. Prog Biophys Mol Biol. 2008;96:3–18. doi: 10.1016/j.pbiomolbio.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comtois P, Nattel S. Interactions between cardiac fibrosis spatial pattern and ionic remodeling on electrical wave propagation. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:4669–4672. doi: 10.1109/IEMBS.2011.6091156. [DOI] [PubMed] [Google Scholar]
- 38.Rickgauer JP, Tank DW. Two-photon excitation of channelrhodopsin-2 at saturation. Proc Natl Acad Sci U S A. 2009;106:15025–15030. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bishop MJ, et al. Synthesis of voltage-sensitive optical signals: application to panoramic optical mapping. Biophys J. 2006;90:2938–2945. doi: 10.1529/biophysj.105.076505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart P, et al. Mathematical models of the electrical action potential of Purkinje fibre cells. Philos Transact A Math Phys Eng Sci. 2009;367:2225–2255. doi: 10.1098/rsta.2008.0283. [DOI] [PubMed] [Google Scholar]
- 41.Vigmond EJ, Clements C. Construction of a computer model to investigate sawtooth effects in the Purkinje system. IEEE Trans Biomed Eng. 2007;54:389–399. doi: 10.1109/TBME.2006.888817. [DOI] [PubMed] [Google Scholar]
- 42.Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat Rev Genet. 2007;8:573–587. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Provost J, Gurev V, Trayanova N, Konofagou EE. Mapping of cardiac electrical activation with electromechanical wave imaging: an in silico-in vivo reciprocity study. Heart Rhythm. 2011;8:752–759. doi: 10.1016/j.hrthm.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gurev V, Lee T, Constantino J, Arevalo H, Trayanova NA. Models of cardiac electromechanics based on individual hearts imaging data: image-based electromechanical models of the heart. Biomech Model Mechanobiol. 2011;10:295–306. doi: 10.1007/s10237-010-0235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie Y, Sato D, Garfinkel A, Qu Z, Weiss JN. So little source, so much sink: requirements for afterdepolarizations to propagate in tissue. Biophys J. 2010;99:1408–1415. doi: 10.1016/j.bpj.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barba-Pichardo R, et al. Ventricular resynchronization therapy by direct His-bundle pacing using an internal cardioverter defibrillator. Europace. 2013;15:83–88. doi: 10.1093/europace/eus228. [DOI] [PubMed] [Google Scholar]
- 47.Salama G, Kanai A, Efimov IR. Subthreshold stimulation of Purkinje fibers interrupts ventricular tachycardia in intact hearts. Experimental study with voltage-sensitive dyes and imaging techniques. Circ Res. 1994;74:604–619. doi: 10.1161/01.res.74.4.604. [DOI] [PubMed] [Google Scholar]
- 48.Boyle PM, Vigmond EJ. An intuitive safety factor for cardiac propagation. Biophys J. 2010;98:L57–L59. doi: 10.1016/j.bpj.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu W, Antzelevitch C. Sodium channel block with mexiletine is effective in reducing dispersion of repolarization and preventing torsade des pointes in LQT2 and LQT3 models of the long-QT syndrome. Circulation. 1997;96:2038–2047. doi: 10.1161/01.cir.96.6.2038. [DOI] [PubMed] [Google Scholar]
- 50.Extramiana F, Antzelevitch C. Amplified transmural dispersion of repolarization as the basis for arrhythmogenesis in a canine ventricular-wedge model of short-QT syndrome. Circulation. 2004;110:3661–3666. doi: 10.1161/01.CIR.0000143078.48699.0C. [DOI] [PubMed] [Google Scholar]
- 51.Glukhov AV, et al. Transmural dispersion of repolarization in failing and nonfailing human ventricle. Circ Res. 2010;106:981–991. doi: 10.1161/CIRCRESAHA.109.204891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Opthof T, et al. The association of abnormal ventricular wall motion and increased dispersion of repolarization in humans is independent of the presence of myocardial infarction. Front Physiol. 2012;3:235. doi: 10.3389/fphys.2012.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Talathi SS, Carney PR, Khargonekar PP. Control of neural synchrony using channelrhodopsin-2: a computational study. J Comput Neurosci. 2011;31:87–103. doi: 10.1007/s10827-010-0296-6. [DOI] [PubMed] [Google Scholar]
- 54.Grossman N, Nikolic K, Toumazou C, Degenaar P. Modeling study of the light stimulation of a neuron cell with channelrhodopsin-2 mutants. IEEE Trans Biomed Eng. 2011;58:1742–1751. doi: 10.1109/TBME.2011.2114883. [DOI] [PubMed] [Google Scholar]
- 55.Zimmermann D, et al. Biophysical characterisation of electrofused giant HEK293-cells as a novel electrophysiological expression system. Biochem Biophys Res Commun. 2006;348:673–681. doi: 10.1016/j.bbrc.2006.07.112. [DOI] [PubMed] [Google Scholar]
- 56.Vigmond EJ, Hughes M, Plank G, Leon LJ. Computational tools for modeling electrical activity in cardiac tissue. J Electrocardiol. 2003;36(1):69–74. doi: 10.1016/j.jelectrocard.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 57.Mahajan A, et al. A rabbit ventricular action potential model replicating cardiac dynamics at rapid heart rates. Biophys J. 2008;94:392–410. doi: 10.1529/biophysj.106.98160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aslanidi OV, Sleiman RN, Boyett MR, Hancox JC, Zhang H. Ionic mechanisms for electrical heterogeneity between rabbit Purkinje fiber and ventricular cells. Biophys J. 2010;98:2420–2431. doi: 10.1016/j.bpj.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyle PM, Veenhuyzen GD, Vigmond EJ. Fusion during entrainment of orthodromic reciprocating tachycardia is enhanced for basal pacing sites but diminished when pacing near Purkinje system end points. Heart Rhythm. 2013;10:444–451. doi: 10.1016/j.hrthm.2012.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.