Abstract
Background
There has been little investigation of gene-by-environment interactions related to sedentary behavior, a risk factor for obesity defined as leisure screen time (ST; i.e., TV, video, and computer games).
Objective
To test the hypothesis that limiting ST use attenuates the genetic predisposition to increased BMI, independent of physical activity.
Design
Using 7,642 wave II participants of the National Longitudinal Study of Adolescent Health, (Add Health; mean=16.4 years, 52.6% female), we assessed the interaction of ST (h/wk) and 41 established obesity SNPs with age- and sex-specific BMI Z scores in 4,788 European-(EA),1,612 African- (AA), and 1,242 Hispanic-American(HA)adolescents.
Results
Nominally significant SNP*ST interaction were found for FLJ35779 in EA, GNPDA2 in AA, and none in HA (EA: beta[SE]=0.016[0.007]), AA: beta[SE]=0.016[0.011]) per 7 h/wk ST and 1 risk allele in relation to BMI Z-score.
Conclusions
While for two established BMI loci, we find evidence that high levels of ST exacerbate the influence of obesity susceptibility variants on body mass, overall we do not find strong evidence for interactions between the majority of established obesity loci. However, future studies with larger sample sizes, or that may build on our current study and the growing published literature, are clearly warranted.
Keywords: Gene-environment interactions, adolescence, obesity, BMI, screen time, African-American, Hispanic-American
Introduction
Adolescence is a risky period for weight gain that can be exacerbated by sedentary behavior, including leisure screen time (ST: TV, videos, and computer games[1, 2]), which are related to obesity independent of physical activity [3, 4]. Individuals at greatest genetic risk for obesity appear to benefit most from physical activity[5–7]. We have recently reported a mitigated effect of obesity variants on BMI in those with increased physical activity[8], yet limited research has investigated whether sedentary behaviors modify a genetic susceptibility to increased BMI. A recent study showed that television viewing in adult women may exacerbate a genetic predisposition to increased BMI[9]. However, whether sedentary behaviors beyond television viewing may also influence a genetic risk for obesity, especially among adolescent populations, is largely unknown. To address this, we examined interactions between ST and established obesity SNPs with age- and sex-specific BMI Z-scores using data from the National Longitudinal Study of Adolescent Health (Add Health).
METHODS
Add Health
Participants
Add Health is a nationally-representative cohort of adolescents (1994–95, n=20,745, mean age=15.7 years; SD=1.8) drawn from a probability sample of 132 US middle and high schools[10]. The cohort was followed in 1996 (Wave II, n = 14,738, mean age=16.2 years; SD=1.6) when respondents were still of middle and high school age. Survey procedures have been described elsewhere [11–13] and were approved by the Institutional Review Board, UNC Chapel Hill.
Outcome Measure: Body Mass Index (BMI)
BMI (kg/m2) was calculated from Wave II measured height and weight. Self-reported height and weight (r=0.95/0.94 with measured weight/height[14])were substituted for those refusing measurement and/or weighing more than scale capacity (n=119). Given the age of our sample, we used age- and sex-specific BMI Z-scores relative to the CDC/NCHS 2000 reference curves [15].
Race/Ethnicity
Race/ethnicity was constructed using ancestral background and family relationship status (i.e., country of origin, ancestry, and adoption): European American (EA), African American (AA), and Hispanic American (HA), with indicators for subpopulation (e.g., Mexican, Cuban) and immigrant status (e.g., US and non-US born).
Screen time
Wave II screen time (ST) was ascertained using a previously validated standard activity recall [16]. The ST items elicited information on hours of TV, video, and computer game use, which were summed into a continuous measure of ST hours/week.
Genetic data
At Wave IV (2008–2009), 59% (n=12,234) of Wave I participants provided saliva samples. DNA was extracted and genotyped from those providing consent (n=12,066). Of the 41 SNPs interrogated, 37 were selected based on confirmed GWA(p < 5×10−8) with BMI [17–19], and 4 with obesity [20] in EA adults. Procedures for genotyping (call rate 95%, discordance 0.3%) have been detailed elsewhere [21]. We excluded 15/41 SNPs (indicated with † in Supplementary Tables) that did not generalize in AA GWAS [22]-[23]. Given the lack of large GWAS in Hispanics, all 41 SNPs were considered. SNPs with genotype counts<10 were also excluded, leaving 41 SNPs in EA, 25 in AA, and 38 in HA.
Analytic sample
We included individuals with phenotype data that had at least 80% of their SNPs genotyped (n=8,646). We excluded: the monozygotic twin with fewer genotyped loci within each pair (n=133), Asian or other race/ethnicity (n=575), pregnant (n=110), disabled (n=44), and individuals missing covariate data (n=150). The final analytic sample included 7,642 individuals measured at Wave II with DNA data (Figure S1).
Statistical analysis
Race/ethnicity stratified SNP-by-ST analyses were conducted using Stata, version 12.1 (Stata Corp, College Station, TX). In additive genetic multivariable models with BMI Z-score as the outcome, we included a SNP-by-ST interaction term and controlled for age, sex, current smoking (≥1 cigarette every day for 30 days), physical activity (moderate to vigorous bouts/week) [16], geographic region, and an indicator for self-reported height and weight (n=55), with within-strata controls: oversampling of highly educated AAs (n=281), Hispanic subpopulation ancestry: Cuban (n=190), Puerto Rican (n=215), Central/South American (n=115), Mexican (n=626), other Hispanic (n=95), and foreign born status (n=264). Sample design effects and relatedness were accounted for using random effects for school and family. Models run excluding participants with self-reported heights and weights found no difference in effect estimates (Tables S5a, S5b, S5c). We performed a race/ethnicity pooled meta-analysis of beta estimates using the inverse standard-error weighted approach in METAL [24]. For nominally significant (p<0.05) SNP-by-ST interactions, we predicted BMI Z-score per risk allele and 7 h/wk of ST. While we examined all nominally significant findings (p<0.05), we corrected for multiple testing: α = 0.05/number of SNPs tested (p≤0.0012 in EA, p≤0.0013 in HA, and p≤0.0019 in AA). In addition, we estimated the main effect for each SNP on BMI Z-score (Table S4).
RESULTS
Sample descriptive statistics are shown in Table 1. ST was positively associated with BMI Z-score in EA and all adolescents combined (p<0.05)but was not significant in HA and AA (Table S1).
Table 1.
Study population characteristics, National Longitudinal Study of Adolescence Wave II respondents with DNA
| European American | African American | Hispanic American | Total | |
|---|---|---|---|---|
| N | 4,788 | 1,612 | 1,242 | 7,642 |
| Age in y, mean (SD) | 16.3 (1.8) | 16.1 (1.6) | 16.4 (1.6) | 16.2 (1.6) |
| Male sex, N (%) | 2,275 (47.5) | 731 (45.4) | 614 (49.3) | 3,620 (47.4) |
| Age- and sex-specific BMI Z-score1, mean (SD) | 0.29 (1.10) | 0.51 (1.08) | 0.46 (1.11) | 0.37 (1.10) |
| Screen time hr/wk2, mean (SD) | 19.1 (16.9) | 27.8 (22.3) | 21.1 (17.6) | 21.2 (18.6) |
| 1st quartile, hr/wk | 0–7 | 0–10 | 0–8 | 0–8 |
| 2nd quartile, hr/wk | 8–14 | 11–22 | 9–15 | 9–15 |
| 3rd quartile, hr/wk | 15–25 | 23–40 | 16–29 | 16–29 |
| 4th quartile, hr/wk | 26+ | 41+ | 30+ | 30+ |
| Bouts3 of physical activity/wk, mean (SD) | 6.3 (3.9) | 5.8 (3.5) | 5.8 (3.8) | 6.1 (3.8) |
| Current smoking, N (%) | 1,938(40.5) | 288 (17.8) | 347 (27.9) | 2,573 (33.7) |
| Region, N (%) | ||||
| West | 714 (14.9) | 238 (14.8) | 497 (40.0) | 1,449 (19.0) |
| Midwest | 1,776 (37.1) | 317 (19.7) | 92 (7.4) | 2,185 (28.6) |
| South | 1,572 (32.8) | 966 (59.9) | 477 (38.4) | 3,015 (39.5) |
| Northeast | 726 (15.2) | 91 (5.7) | 176 (14.2) | 993 (13.0) |
BMI: Body mass index, calculated as weight in kilograms divided by height in meters squared and scaled for sex and age [Ogden, C.L., et al., Centers for Disease Control and Prevention 2000 growth charts for the United States: improvements to the 1977 National Center for Health Statistics version. Pediatrics, 2002. 109(1): p. 45–60.]
Screen time is measured as the total sum of hours of screen time from television, video, and computer games per week.
Bouts of physical activity include sessions of exercise or sport where intensity was enough to sweat.
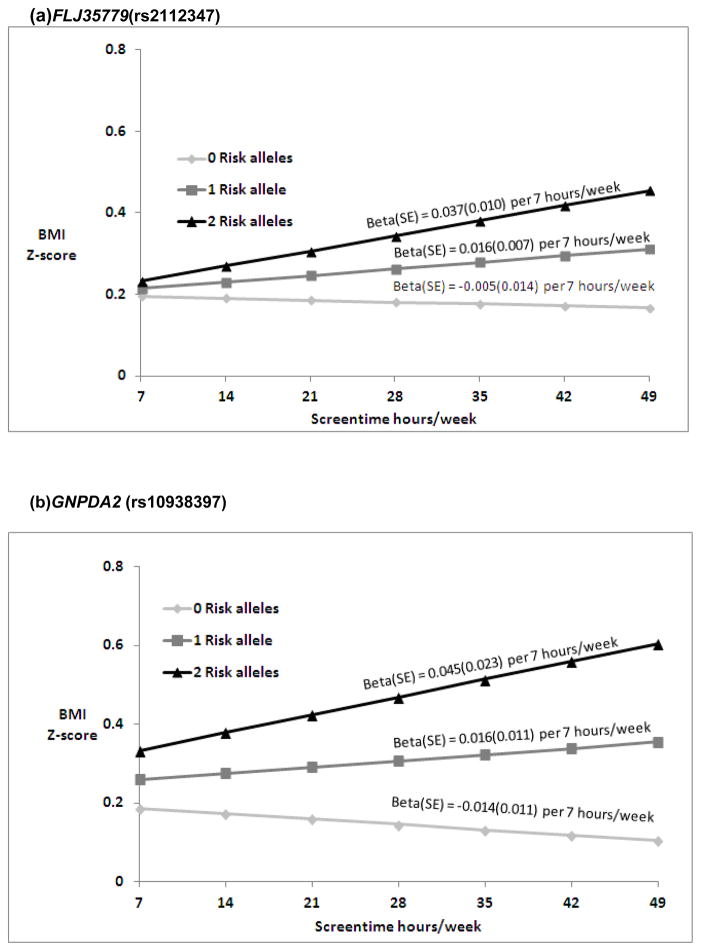
Nominally significant SNP-by-ST interactions were found in EA for FLJ35779 (rs2112347)and in AA for GNPDA2 (rs10938357)(Table S2a and S2b). In EA, predictions for BMI Z-score (Figure 1a) suggest a change of −0.005, 0.016 and 0.037 in BMI Z-score per 7 hours/week of ST with 0, 1 and 2 risk (T) alleles(FLJ35779). For AA, the ST-by-GNPDA2 interaction suggests a change of −0.014, 0.016, and 0.045 in BMI Z-score per 7 hours/week of ST with 0, 1 and 2 risk (A) alleles (Figure 1b). None of the interactions tested in HA (Table S2c) or the combined pooled meta-analyses (Table S3) were significant after multiple testing correction.
Figure 1.
Predicated BMI Z-score from model based coefficients1 per 7 hours/week of ST in the presence of 0, 1 and 2 risk (T) FLJ35779 (rs2112347) alleles, respectively (p for interaction = 0.02) in EA (a), and 0, 1 and 2 risk (A) GNPDA2 (rs10938397) alleles, respectively (p for interaction = 0.03) in AA (b).
Abbreviations: BMI (body mass index), ST (hours per week of screen time), EA (European American), AA (African American)
1Beta estimates are presented for the interaction model: Multivariable linear models of adolescent BMI Z scores regressed on SNP and ST (hr/wk), with SNP by ST interaction term, controlling for age, sex, current smoking (at least one cigarette every day for 30 days), geographic region, and self-reported heights and weights (n=39 EA, n= 12 AA), oversampling of highly educated African Americans (n=281; AA stratum only). Random intercepts allowed for individual, family and school with no sample weighting.
2Ogden, C.L., et al., Centers for Disease Control and Prevention 2000 growth charts for the United States: improvements to the 1977 National Center for Health Statistics version. Pediatrics, 2002. 109(1): p. 45–60.
DISCUSSION
Our findings provide tentative support for interaction between established obesity SNPs and ST with BMI Z-score in a nationally-representative adolescent sample. We found nominally significant SNP-by-ST interactions in EA (rs2112347, near FLJ35779) and AA (rs10938357 near GNPDA2).
Our analysis of SNP-by-ST interaction in HA was limited by a lack of established SNPs in this diverse population and small sample size. We focused on SNPs with well-established main effects in EA populations and generalization in AA populations, a strategy which could have introduced some bias. However, our focus on middle and high school ages reduces heterogeneity due to lifecycle changes. The majority of our sample was post-pubertal, so weight changes likely reflect changes in fat mass. We did not adjust for sexual maturation, since several loci associated with menarche and BMI have pleiotropic effects [25], thus adjusting for sexual development might diminish true BMI associations. Finally, ST exposure can also subject individuals to content that might influence lifestyle behaviors that are also associated with obesity, such as consuming advertised foods or smoking as a result of exposure to smoking messages.
Conclusions
Although we find suggestive evidence that ST may exacerbate the influence of FLJ35779 in EA and GNPDA2 in AA on BMI Z-score, strong evidence for interaction between the majority of established obesity loci and ST on BMI is lacking. However, our study was powered to detect fairly large interactions, which we did not observe. These estimates are an important resource that should pooled with other samples for meta-analyses. Indeed, our findings are relevant to the larger discussion of obesity susceptibility and modifiable behaviors influencing BMI.
Supplementary Material
Acknowledgments
We thank Amy Perou of the BioSpecimen Processing facility and Jason Luo of the Mammalian Genotyping Core at University of North Carolina at Chapel Hill. This work was funded by National Institutes of Health grant R01HD057194. P.G.L., K.E.N., E.M.L., L.A.L., A.S.R., K.M.Y., and M.G. contributed to study design, M.G. to data analysis, and M.G., K.E.N., and P.G.L. contributed to writing of the manuscript, all other authors provided critical evaluation of the manuscript. M.G., K.E.N., and P.G.L. had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. This research uses data from Add Health, a program project directed by Kathleen Mullan Harris and designed by J. Richard Udry, Peter S. Bearman, and Kathleen Mullan Harris at the University of North Carolina at Chapel Hill, and funded by grant P01-HD31921 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, with cooperative funding from 23 other federal agencies and foundations. Special acknowledgment is due Ronald R. Rindfuss and Barbara Entwisle for assistance in the original design. Information on how to obtain the Add Health data files is available on the Add Health website (http://www.cpc.unc.edu/addhealth). No direct support was received from grant P01-HD31921 for this analysis. We are grateful to the Carolina Population Center (R24 HD050924) for general support.
ABBREVIATIONS
- ST
screen time
- SNP
Single Nucleotide Polymorphism
- GWA
Genome-Wide Association
- EA
European American
- AA
African American
- HA
Hispanic American
Footnotes
There were no potential or real conflicts of financial or personal interest with the financial sponsors of the scientific project.
References
- 1.Gordon-Larsen P, Adair LS, Nelson MC, PBM Five-year obesity incidence in the transition period between adolescence and adulthood: the National Longitudinal Study of Adolescent Health. American Journal of Clinical Nutrition. 2004;80(3):569–575. doi: 10.1093/ajcn/80.3.569. [DOI] [PubMed] [Google Scholar]
- 2.Must A, Tybor DJ. Physical activity and sedentary behavior: a review of longitudinal studies of weight and adiposity in youth. Int J Obes (Lond) 2005;29(Suppl 2):S84–96. doi: 10.1038/sj.ijo.0803064. [DOI] [PubMed] [Google Scholar]
- 3.Hu FB, Li TY, Colditz GA, Willett WC, Manson JE. Television watching and other sedentary behaviors in relation to risk of obesity and type 2 diabetes mellitus in women. JAMA. 2003;289(14):1785–91. doi: 10.1001/jama.289.14.1785. [DOI] [PubMed] [Google Scholar]
- 4.Jeffery RW, French SA. Epidemic obesity in the United States: are fast foods and television viewing contributing? Am J Public Health. 1998;88(2):277–80. doi: 10.2105/ajph.88.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vimaleswaran KS, Li S, Zhao JH, et al. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene. Am J Clin Nutr. 2009;90(2):425–8. doi: 10.3945/ajcn.2009.27652. [DOI] [PubMed] [Google Scholar]
- 6.Kilpelainen TO, Qi L, Brage S, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8(11):e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mustelin L, Silventoinen K, Pietilainen K, Rissanen A, Kaprio J. Physical activity reduces the influence of genetic effects on BMI and waist circumference: a study in young adult twins. Int J Obes (Lond) 2009;33(1):29–36. doi: 10.1038/ijo.2008.258. [DOI] [PubMed] [Google Scholar]
- 8.Richardson AS, North KE, Graff M, et al. Moderate to vigorous physical activity interactions with genetic variants and body mass index in a large US ethnically diverse cohort. Pediatric Obesity. 2013 doi: 10.1111/j.2047-6310.2013.00152.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi Q, Li Y, Chomistek AK, et al. Television watching, leisure time physical activity, and the genetic predisposition in relation to body mass index in women and men. Circulation. 2012;126(15):1821–7. doi: 10.1161/CIRCULATIONAHA.112.098061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris KM, Halpern CT, Smolen A, Haberstick BC. The National Longitudinal Study of Adolescent Health (Add Health) twin data. Twin Res Hum Genet. 2006;9(6):988–97. doi: 10.1375/183242706779462787. [DOI] [PubMed] [Google Scholar]
- 11.Harris KM. An integrative approach to health. Demography. 2010;47(1):1–22. doi: 10.1353/dem.0.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller WC, Ford CA, Morris M, et al. Prevalence of chlamydial and gonococcal infections among young adults in the United States. JAMA. 2004;291(18):2229–36. doi: 10.1001/jama.291.18.2229. [DOI] [PubMed] [Google Scholar]
- 13.Resnick MD, Bearman PS, Blum RW, et al. Protecting adolescents from harm. Findings from the National Longitudinal Study on Adolescent Health. JAMA. 1997;278(10):823–32. doi: 10.1001/jama.278.10.823. [DOI] [PubMed] [Google Scholar]
- 14.Goodman E, Hinden BR, Khandelwal S. Accuracy of teen and parental reports of obesity and body mass index. Pediatrics. 2000;106(1 Pt 1):52–58. doi: 10.1542/peds.106.1.52. [DOI] [PubMed] [Google Scholar]
- 15.Ogden CL, Kuczmarski RJ, Flegal KM, et al. Centers for Disease Control and Prevention 2000 growth charts for the United States: improvements to the 1977 National Center for Health Statistics version. Pediatrics. 2002;109(1):45–60. doi: 10.1542/peds.109.1.45. [DOI] [PubMed] [Google Scholar]
- 16.Sallis JF, Strikmiller PK, Harsha DW, et al. Validation of interviewer- and self-administered physical activity checklists for fifth grade students. Med Sci Sports Exerc. 1996;28(7):840–51. doi: 10.1097/00005768-199607000-00011. [DOI] [PubMed] [Google Scholar]
- 17.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorleifsson G, Walters G, Gudbjartsson D, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nature Genetics. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 19.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyre D, Delplanque J, Chevre JC, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41(2):157–9. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 21.Graff M, North KE, Mohlke KL, et al. Estimation of genetic effects on BMI during adolescence in an ethnically diverse cohort: The National Longitudinal Study of Adolescent Health. Nutrition and Diabetes. 2012;2:e47. doi: 10.1038/nutd.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monda KL, Chen CH, Taylor K, et al. Three Novel Loci Identified with BMI in a Genome-wide Association Study of 47,098 Men and Women of African Ancestry. Human Molecular Genetics. 2012 [Google Scholar]
- 23.Kang SJ, Chiang CW, Palmer CD. Genome-wide association of anthropometric traits in African- and African-derived populations. Human Molecular Genetetics. 2010;19(13):2725–38. doi: 10.1093/hmg/ddq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elks CE, Perry JR, Sulem P, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42(12):1077–85. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.