Abstract
The production of animals by somatic cell nuclear transfer (SCNT) is inefficient, with approximately 2% of micromanipulated oocytes going to term and resulting in live births. However, it is the most commonly used method for the generation of cloned transgenic livestock as it facilitates the attainment of transgenic animals once the nuclear donor cells are stably transfected and more importantly as alternatives methods of transgenesis in farm animals have proven even less efficient. Here we describe piggyBac-mediated transposition of a transgene into porcine primary cells and use of these genetically modified cells as nuclear donors for the generation of transgenic pigs by SCNT. Gene transfer by piggyBac transposition serves to provide an alternative approach for the transfection of nuclear donor cells used in SCNT.
Introduction
A transgenic animal can be described as one whose genome has been altered by the introduction of foreign genetic material. The term transgenic was introduced by the developers of the initial pronuclear microinjection (PNI) procedure, a method whereby linear transgene DNA is injected into the male pronucleus of a zygote. (Gordon et al. 1980). Several alternative procedures for animal transgenesis have been developed in the last thirty-two years with the aim of improving the efficiency and ease of delivery for the transgenic DNA (Marh et al. 2012; Lavitrano et al. 1989; Lois et al. 2002; Perry et al. 1999; Urschitz et al. 2010). However, the preferred and most widely used method to generate transgenic animals is PNI, due to ease of PNI’s implementation and the lack of specialized requirements such as containment facilities necessary for lentiviral mediated transgenesis (Lois et al. 2002). However, PNI is impeded by low efficiency, concatemerized transgene insertion into the genome, and unpredictable levels and patterns of transgene expression (Wall 2001). The implementation of PNI has proven more difficult in livestock such as cattle, sheep and pig where the high lipid content of the oocyte or the one cell embryo makes it impossible to visualize the nuclear structures using conventional light microscopy (Wall et al. 1985).
An alternative transgenesis approach known as intracytoplasmic sperm injection transgenesis (ICSI-Tr), relies on spermatozoa as transgene DNA vectors (Perry et al. 1999). While efficient in mice, this technique has proven inefficient in pigs and appears therefore unsuitable for routine transgenic production in these animals (Garcia-Vazquez et al. 2011; Garcia-Vazquez et al. 2010; Wu et al. 2009). Therefore, investigators have been exploring other methods and one method in particular, somatic cell nuclear transfer (SCNT) has achieved wide acceptance. This approach has been extended to the generation of genetically modified pigs by genomic modification of somatic cells prior to SCNT (Whyte and Prather 2011). A variety of vectors have been used for the modification of somatic cells used in SCNT, for example non-insertional plasmid DNA, virally delivered constructs (Park et al. 2001), and transposase-based vectors such as Sleeping Beauty (Al-Mashhadi et al. 2013; Carlson et al. 2011; Staunstrup et al. 2012; Jakobsen et al. 2011). An alternative transposase that has emerged as mediator for gene transfer in somatic cells is piggyBac(pB), originally isolated from the moth Trichoplusia ni (Cary et al. 1989). We have demonstrated that it is very efficiently in the transposition of reporter genes into the genome of mammalian cell lines in the trans configuration in which the helper contains the transposase gene and the donor harbors the transposon (Wu et al. 2006). The pB transposase was eventually modified to a mammalian codon bias form of the transposase (mPBase), has been shown to be even more efficient in mediating transposition (Cadinanos and Bradley 2007). Recent improvements of the mPBase have rendered it hyperactive (Yusa et al. 2011), and it has proven very effective in the production of transgenic mice (Marh et al. 2012). In this manuscript we describe the production of EGFP expressing transgenic piglets by SCNT with fetal fibroblasts (FFs) isolated from a 25 day-old male Duroc fetus. The transposition efficiency of FFs with mPBase by colony count was 30 fold higher when compared to FFs transfected with the linearized plasmid pEGFP-N1. A single mPBase transfected cellular clone, when used with the classical elecrofusion SCNT method resulted in 20 live transgenic EGFP expressing piglets. Modification of this fetal fibroblasts clone by pB transposition did not result in a decrease in the efficiency of cloning compared to non-transposed cells.
Materials and Methods
Animals
Hybrid LY (male Landrace × female Yorkshire) sows in parity 2-5 were used as embryo recipients. All animals in this study were maintained in a swine farm located in Yunfu City, Guangdong Province, China. The protocol of animal handling and treatment was reviewed and approved by the Animal Care and Use Committee of the South China Agricultural University.
Vector construction
Plasmid pEGFP-N1 was purchased from Invitrogen (Life Technologies, CA, USA). Helper vector mPBase, also designated as mPB by Cadiñanos and Bardley (Cadinanos and Bradley 2007), and donor vector pCyL50, Wang et al., (Wang et al. 2008), was a kind gift from The Wellcome Trust Sanger Institute (Cambridgeshire, UK). The neo-2a-EGFP fusion DNA fragment flanked by EcoRI and XbaI was synthesized and inserted into plasmid pcDNA3.1 (Invitrogen, Life Technologies, CA, USA) to construct pcDNA3.1-neo-EGFP. The CMV-neo-EGFP fragment containing the CMV promoter, neo-2a-EGFP fusion gene and polyA sequence were amplified from pcDNA3.1-neo-EGFP by PCR, with the SalI restriction site added to both ends of the fragment during the PCR amplification. The CMV-neo-EGFP fragment was then inserted into the SalI site in the donor vector pCyL50 by restriction digestion and ligation to generate pB CMV-neo-EGFP donor plasmid.
Cell transfection and transgenic cell colony selection
FFs were isolated from a 25 day-old male Duroc fetus, and were cultured in 6 cm dishes until they reached 90% confluence. The cells were then either transfected with linearized pEGFP-N1, to test for the frequency of non-homologous recombineering (NHR) of a linearized plasmid into the genome of the FF cells. As a comparison to linear plasmid integration, another batch of FF cells were alternatively co-transfected with 2100 ng of donor vector pB CMV-neo-EGFP, and 730 ng of helper vector mPBase in a molar ratio of 3:1 in a 6 cm dish by lipofectamine. At 24 h post-transfection, the cells were subjected to antibiotic selection with 1 mg/ml G418 (Gibco, Life Technologies, CA, USA) for 14 days. Surviving cell colonies expressing EGFP were counted using fluorescence microscopy. Photographs of EGFP-expressing cell colonies were taken under blue light by a camera with light filters.
Somatic cell nuclear transfer
A G418-resistant and EGFP positive cell colony was randomly selected and isolated by using a cloning cylinder, and then cultured in a single well of a 48-well plate for 3 days until it reached 90% confluence. The cells were transferred to a single well of a 24-well plate and cultured for another 5 days. Subsequently the transgenic fetal fibroblasts were collected for SCNT. Wild-type male Duroc FFs at 3rd passage were used as a control for SCNT. The SCNT experiments for both transgenic and wild-type male Duroc fetal fibroblasts were performed under the same condition and following the same protocol as previously described (Li et al. 2013).
Porcine ovaries were collected from a local slaughterhouse. Cumulus-oocyte complexes (COCs) were aspirated from the ovaries and matured in vitro for 42–44 h following the protocol described by Deng et al., (Deng et al. 2011). Matured COCs were freed from cumulus cells by repeated pipetting in 0.1% hyaluronidase. Subsequently, matured oocytes with a first polar body were selected for cloning.
The first polar body and adjacent cytoplasm containing all the maternal chromosomes were extruded from the selected mature oocyte by squeezing with a fine glass needle. A single fibroblast cell was microinjected into the perivitelline space of the oocytes. The oocyte-donor cell complexes were cultured in PZM3 medium at 39℃ for 1.5 h and then activated to fuse by two successive DC pulses of 1.2 kv/cm for 30 μs, using an electro-fusion instrument (model: CF150/B, BLS).
The reconstructed embryos were cultured in PZM3 medium at 39℃ with 5% CO2, 7% O2, 88% N2 and 100% humidity for 20h, and then loaded into a transfer tube and kept in a portable incubator (Minitube) during transportation to the farm where the recipient sow was housed. Estrous-synchronized hybrid sows were used as embryo recipients. They were anesthetized with ketamine and xylazine for induction and 3% of isofluorane for maintenance. One oviduct was exposed by surgery. The cloned embryos (200-250 embryos per recipient) were delivered directly into the oviduct of the recipient sow using a syringe. The pregnancy status of the recipient sows was monitored using an ultrasound machine equipped with a convex transducer at approximately one month after embryo transfer.
Reporter gene expression in transgenic pigs
Transgenic pigs, their organs and tissues were analyzed for EGFP expression by fluorescence under blue light. Photographs of transgenic pigs, wild-type pigs, and their organs and tissues were taken under blue light or normal light by a camera with or without light filters.
PCR analysis
Genomic DNA was isolated from tail biopsies of all 20 transgenic and 2 wild-type control piglets using Tissue DNA extraction kit (Omega Bio-Tek, Norcross, GA, USA). A 609 bp fragment of the EGFP gene, a 200 bp fragment extending from the 5’-TRE into the plasmid backbone, a 90 bp fragment extending from the 3’-TRE into the backbone, a 3.94 kb fragment covering the whole piggyBac transposase gene expression cassette, and a 140 bp fragment of the internal control β-actin gene were amplified by PCR using primer sets P1+P2, P19+P20, P17+P18, P3+P4, and P5+P6, respectively (for primer sequences see Table 1, for primer locations on the plasmids see Figure 1A and 1B). The PCR products were sequenced to confirm their identity.
Table 1.
Primer information.
| Name | Sequences (5’-3’) |
|---|---|
| P1 (PCR EGFP-F) | CGTGCAGTGCTTCAGCCGCTACCCCGACC |
| P2 (PCR EGFP-R) | CAAGGAAGGCACGGGGGAGGGGCAAACAA |
| P3 (PCR Transposase-F) | CAGTATCTGCTCCCTGCTTGTG |
| P4 (PCR Transposase-R) | AGCCGATTGTCTGTTGTGCC |
| P5 (PCR (β-actin-F) | CCACGAAACTACCTTCAACTC |
| P6 (PCR (β-actin-R) | TGATCTCCTTCTGCATCCTGT |
| P7 (Real time PCR EGFP-F) | GAGCGCACCATCTTCTTCAAG |
| P8 (Real time PCR EGFP-R) | TGTCGCCCTCGAACTTACA |
| P9 (Real time PCR PB 5’TR-F) | CTAAATAGCGCGAATCCGTC |
| P10 (Real time PCR PB 5’TR-R) | TCATTTTGACTCACGCGG |
| P11 (Real time PCR β-actin-F) | CTCGATCATGAAGTGCGACGT |
| P12 (Real time PCR β-actin-R) | GTGATCTCCTTCTGCATCCTG |
| P13 (Inverse PCR PB5’TR) | CTTACCGCATTGACAAGCAC |
| P14 (Inverse PCR neo) | CACTTCGCCCAATAGCAG |
| P15 (Inverse PCR PB3’TR) | ATACAGACCGATAAAACACATG |
| P16 (Inverse PCR CMV) | GGCTAACTAGAGAACCCACTG |
| P17(PCR PB3’TR) | TCGGTATTCACGACAGCAG |
| P18(PCR PB3’TR) | ATCTTTAACGTACGTCACAATAT |
| P19(PCR PB5’TR) | ACGGATTCGCGCTATTTAGA |
| P20(PCR PB5’TR) | TGATCTCCTTCTGCATCCTGT |
Fig.1. Generation of transgenic porcine fetal fibroblast colonies by piggyBac transposon-mediated gene transfer.
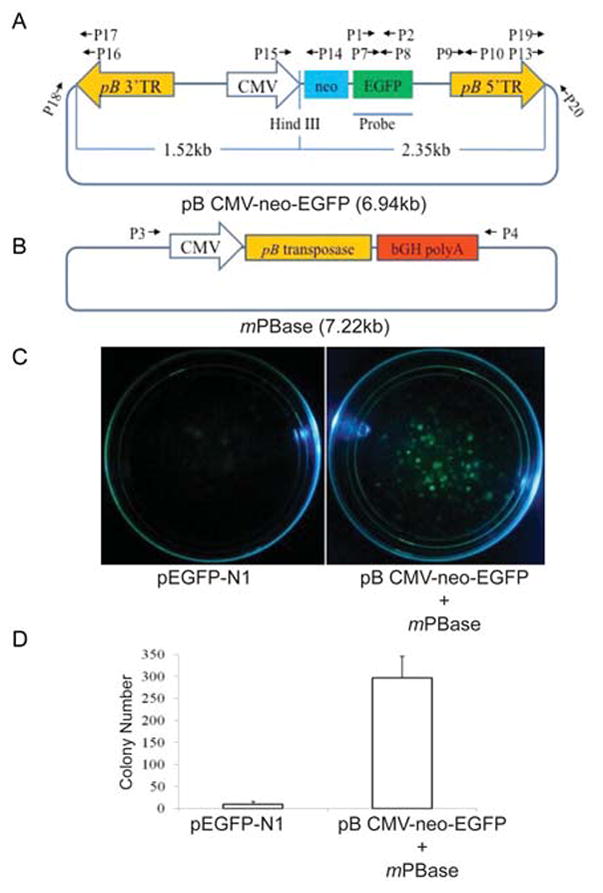
Structure of plasmid vectors, donor pB CMV-neo-EGFP (A) and helper mPBase (B); pB 5’TR, pB transposon 5’ terminal repeat element. pB 3’TR, pB transposon 3’ terminal repeat element. CMV, CMV promoter. neo, neomycin-resistant gene. EGFP, enhanced green fluorescence protein gene. Probe, the probe used for Southern blot. bGH polyA, bovine growth hormone gene polyA. (C). Surviving fetal fibroblast colonies in 10 cm dishes after transfection with linearized pEGFP-N1 or co-transfection with donor vector pB CMV-neo-EGFP and helper vector mPBase, and 2 week selection with G418. The pictures of the dishes with G418-resistant cell colonies were taken under blue light. (D). Colony count of G418-resistant cells in the dishes. The experiment was performed in triplicate and the data is presented as mean ± SD. A mean of 10 and 309 colonies was observed in cultures transfected with linearized plasmid pEGFP-N1, or co-transfected with donor vector pB CMV-neo-EGFP and helper vector mPBase, respectively.
Southern Blot analysis
Ten microgram of tail genomic DNA from each of the 20 transgenic and 2 wild-type piglets was restriction digested with Hind III (New England Biolabs, Ipswich, MA) and separated by electrophoresis in a 0.8% agarose gel. The DNA was then transferred to a nylon membrane (Amersham Biosciences, UK) by the capillary transfer method. During transfer, the DNA was nicked and denatured. The membrane was prehybridized overnight at 42℃ and then hybridized with an 800 bp EGFP gene probe labeled with digoxigenin by using a PCR DIG Probe Synthesis Kit (Roche, China). Hybridization and washing were performed with DIG-High Prime DNA Labeling and Detection Starter KitⅡ(Roche). After hybridization, the membrane was incubated for 30 min in blocking solution and further incubated for 30 min in Anti-Digoxigenin-AP antibody solution. The membrane was then exposed for 5-20 min after incubation with 1ml of CSPD ready-to-use, and the Southern photograph was captured with the EC3 imaging system (UAP, CA, USA).
Real-time quantitative PCR analysis
A real-time PCR method previously described by Grandjean et al., (Grandjean et al. 2011) was used to determine the copy number of EGFP transgenes and pB transposons integrated into the genome of transgenic pigs. Twenty nanogram of genomic DNA was analyzed by real-time quantitative PCR using the SYBR Green-Taq polymerase kit and ABI Prism 7700 PCR machine. Primer sets P7+P8, P9+P10 and P11+P12 (for primer sequences see Table 1) were used to amplify the EGFP transgene, pB transposon 5’ terminal repeat (pB 5’-TRE) and the reference gene β-actin, respectively. The copy number of β-actin was used as reference to estimate the copy number of EGFP transgene and pB 5’-TRE’s in the genome. The sequence of the 114 bp fragment of porcine β-actin gene amplified by P11 and P12 primers was blasted against the Sus scrofa (pig) genomic DNA sequence database (Build Sscrofa10.2) on NCBI BLAST website. A number of 2 blast hits was found per haploid genome. Since the DNA sample used for real-time PCR was isolated from the diploid cells of transgenic pigs, we estimated that there are 4 copies of 114 bp β-actin amplicon in each diploid genome of transgenic pigs. This number was used as a normalization standard for calculation of copy number of EGFP transgenes and pB 5’-TRE in the genome of transgenic pigs, by the 2-ΔCt method based on the threshold cycle (Ct) values, as described by Livak and Schmittgen (Livak and Schmittgen 2001).
Inverse PCR analysis
To analyze transposon insertion sites, one milligram of transgenic pig’s genomic DNA was restriction digested with Hind III, purified by a DNA purification column (Qiagen, China) and eluted with 100 μl of ddH2O. After adjustment with ddH2O to final volume, T4 ligase was added to a final concentration of 10U/μl, and the 1000 μl ligation mix was incubated at 16℃ overnight. Ligated DNA was purified with a Qiagen DNA purification column and eluded from it with 100 μl of ddH2O. A 2 μl aliquot was used as template for the PCR reaction with primer set: P13+P14, or P15+P16 (for primer sequences see Table 1, for primer location see Figure 1A). The resulting PCR products were cloned by ligation into a TA vector (Life Technologies, CA, USA) and sequenced. The obtained sequences were aligned to the sequence of the donor vector pB CMV-neo-EGFP and the Sus scrofa (pig) genomic DNA sequence database (Build Sscrofa10.2) using NCBI BLAST.
Results
Efficient generation of transgenic porcine fetal fibroblast colonies by piggyBac transposon-mediated gene transfer
To investigate the potential use of pB vectors in pig transgenesis, we constructed a pB donor vector transposon plasmid carrying a human cytomegalovirus (CMV) promoter, a neomycin-resistant gene (neo) linked to an enhanced green fluorescent protein gene (EGFP) by a 2A peptide. This plasmid was named as donor vector pB CMV-neo-EGFP (Figure 1A). Pig fetal fibroblasts were co-transfected with donor vector pB CMV-neo-EGFP and a pB transposase expression helper vector, mPBase (Figure 1B) to select G418-resistant EGFP cell colonies. FFs transfected with the linearized pEGFP-N1 plasmid served as a control. The number of surviving EGFP positive colonies resulting from co-transfections of donor and pB-helper plasmids was 30 fold higher than those only transfected with linearized plasmid pEGFP-N1 (Figure 1C and 1D). This data clearly demonstrates that the pB transposition system can very efficiently mediate integration of transgenes into the genome of porcine fetal fibroblasts (FFs) by transposition.
pB vector-transfected fetal fibroblasts as donor cells for transgenic pigs production by SCNT
To test whether the transgenic fibroblasts generated by transfection with pB vectors could be used as nuclear donor cells for efficient production of transgenic pigs by the SCNT technique, we randomly picked a G418-resistant EGFP positive FF cell colony and used it for SCNT. A total of 1542 transgenic cloned embryos were produced and transferred to 7 surrogate sows at approximately 2-cell stage. Four of the recipient sows maintained pregnancy to term and in total delivered 20 live transgenic cloned piglets (Table 2). The cloning efficiency resulting from using pB vector-transfected transgenic FFs as donor cells was 1.3% (= 20/1542, the number of live born cloned piglets divided by the number of transferred cloned embryos). In contrast the cloning efficiency using wild-type FFs as donor cells for SCNT was significantly lower (0.605% =31/5128) (Table 2). PB transposon-mediated transgene integration into pig genome does therefore appear not to impair the function of developmentally important genes and the use of pB vector transfection-derived transgenic cells as donors for pig cloning does not decrease SCNT efficiency.
Table 2.
Comparison of cloning efficiency resulted from using wild-type and PB vectors-transfected transgenic fetal fibroblast as donor cells for SCNT.
| Donor fetal fibroblast genotype | Transgenic | Wild-type |
|---|---|---|
| No. of transferred cloned embryos | 1542 | 5128 |
| No. of recipients | 7 | 23 |
| No. of embryos/recipient | ≈220 | ≈226 |
| No. of pregnant recipients (pregnancy rate) # | 4 (57.14%=4/7) |
11 (47.83%=11/23) |
| No. of farrowed recipients (farrowing rate) | 4 (57.14%=4/7) |
9 (39.13%=9/23) |
| No. of delivered cloned piglets (birth rate) | 20* (1.30%=20/1542) |
31◎ (0.60%=31/5128) |
| No. of delivered live normal cloned piglets (birth rate) | 18 (1.17%=18/1542) |
24 (0.47%=24/5128) |
Pregnancy of recipients was examined at one month after embryo transfer.
All these 20 piglets were born alive and 2 of them showed abnormalities.
Three of these 31 piglets were stillborn, and 4 of the remaining live piglets showed abnormalities.
All live born 20 cloned transgenic piglets were EGFP positive, with the fluorescent signal particularly strong on their noses and hooves (Figure 2A). Two of the new born piglets displayed abnormalities by phenotype. One suffered from Enchephalocele and the other demonstrated joint contracture. Both of these EGFP positive animals were sacrificed, together with two healthy ones for determination of transgene expression in their internal organs and tissues. The tongue, liver, heart, intestine, lung, stomach, testis, spleen, kidney and muscle were dissected for EGFP observation. Green fluorescence was visualized on all the analyzed organs and tissues. Figure 2B shows a representative picture displaying strong green fluorescence observable on the organs of transgenic pigs.
Fig.2. EGFP expression in transgenic pigs produced by pB transposon-mediated gene transfer.
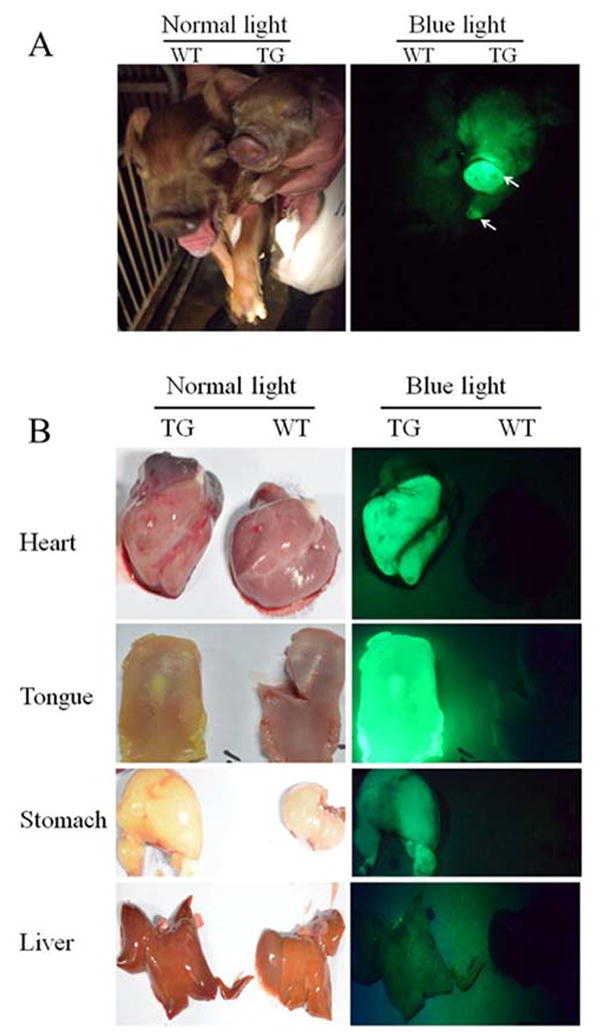
(A). A representative picture showing green florescence was not visualized on wild-type (WT) piglets, but was clearly observed on transgenic (TG) piglets, especially on their noses and hooves (indicated by the arrows) under blue light. (B). A representative picture showing green florescence was also clearly observed on the organs of TG piglets but not on that of WT piglets under blue light.
Genetic analyses of transgenic pigs produced by pB transposon-mediated gene transfer
From the genomic DNA of the transgenic pigs we obtained EGFP PCR products, while the PCR analysis of the wild type pig DNA did not result in any amplification (Figure 3A), demonstrating that all transgenic pigs carry the EGFP transgene. In contrast, we were unable to detect any amplification from the pB transposase gene expression cassette or the 5’- and 3’-TREs in the genomic DNA of transgenic cloned pigs (Figure 3A), suggesting the absence of donor and helper plasmid fragments in the genome of cloned transgenic pigs.
Fig.3. Genetic analyses of transgenic pigs and FF clones produced by pB transposon-mediated gene transfer.
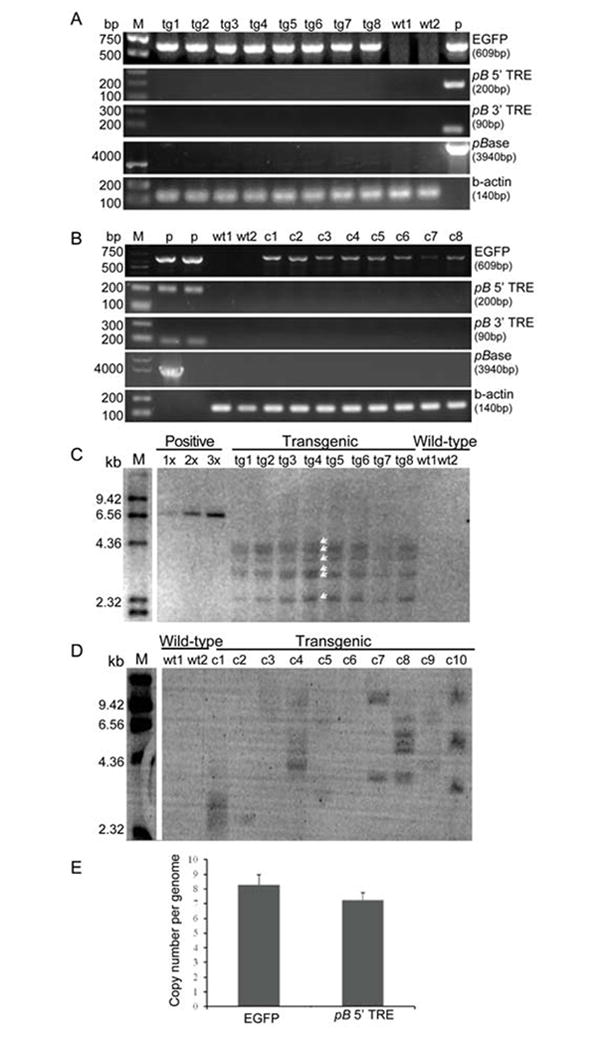
(A). PCR analysis of EGFP, pB 5’-TRE, pB 3’-TRE and pB transposase expression gene cassette in the genome of transgenic pigs. A 609 bp fragment of EGFP gene was amplified from the genomic DNA of all the transgenic pigs but not from wild-type pigs. PCR products for pB 5’-TRE, pB 3’-TRE and pB transposase were not detected. P, positive control using plasmid DNA of donor vector pB CMV-neo-EGFP as template for amplification of EGFP gene, or using plasmid DNA of helper vector mPBase as template for amplification of pB transposase gene expression cassette. (B). PCR analysis of EGFP, pB 5’-TRE, pB 3’-TRE and pB transposase expression gene cassette in the genome of eight individual FF clones. Again we detected EGFP, but no PCR products for pB 5’-TRE, pB 3’-TRE and pB transposase. (C). Southern blot analysis of EGFP in transgenic pigs. 1x, 2x and 3x, represents plasmid positive control respectively harboring 1 copy, 2 copies and 3 copies of transgene per pig genome. Southern blot analysis of positive control was carried out with a mixture of 10 μg of wild-type pigs’ genomic DNA and 2.57×10-5 μg (1 copy), or 5.14×10-5 μg (2 copies), or 7.71×10-5 μg (3 copies) of donor vector pB CMV-neo-EGFP plasmid DNA as total starting DNA. The amount of plasmid DNA used to mix with wild-type pigs’ genomic DNA for Southern blot analysis of positive controls was calculated based on the following equation: copy number (1, or 2, or 3) × plasmid length (6.94×103 bp) / pig genome whole length (27×108 bp) = amount of plasmid DNA / amount of WT pig genomic DNA (10 μg). Six bands (indicated by white arrows) were detected from the genome of transgenic pigs, and the intensity of each band was similar with that of 1 copy positive control, suggesting the transgene was integrated in at least 6 sites of each transgenic pig genome, and that each insertion site contains only 1 copy of transgene. (D). Southern blot analysis of EGFP in ten individual transgenic and two wild-type FF clones. (E). Real-time quantitative PCR analysis of transgene copy number per transgenic pig genome. Data is presented as mean ± SD (n=4).
To analyze the average number of pB transpositions in a larger number of FFs and to assess the extent of non-transpositional transgenesis, we performed an additional Southern analysis with the DNA from ten independent cellular clones. PCR analysis of DNA from eighth of these FFs was carried out to determine if broken plasmids had been integrated into their genome non-transpositionally. We obtained amplification products (Figure 3B) for EGFP from the genomic DNA of eight FFs but not from non-transfected cells, demonstrating that these transgenic FFs carry the EGFP transgene. In contrast, we were unable to detect any amplification from the backbone regions flanking the 5’- and 3’-TREs of the donor plasmid, or the pB transposase gene expression cassette from the helper plasmid. This data confirms that indeed transpositional insertion of the transposon is responsible for the integration of the transgene.
The Southern blot analysis of the transgenic pig DNA indicated that at least six individual copies of the EGFP transgene were integrated in each of the pigs (Figure 3C). Southern blots performed on the DNA of the ten additional individual FF clones indicated that the integrated transgene copy number ranged from one to six copies per clone (Figure 3D).
An analysis by real-time quantitative PCR indicated that each transgenic pig’s genome carries approximately 8 copies (8 insertion sites) of the transgene (Figure 3E). It is possible that in the Southern blot analysis, some of the EGFP probe-reactive bands detected actually represent 2 different fragments of similar size, undistinguishable by this type of analysis. Such bands usually demonstrate intensity in Southern blots stronger than that of the single copy positive control, but weaker than that of the 3 copy positive control (Figure 3C). Therefore, each transgenic pig’s genome could actually contain more than the visible 6 copies (6 integration sites) of transgene, some migrating together at the same molecular weight.
Identification of pB transposon integration sites in transgenic pigs genome
Inverse PCR was used to identify the pB transposon and transgene insertion sites in the genome of transgenic pigs expressing EGFP. Two integration sites were identified (Figure 4), neither of them located near known functional genes as determined by alignment to the NCBI Sus scrofa (pig) genomic DNA database. We therefore assumed that these two pB transposon insertion events did not interfere with the activity of any known gene of the host cell genome.
Fig.4. Identification of pB transposon integration sites in transgenic pig’s genome.
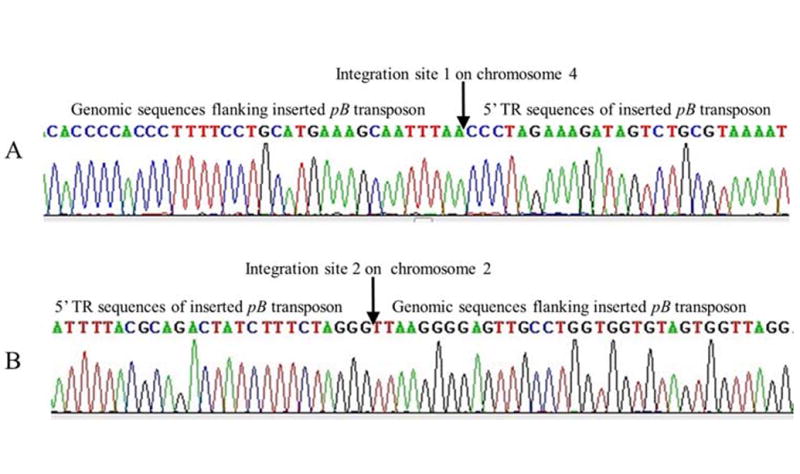
Chromosomal DNA sequences flanking the inserted pB transposon were identified by sequencing the inverse PCR products. The obtained flanking DNA sequences were then used to blast against Sus scrofa (pig) genomic DNA database to find out their location in pig genome. All identified insertion site sequences contain the TTAA sequences which is indicative of the insertion border between the pB transposon and flanking genomic DNA. (A). An identified integration site of pB transposon on chromosome 4. The pB transposon was inserted in between 716 bp and 717 bp of GenBank sequence (GenBank number NW_003534687.2, length=657315 bp). (B). An identified integration site of pB transposon on chromosome 2. The pB transposon was inserted in between 96558 bp and 96559 bp of GenBank sequence (GenBank number NW_003299559.3, length=172486 bp).
Conclusion
SCNT is commonly used for the production of transgenic pigs (Whyte and Prather 2011). However, efficiency rates have not been significantly improved since the introduction of this method more than 10 years ago (Wakayama et al. 1998; Wilmut et al. 1997). Oocytes used for pig SCNT are matured in vitro and are considered to be the limiting factor post enucleation for the attainment of greater live birth efficiencies (Betthauser et al. 2000). At the same time, in vivo maturation of oocytes have not proven to be more effective in terms of overall SCNT cloning efficiency (Onishi et al. 2000). The reprogramming of the transferred nucleus, required during SCNT therefore appears to be very important for success of the procedure. Such observations and transcriptional activities analyzed in human trophoblast cells indicate that there must be as yet unknown factors regulating gene expression which have to be elucidated before efficiency improvements in SCNT by reprogramming of donor nuclei can be expected (Wu et al. 2001).
Another factor influencing the efficiency of transgenic pig production by SCNT is the method used to transfect the nuclear donor cells. For example, virus transfection is highly efficient and has resulted in the first transgenic cloned pigs (Park et al. 2001). However, many laboratories working in animal transgenesis are not equipped to safely use viruses. Therefore, alternative cell transfection mechanisms are necessary for implementation of animal transgenesis by SCNT. Transposase’s have been shown to be very effective for the genetic modification of mammalian cell lines. These efficient transposition molecules can be propagated in plasmids and then transferred into cells by either lipofection or electroporation mediated transfection. Once inside the nucleus of the cell, they insert the transgene containing transposon into the genome of the host cell via a cut-and-paste mechanism. The Sleeping Beauty(SB) transposon/transposase system has previously been used to deliver transgenes to neonatal porcine fibroblasts (NPFs) for use in SCNT experiments (Jakobsen et al. 2011) and was also effective in the development of transgenic cloned pigs for a skin inflammation model expressing the human β1 and α2 integrin genes (Staunstrup et al. 2012).
In the current manuscript we describe the piggyBac transposase system in the trans, two component donor-helper configuration, for the genetic modification of FFs. These cells were then used for SCNT-based production of transgenic pigs. We were able to demonstrate non-concatemerised insertions of single-copy EGFP transgenes. As we transferred embryos to surrogate mother sows at the two cell stage, a correct estimation of micromanipulated embryos and their relationship to transgenic offspring’s produced can be calculated. Additionally, transfer at this early developmental stage avoids in vitro culturing of these embryos and the well documented media effects on their development (Cao et al. 2012; Rinaudo and Schultz 2004). The lack of high quality blastocyst selection with this procedure prevents the biasing of the efficiency for transgenic animal production. We generated 20 full transgenic piglets using piggyBac-modified FFs representing an overall cloning efficiency of 1.3% (20/1542 embryos transferred). Four piglets were sacrificed for organ analysis and the remaining 16 piglets survived weaning and are alive today. In summary, here we report piggyBac mediated transposition for the genetic modification of FFs for SCNT. We have demonstrated that our piggyBac transposase/transposon two plasmid approach for transfection of nuclear donor cells resulted in the production of mostly healthy piglets. As transposon-based non-viral vectors offer a more easily implemented and potentially safer method to genetically modify primary cells, this approach represents an attractive alternative to viral vector-mediated transgenesis.
Acknowledgments
This study was supported by a grant from the National Science Foundation for Young Scholars of China (grant number: 31101689), a grant from the National High Technology Research and Development Program of China (863 Program, grant number: 2011AA100304), a grant from Department of Science and Technology of Guangdong, China (grant number: 2011A020901001) and by National Institutes of Health Grants 5P20RR024206 and R01 GM083158-01A1 (to S.M.).
References
- Al-Mashhadi RH, Sorensen CB, Kragh PM, Christoffersen C, Mortensen MB, Tolbod LP, Thim T, Du Y, Li J, Liu Y, Moldt B, Schmidt M, Vajta G, Larsen T, Purup S, Bolund L, Nielsen LB, Callesen H, Falk E, Mikkelsen JG, Bentzon JF. Familial hypercholesterolemia and atherosclerosis in cloned minipigs created by DNA transposition of a human PCSK9 gain-of-function mutant. Science translational medicine. 2013;5(166):166ra–161. doi: 10.1126/scitranslmed.3004853. [DOI] [PubMed] [Google Scholar]
- Betthauser J, Forsberg E, Augenstein M, Childs L, Eilertsen K, Enos J, Forsythe T, Golueke P, Jurgella G, Koppang R, Lesmeister T, Mallon K, Mell G, Misica P, Pace M, Pfister-Genskow M, Strelchenko N, Voelker G, Watt S, Thompson S, Bishop M. Production of cloned pigs from in vitro systems. Nature biotechnology. 2000;18(10):1055–1059. doi: 10.1038/80242. [DOI] [PubMed] [Google Scholar]
- Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic acids research. 2007;35(12):e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Sui L, Li Y, Ji S, Zhang X, Zhang Y. Effects of chemically defined medium on early development of porcine embryos derived from parthenogenetic activation and cloning. Zygote. 2012;20(3):229–236. doi: 10.1017/S0967199411000153. [DOI] [PubMed] [Google Scholar]
- Carlson DF, Garbe JR, Tan W, Martin MJ, Dobrinsky JR, Hackett PB, Clark KJ, Fahrenkrug SC. Strategies for selection marker-free swine transgenesis using the Sleeping Beauty transposon system. Transgenic Res. 2011;20(5):1125–1137. doi: 10.1007/s11248-010-9481-7. [DOI] [PubMed] [Google Scholar]
- Cary LC, Goebel M, Corsaro BG, Wang HG, Rosen E, Fraser MJ. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172(1):156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- Deng W, Yang D, Zhao B, Ouyang Z, Song J, Fan N, Liu Z, Zhao Y, Wu Q, Nashun B, Tang J, Wu Z, Gu W, Lai L. Use of the 2A peptide for generation of multi-transgenic pigs through a single round of nuclear transfer. PLoS One. 2011;6(5):e19986. doi: 10.1371/journal.pone.0019986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vazquez FA, Ruiz S, Grullon LA, de Ondiz A, Gutierrez-Adan A, Gadea J. Factors affecting porcine sperm mediated gene transfer. Research in veterinary science. 2011;91(3):446–453. doi: 10.1016/j.rvsc.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Garcia-Vazquez FA, Ruiz S, Matas C, Izquierdo-Rico MJ, Grullon LA, De Ondiz A, Vieira L, Aviles-Lopez K, Gutierrez-Adan A, Gadea J. Production of transgenic piglets using ICSI-sperm-mediated gene transfer in combination with recombinase RecA. Reproduction. 2010;140(2):259–272. doi: 10.1530/REP-10-0129. [DOI] [PubMed] [Google Scholar]
- Gordon JW, Scangos GA, Plotkin DJ, Barbarosa JA, Ruddle FH. Genetic Transformation of Mouse Embryos by Microinjection of Purified DNA. PNAS. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean M, Girod PA, Calabrese D, Kostyrko K, Wicht M, Yerly F, Mazza C, Beckmann JS, Martinet D, Mermod N. High-level transgene expression by homologous recombination-mediated gene transfer. Nucleic acids research. 2011;39(15):e104. doi: 10.1093/nar/gkr436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen JE, Li J, Kragh PM, Moldt B, Lin L, Liu Y, Schmidt M, Winther KD, Schyth BD, Holm IE, Vajta G, Bolund L, Callesen H, Jorgensen AL, Nielsen AL, Mikkelsen JG. Pig transgenesis by Sleeping Beauty DNA transposition. Transgenic Res. 2011;20(3):533–545. doi: 10.1007/s11248-010-9438-x. [DOI] [PubMed] [Google Scholar]
- Lavitrano M, Camaioni A, Fazio VM, Dolci S, Farace MG, Spadafora C. Sperm cells as vectors for introducing foreign DNA into eggs: genetic transformation of mice. Cell. 1989;57(5):717–723. doi: 10.1016/0092-8674(89)90787-3. [DOI] [PubMed] [Google Scholar]
- Li Z, Shi J, Liu D, Zhou R, Zeng H, Zhou X, Mai R, Zeng S, Luo L, Yu W, Zhang S, Wu Z. Effects of donor fibroblast cell type and transferred cloned embryo number on the efficiency of pig cloning. Cellular reprogramming. 2013;15(1):35–42. doi: 10.1089/cell.2012.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Science. 5556. Vol. 295. New York, NY: 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors; pp. 868–872. [DOI] [PubMed] [Google Scholar]
- Marh J, Stoytcheva Z, Urschitz J, Sugawara A, Yamashiro H, Owens JB, Stoytchev I, Pelczar P, Yanagimachi R, Moisyadi S. Hyperactive self-inactivating piggyBac for transposase-enhanced pronuclear microinjection transgenesis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(47):19184–19189. doi: 10.1073/pnas.1216473109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi A, Iwamoto M, Akita T, Mikawa S, Takeda K, Awata T, Hanada H, Perry AC. Science. 5482. Vol. 289. New York NY: 2000. Pig cloning by microinjection of fetal fibroblast nuclei; pp. 1188–1190. [DOI] [PubMed] [Google Scholar]
- Park KW, Cheong HT, Lai L, Im GS, Kuhholzer B, Bonk A, Samuel M, Rieke A, Day BN, Murphy CN, Carter DB, Prather RS. Production of nuclear transfer-derived swine that express the enhanced green fluorescent protein. Animal biotechnology. 2001;12(2):173–181. doi: 10.1081/abio-100108344. [DOI] [PubMed] [Google Scholar]
- Perry AC, Wakayama T, Kishikawa H, Kasai T, Okabe M, Toyoda Y, Yanagimachi R. Science. 5417. Vol. 284. New York NY: 1999. Mammalian transgenesis by intracytoplasmic sperm injection; pp. 1180–1183. [DOI] [PubMed] [Google Scholar]
- Rinaudo P, Schultz RM. Effects of embryo culture on global pattern of gene expression in preimplantation mouse embryos. Reproduction. 2004;128(3):301–311. doi: 10.1530/rep.1.00297. [DOI] [PubMed] [Google Scholar]
- Staunstrup NH, Madsen J, Primo MN, Li J, Liu Y, Kragh PM, Li R, Schmidt M, Purup S, Dagnaes-Hansen F, Svensson L, Petersen TK, Callesen H, Bolund L, Mikkelsen JG. Development of transgenic cloned pig models of skin inflammation by DNA transposon-directed ectopic expression of human beta1 and alpha2 integrin. PLoS One. 2012;7(5):e36658. doi: 10.1371/journal.pone.0036658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urschitz J, Kawasumi M, Owens J, Morozumi K, Yamashiro H, Stoytchev I, Marh J, Dee JA, Kawamoto K, Coates CJ, Kaminski JM, Pelczar P, Yanagimachi R, Moisyadi S. Helper-independent piggyBac plasmids for gene delivery approaches: strategies for avoiding potential genotoxic effects. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(18):8117–8122. doi: 10.1073/pnas.1003674107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394(6691):369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- Wall RJ. Pronuclear microinjection. Cloning Stem Cells. 2001;3(4):209–220. doi: 10.1089/15362300152725936. [DOI] [PubMed] [Google Scholar]
- Wall RJ, Pursel VG, Hammer RE, Brinster RL. Development of porcine ova that were centrifuged to permit visualization of pronuclei and nuclei. Biol Reprod. 1985;32(3):645–651. doi: 10.1095/biolreprod32.3.645. [DOI] [PubMed] [Google Scholar]
- Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, Wang X, Bradley A, Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(27):9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte JJ, Prather RS. Genetic modifications of pigs for medicine and agriculture. Molecular reproduction and development. 2011;78(10-11):879–891. doi: 10.1002/mrd.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385(6619):810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Wu D, Luo S, Wang Y, Zhuang L, Chen Y, Peng C. Smads in human trophoblast cells: expression, regulation and role in TGF-beta-induced transcriptional activity. Molecular and cellular endocrinology. 2001;175(1-2):111–121. doi: 10.1016/s0303-7207(01)00397-5. [DOI] [PubMed] [Google Scholar]
- Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, Kaminski JM. piggyBac is a flexible and highly active transposon as compared to Sleeping Beauty, Tol2, and Mos1 in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(41):15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liu CJ, Wan PC, Hao ZD, Zeng SM. Factors affecting the efficiency of producing porcine embryos expressing enhanced green fluorescence protein by ICSI-mediated gene transfer method. Anim Reprod Sci. 2009;113(1-4):156–166. doi: 10.1016/j.anireprosci.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(4):1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]


