Abstract
The P1B-ATPases are a ubiquitous family of metal transporters. These transporters are classified into subfamilies on the basis of substrate specificity, which is conferred by conserved amino acids in the last three transmembrane domains. Five subfamilies have been identified to date, and representative members of four (P1B-1 to P1B-4) have been studied. The fifth family (P1B-5), of which some members contain a C-terminal hemerythrin (Hr) domain, is less well characterized. The S. meliloti Sma1163 gene encodes for a P1B-5-ATPase, denoted Nia (Nickel/iron ATPase), that is induced by exogenous Fe2+ and Ni2+. The nia mutant accumulates nickel and iron, suggesting a possible role in detoxification of these two elements under free-living conditions, as well as in symbiosis, when the highest expression levels are measured. This function is supported by an inhibitory effect of Fe2+ and Ni2+ on the pNPPase activity, and by the ability of Nia to bind Fe2+ in the transmembrane domain. Optical and X-ray absorption spectroscopic studies of the isolated Hr domain confirm the presence of a dinuclear iron center and suggest that this domain might function as an iron sensor.
Introduction
The P-type ATPases are a family of integral membrane proteins that use the energy of ATP hydrolysis to transport cations across cell membranes.1 This superfamily is classified into groups on the basis of substrate specificity, which is conferred by specific transmembrane topologies and amino acid sequences in the transmembrane metal binding sites (TM-MBSs). The P1B-type ATPases are the most widely distributed group and confer diverse heavy metal tolerance to microorganisms.2 They are characterized by a shared core architecture, consisting of six to eight transmembrane (TM) helices, a soluble cytosolic ATP binding domain (ATPBD) present between the second and third-to-last transmembrane regions and a preceding cytosolic actuator domain (A). The P1B-ATPases are responsible for transition metal efflux out of the cytosol into either the extracellular space or to the lumen of intracellular organelles.3,4 Efflux results in either transition metal detoxification or cofactor delivery to the apo forms of metalloproteins.
P1B-ATPase substrate specificity is proposed to arise from the identities and positions of conserved metal-coordinating residues in the TM-MBSs within the last three TM helices.5 Mutation of these residues results in the loss of transmembrane metal binding capabilities, and consequently transport.3,6-9 In particular, a three-residue cysteine-containing motif in TM4 is important for metal transport activity. The study of these transmembrane signature sequence motifs serves as the basis for subdivision of the P1B-ATPases into five substrate-specific subfamilies designated P1B-1 to P1B-5,2,5 in which the P1B-1-ATPases transport Cu+/Ag+,10 the P1B-2-ATPases transport Zn2+/Cd2+/Pb2+,11-14 the P1B-3-ATPases transport Cu2+,15 and the P1B-4-ATPases are activated by Co2+/Ni2+.16,17
The fifth subfamily, the P1B-5-ATPases, is less well characterized and contained only five sequences distinct from all other subfamilies when this classification scheme was established in 2003.5 None of these P1B-5-ATPases were biochemically examined beyond their sequence classification. The sequence motif in TM4 was defined as TPCP, and several conserved oxygen-containing residues in TM5 and TM6 were proposed as possible ligands for an undetermined metal substrate. In 2010, bioinformatic analysis identified 200 additional P1B-5-ATPases, enabling further definition of the characteristics of the subfamily.18 Membrane topology predictions show that the P1B-5-ATPases possess six or seven TM helices, with a signature sequence of (T/S)PCP in TM4. The final helix TM6 houses a QEXXD motif that is almost 100% conserved and is proposed to contribute key metal binding residues, along with the (T/S)PCP motif.
Interestingly, a C-terminal soluble domain was also identified in ~20% of the P1B-5-ATPase sequences. This domain has sequence similarity to hemerythrin (Hr) proteins, a group of soluble proteins that can reversibly bind oxygen utilizing a diiron active site.18 Sequence alignments indicated that these P1B-5-ATPase Hr domains (P1B-5-Hr) do not have the conserved metal binding residues found in eukaryotic and prokaryotic Hr proteins. In structurally characterized eukaryotic Hr proteins, the two iron ions are coordinated by three histidines and bridged by an aspartic acid and a glutamic acid.19 Recent structural characterization of prokaryotic Hr domains shows that the bridging amino acids are conserved, but only two histidines coordinate the diiron site.20 In P1B-5 Hr domains, the missing histidine is replaced by a glutamic acid and the bridging amino acids can be switched, leading to the consensus motif HHX3(D/E)-HXE-HX4(E/D).18 The presence of this Hr domain in some P1B-5-ATPases suggests that they could be involved in iron regulation or O2 sensing coupled to transport of iron or another metal such as Ni2+.18
Biochemical studies of an intact P1B-5-ATPase could provide insight into these proposed functions. Towards this goal, the Sinorhizobium meliloti Sma1163 gene, encoding for a P1B-5-ATPase with a C-terminal Hr domain, was studied. S. meliloti is an aerobic microorganism that has to adapt from 20% O2 environments when free-living to 1% O2 when fixing nitrogen in its host legume root nodules.21 In this situation, an oxygen sensing metal transporter might be advantageous. Moreover, the microaerobic environment in the nodule coincides with a need for intense metal transport to synthesize the key enzymes of symbiotic nitrogen fixation. Our results indicate that Sma1163 is induced by different transition metals, but the highest expression levels are achieved in the nodule under microaerobic conditions. Mutation of Sma1163 results in defects in iron and nickel efflux capabilities. These results suggest a role in Ni2+ and Fe2+ detoxification and led to naming Sma1163 as Nia (nickel/iron-ATPase). Biochemical characterization of recombinant purified Nia constructs, including wild type protein (Nia), protein with the C-terminal Hr domain deleted (ΔHr-Nia), and the isolated C-terminal Hr domain (Nia-Hr) were carried out in an attempt to establish the metal specificity of the P1B-5-ATPase subfamily. The combined results of these studies suggest that Ni2+ and Fe2+ are substrates of Nia.
Materials and methods
S. meliloti strains and growth conditions
Wild type and nia (Sma1163::miniTn5) S. meliloti 2011 strains were obtained from Dr. Anke Becker (University of Bielefeld, Germany). Strains were grown in complex tryptone yeast extract (TY) medium22 at 30 °C supplemented with 50 μg/mL kanamycin. Medicago truncatula was infected with S. meliloti following standard protocols.23 The day of first nodule and the number of nodules at 10 and 21 days post infection (dpi) were monitored.
Metal sensitivity and accumulation tests
A 36 h old culture was used to inoculate fresh TY cultures at OD600 0.1. These were supplemented with the desired metal concentrations as indicated. The cells were grown for approximately 36 h and turbidity at 600 nm was measured. Metal concentrations were determined by Furnace Atomic Absorption Spectroscopy (Varian SpectrAA 880/GTA 100). Briefly, cells were grown in TY medium supplemented or not with 1 mM NiSO4 or 0.5 mM FeCl2 for 36 h and pellets mineralized with HNO3 for 1 h at 80 °C and incubated overnight at room temperature. Protein concentrations were determined by Bradford’s method.24
RNA extraction and qPCR
RNA from free-living S. meliloti was isolated from 5 mL liquid TY cultures, grown till late exponential phase at 30°C in either 20 or 1 % O2 and supplemented when indicated with 0.3 mM CoCl2, 2 mM CuCl2, 1.25 mM NiSO4 or 0.5 mM FeCl2. RNA was stabilized with RNA Protect Bacteria reagent (Qiagen, Valencia, CA) and isolated with RNeasy Minikit (Qiagen). Nodule RNA was obtained from 21 dpi nodules using Trizol (Invitrogen, Carlsbad, CA). In all cases, DNase treatments were carried out until no contaminant DNA was detected by PCR on the RNA extraction using S. meliloti 16S primers (Table 1). cDNA was obtained from 1 μg of DNA-free RNA using SuperScript III reverse transcriptase (Invitrogen).
Table 1.
Summary of primers
| Name | Use | Sequence |
|---|---|---|
| 5Sma1163-1166 | Mapping the Nia polycistron | AGGTCCACAGCCTCACCTCC |
| 3Sma1163-1166 | Mapping the Nia polycistron | CCTGCACCGTGGATGGACAG |
| 5Sma1166-1168 | Mapping the Nia polycistron | AGCCATATTCGCCGCCAGGAGCA |
| 3Sma1166-1168 | Mapping the Nia polycistron | TCCTCTCGGGCCTGCTCCAGG |
| Sma1163 qF | qPCR of Nia | GACCTCCCGGAAGACGAGGT |
| Sma1163 qR | qPCR of Nia | CTCATCCCGGCAAAAGCATC |
| 16S-F | qPCR of 16S rRNA | GATAAGCCGAGAGGAAGGTG |
| 16S-R | qPCR of 16S rRNA | GTGTAGCCCAGCCCGTAAG |
| 5Sma1163 | Cloning of Nia and ΔHr-Nia in pBAD-TOPO |
ATGCGGGATATTGCGCAGA |
| 3Sma1163 | Cloning of Nia in pBAD- TOPO |
CTCGTGCTCGAGCGATCGAT |
| 3ΔHrSma1163 | Cloning of ΔHr-Nia in pBAD- TOPO |
CGCATGCTCCAAACGTGTTGTTTT |
| 5Nia-Hr | Cloning of Nia-Hr in pPR- IBA1 |
ATGGTAGGTCTCAAATGGGCGATCCCATGCG CGGCTG |
| 3Nia-Hr | Cloning of Nia-Hr in pPR- IBA1 |
ATGGTAGGTCTCAGCGCTCTCGTGCTCGAGC GATCGATAG |
Gene expression was studied by quantitative real-time RT-PCR (iCycler iQ, BioRad, Hercules, CA) with primers Sma1163qF and Sma1163qR (Table 1) and standardized with the 16S primers. Real time cycler conditions have been described previously.25 Determinations were carried out with RNA extracted from three independent biological samples, with the threshold cycle (Ct) determined in triplicate. The relative levels of transcription were calculated by using the 2−ΔΔct method.26 In all RT-PCR reactions, a non-RT control was used to detect any possible DNA contamination.
Cloning of Nia constructs
Nia (Sma1163) and ΔHr-Nia cDNA were amplified with the primers listed in Table 1 and cloned in pBAD-TOPO (Invitrogen) to add a C-terminal 6xHis tag. Both plasmids were transformed into E. coli Top10 chemically competent cells and spread on Luria-Bertani (LB) agar plates containing 100 μg/mL ampicillin. The C-terminal domain of Nia (amino acids 645-779) was determined by modeling using TMHMM 2.027 to identify the last amino acid of the last TM segment. The remaining residues were then aligned with the sequence of DcrH-Hr.20,28 This Nia-Hr domain was amplified from pBAD-TOPO::Nia using the primers listed in Table 1, introducing BsaI sites. BsaI digested PCR product was cloned in pPR-IBA1 (IBA) to create constructs containing an eight-residue streptactin tag (SAWSHPQFEK) fused to the C-terminus of the Nia-Hr domain. The vector was transformed into E. coli 10G chemically competent cells (Lucigen) and spread on LB agar plates containing 100 μg/mL ampicillin. The plasmid was then transformed into BL21 Star competent cells (Invitrogen). Gene sequences were confirmed by DNA sequencing (ACGT Inc.).
Protein expression and purification of Nia and ΔHr-Nia
Overnight cultures were used to inoculate 1 L LB media supplemented with100 μg/ml ampicillin and grown at 37 °C. Protein expression was induced by addition of 0.02% (w/v) L-arabinose at an OD600 ≈ 0.6. Cells were harvested 3 h post-induction by centrifugation at 4800 g for 10 min at 4 °C. They were then resuspended in a buffer containing 25 mM Tris, pH 7.0, 100 mM sucrose, and 1 mM PMSF and stored at −80 °C.
All purification procedures were carried out at 4 °C. Thawed cells were lysed by three passes through a pneumatic M-110L microfluidizer processor (Microfluidics). Lysed cells were centrifuged at 8000 g for 30 min. The supernatant was then ultracentrifuged at 163000 g for 1 h. Pelleted membranes were resuspended in fresh buffer (25 mM Tris, pH 7.0, 100 mM sucrose, 1 mM PMSF) and centrifuged at 163000 g for 45 min. The pelleted washed membranes were then resuspended and homogenized in 25 mM Tris, pH 8.0, 100 mM sucrose, 500 mM NaCl, 1 mM PMSF. Protein concentration in the membrane fractions was determined by the detergent-compatible Lowry assay (Bio-Rad) and adjusted to 3 mg/mL. Protein solubilization was performed by dropwise addition of n-dodecyl-β-D-maltopyranoside (DDM) to 1%, followed by incubation at 4 °C with gentle stirring for 1 h. Insoluble material was removed by ultracentrifugation at 163000 g for 45 min.
The supernatant was then applied to a Ni2+-loaded HiTrap Chelating HP column (GE Healthcare) and washed with four column volumes of 25 mM Tris pH 8.0, 100 mM sucrose, 500 mM NaCl, 0.05% DDM (buffer A) followed by a wash of six column volumes of buffer A with 50 mM imidazole. Pure proteins were eluted with 150 mM imidazole in buffer A. Fractions containing pure protein were pooled, concentrated and exchanged into 25 mM Tris pH 8.0, 100 mM sucrose, 50 mM NaCl, 1 mM DTT, 0.01% DDM by filtration through 50 kD cut-off Amicon spin concentrators. Protein concentrations were determined by the Pierce 660 nm protein assay and purity was evaluated using SDS-PAGE.
Expression and purification of Nia-Hr
LB media supplemented with 100 μg/mL ampicillin was inoculated with an overnight cell culture and incubated at 37 °C. When cells reached an OD600 ≈ 0.6, the cultures were moved to 4 °C for ~2 h. Following cold shock, protein expression was induced by addition of 1 mM IPTG and cells were grown overnight (16-18 h) at 18 °C. This procedure (cold shock/overnight induction) reduced inclusion body formation. Cells were harvested by centrifugation at 4800 g for 10 min at 4 °C, resuspended in 50 mM Tris, pH 7.5, 200 mM NaCl, 500 mM sucrose, 1 mM PMSF and stored at −80 °C.
All purification steps were carried out at 4 °C. Thawed cells were lysed by sonication for 12 min using 15 s pulses with 30 s rest period between each pulse and then ultracentrifuged at 163000 g for 45 min. The supernatant was applied to a Tricorn column packed with Strep-tactin Superflow Plus resin (Qiagen) using an ÄKTA chromatography system. The column was washed with 5 column volumes of 50 mM Tris, pH 7.5, 200 mM NaCl, 10% glycerol (wash buffer). Pure protein was eluted with 2 column volumes of wash buffer supplemented with 2.5 mM desthiobiotin. Fractions containing pure protein were concentrated using 10 kDa Amicon spin concentrators. Protein concentration was determined as described above, and purity was evaluated using SDS-PAGE.
Metal Loading
Metal loading was performed by the anaerobic addition of 10 molar equivalents of an anaerobic stock solution of 10 mM Fe(NH4)2(SO4)•6H2O stabilized with 5 mN H2SO4. The protein was then exchanged anaerobically into fresh degassed buffer using a 10DG desalting column (Bio-Rad). All anaerobic procedures were carried out in a Coy anaerobic chamber. Nickel loading was performed by aerobic addition of ten molar equivalents of a stock solution of 1 mM NiCl2 to the protein solution followed by the same desalting procedure. In the case of Nia-Hr, apo samples were prepared by the addition of 10 molar equivalents of desferrioxamine (Sigma Aldrich) to the as isolated Nia-Hr. Samples were incubated for 1 h and passed through a 10DG desalting column (Bio-Rad). Two molar equivalents of Fe(NH4)2(SO4)2 or NiCl2 were added to apo Nia-Hr, followed by desalting.
Samples for metal content determinations were prepared by digestion of protein in 5 mL of 5% TraceSelect nitric acid (Sigma Aldrich) in Chelex-treated water followed by filtration through a 22 μm sterile filter. Iron standards for calibration (Sigma Aldrich) were prepared in 5% nitric acid as well. Protein concentrations were measured by the Pierce 660 nm protein assay. Metal concentrations were measured by inductively coupled plasma optical emission spectroscopy (ICP-OES) (Varian Vista MPX) at the Integrated Molecular Structure Education and Research Center (IMSERC) of Northwestern University. Reported ICP-OES results are the average of three replicate experiments performed for at least two independent protein sample preparations (Table 2).
Table 2.
Metal binding properties of Nia and Nia-derived constructsa
| Protein | Fe/monomer | Ni/monomer |
|---|---|---|
| Nia | 5.6 ± 0.8 a | 1.12 ± 0.02 |
| ΔHr-Nia | 3.7 ± 0.2 | 0.46 ± 0.02 |
| Nia-Hr | 2.2 ± 0.1 | 1.11 ± 0.02 |
Data are the average of at least three independent experiments
Optical spectroscopy of Nia-Hr
The UV-visible spectra of Nia-Hr were recorded using a Perkin Elmer LAMBDA 1050 spectrophotometer at room temperature. To produce the azide adduct, 50 molar equivalents of NaN3 were added from a 100 mM stock solution and incubated for 30 min prior to spectroscopic measurements. Reduced samples were obtained by adding 10 molar equivalents of NaS2O4 from a 100 mM stock solution and gently mixing for 30 min before spectroscopic measurements.
X-ray absorption spectroscopy (XAS)
Samples for XAS were prepared in 25 mM Tris, pH 7.5, 100 mM NaCl, 40% glycerol. Iron concentrations were in the 1-1.5 mM range. Two independent replicates of iron-loaded Nia-Hr and Nia-Hr azide adduct were prepared, loaded into Lucite cells wrapped with Kapton tape, and frozen in liquid nitrogen. XAS data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) on beamline 7-3, equipped with a single rhodium coated silicon mirror and a Si[220] double crystal monochromator detuned 50% for harmonic rejection. Samples were maintained at 10K using an Oxford Instruments continuous-flow liquid helium cryostat. Protein fluorescence excitation spectra were collected using a 30-element Ge solid-state array detector. An iron foil absorption spectrum was simultaneously collected with the protein sample for calibrating X-ray energies. XAS spectra were measured as described previously.29 Data were processed using the Macintosh OS X version of the EXAFSPAK program suite integrated with the Feff version 7.2 software for theoretical model generation.30,31 XAS data reduction and analysis were performed following previously reported protocols.32 Extended X-ray absorption fine structure (EXAFS) data were simulated over a k range of 1 to 14.25 Å−1 for a spectral resolution of 0.12 Å. Data were fit using a scale factor of 0.95 and Eo values for Fe-O/N/C and Fe-Fe interactions of −10 and −15, respectively. Simulation parameters for fitting the raw unfiltered data are given along with the number of degrees of freedom weighted simulation “goodness of fit” (F’) parameter in Table 3.
Table 3.
Summary of EXAFS simulation analysisa
| Fe-nearest neighbor ligandsb |
Fe-long-range ligandsb |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| sample | atomc | R (Å)d | CNe | σ 2f | atomc | R (Å)d | CNe | σ 2f | F’g |
| Native | O/N | 1.96 | 2.5 | 4.77 | C | 3.11 | 3.0 | 4.64 | 0.84 |
| O/N | 2.08 | 2.0 | 4.30 | Fe | 3.43 | 0.5 | 2.68 | ||
| C | 4.01 | 2.0 | 1.55 | ||||||
| Azide | O/N | 1.96 | 2.5 | 5.12 | C | 3.10 | 3.0 | 4.37 | 0.37 |
| O/N | 2.08 | 2.5 | 5.70 | Fe | 3.44 | 0.5 | 3.63 | ||
| C | 4.07 | 2.0 | 3.08 | ||||||
Values given in table represent the best-fit simulation parameters.
Independent metal-ligand scattering environment.
Scattering atoms: O(oxygen), N(nitrogen) and C(carbon).
Metal-ligand bond length (all standard deviations < 0.03 Å).
Metal-ligand coordination number (all standard deviations < 1.0).
Debye-Waller factor given in Å2 × 103 (all standard deviations < 0.9 Å).
Number of degrees of freedom weighted mean square deviation between empirical and theoretical data.
Para-nitrophenylphosphatase (pNPPase) activity assay
In the pNPPase activity assay, enzyme-mediated hydrolysis of pNPP to p-nitrophenol is determined by monitoring the increase of the p-nitrophenol absorption peak at 410 nm.33-35 The pNPPase assay was performed in a 1 mL solution containing 20-100 μg of a single purified P1B-5-ATPase construct, 50 mM Tris pH 7.5, 50 mM NaCl, 5 mM MgSO4, 0.01% DDM, 0.01% asolectin, and 60 mM pNPP. Negative controls with identical components but no protein were also prepared, as well as samples containing 100 μM metal salts (CoCl2, NiCl2, Fe(NH4)2(SO4)). The reaction solution was incubated at 37 °C for 10 min, at which point a 200 μL aliquot of the reaction was removed and added to stopping solution of 800 μL 0.5 M NaOH and mixed. The absorbance of p-nitrophenol in the mixture was measured at 410 nm, and the concentration was calculated using a molar extinction coefficient at 410 nm of 17000 M−1 cm−1.33 All samples were tested in duplicate.
Results and discussion
Nia is required for Fe2+ and Ni2+ detoxification
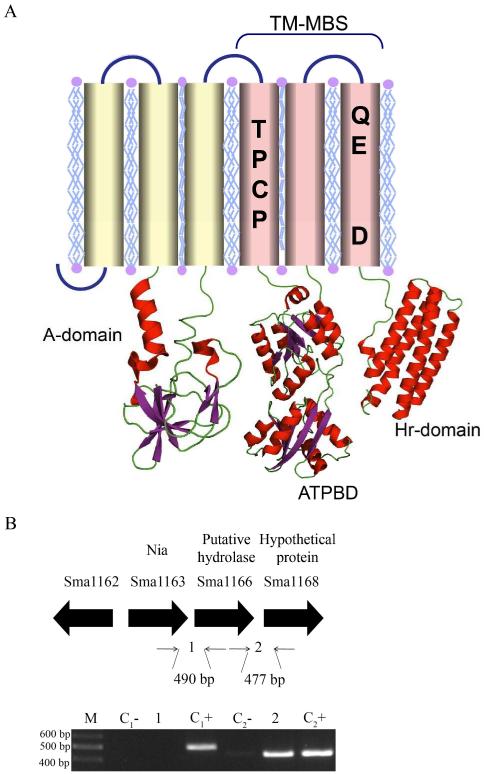
Nia is a putative P1B-5-ATPase with six predicted TM helices and signature motifs in TM 4-6 (Fig. 1A). It is encoded by the gene Sma1163 present in the symbiotic plasmid A of S. meliloti. Bioinformatics analysis of its genetic environment indicates that Sma1163 might be part of the same operon as Sma1166 (putative hydrolase) and Sma1168 (hypothetical protein). However, RT-PCR studies indicate that it is an operon formed by just Sma1163 with no apparent neighboring regulator (Fig. 1B).
Fig. 1.
Nia structure and genetic enviroment. (A) Topology diagram with conserved amino acids in H4 and H6 indicated. (B) Genetic environment of Nia (Sma1163) cistron and determination of its constituents by RT-PCR of the intergenic sequences (1-2). Putative function is indicated above the accession number. Cx-indicates the PCR of the X intergenic sequence using as template RNA without being retrotranscribed (negative control), while the PCR of the Cx+ lane used genomic DNA as template (positive control).
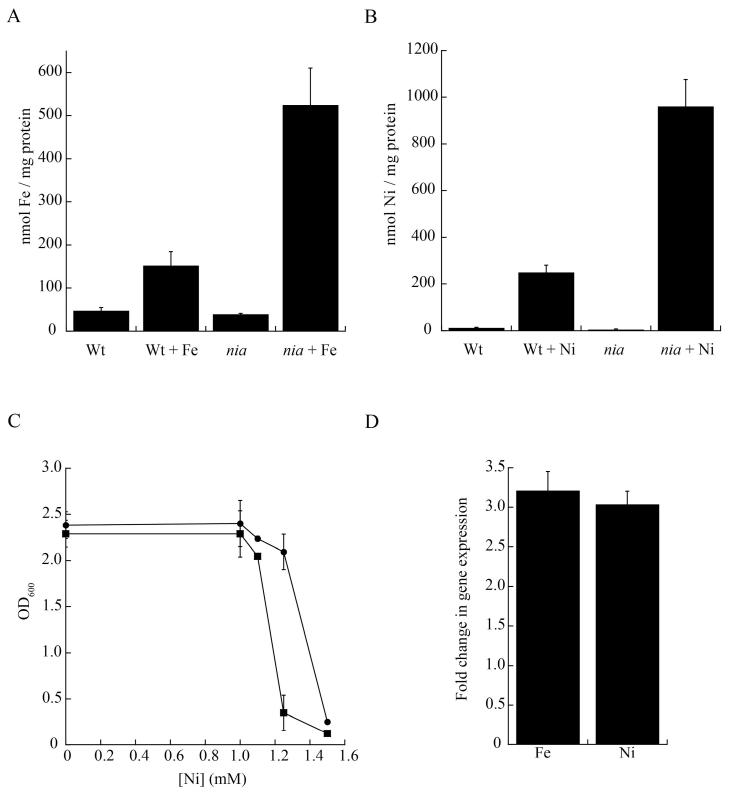
Bacterial P1B-type ATPases have been frequently associated with metal detoxification and tolerance.3,25 To test this possibility, wild type and nia mutant S. meliloti strains were grown in TY supplemented with different concentrations of metals. Mutant strains appear to accumulate Fe and Ni2+ (Fig. 2A, B), and no significant accumulation of other metals was observed (Supplemental Tables S1-S3). Since these data indicate a role for Nia in the detoxification of these metals, it might be expected that Ni2+ or Fe2+ would hamper bacterial proliferation. Growth curves were obtained from wild type and nia strains grown in TY supplemented with increasing concentrations of Ni2+ and Fe2+. The results indicate a higher sensitivity to Ni2+ in mutant strains (Fig. 2C), with very little or no increased toxicity to Fe2+ (data not shown). The latter could be the result of efficient intracellular Fe-chelation machinery, and could also be related to the fact that no concentrations higher than 1 mM FeCl2 could be tested due to metal precipitation. The presence of high levels of other metals (Cu2+, Zn2+) in the culture media did not affect the mutant strain growth (data not shown). Further supporting the idea of Nia playing a role in the transport of Fe2+ and Ni2+, the expression of nia was induced by both metal ions (Fig. 2D), whereas others had no effect on nia transcription (2.03 +/−0.27 induction fold by Co2+; 1.25 +/−0.22 induction fold by Cu2+).
Fig. 2.
Comparison of wild type (WT) and nia mutant S. meliloti strains. (A) Iron contents when grown in TY and in TY medium supplemented with 0.5 mM FeCl2. (B) Nickel contents of wild when grown in TY and in TY medium supplemented with 1.0 mM NiSO4. (C) Effect of increasing concentrations of NiSO4 on the growth of Wt (•) and nia (■) strains. (D) Iron and nickel-dependent regulation of Nia expression. Gene expression levels were determined by qPCR on RNA extracted from cultures grown in the presence of either 0.5 mM FeCl2 or 1 mM NiSO4, standardized to the expression levels of 16S rRNA, and referred to the expression in unamended TY medium. Data are the mean ± SE of three independent experiments.
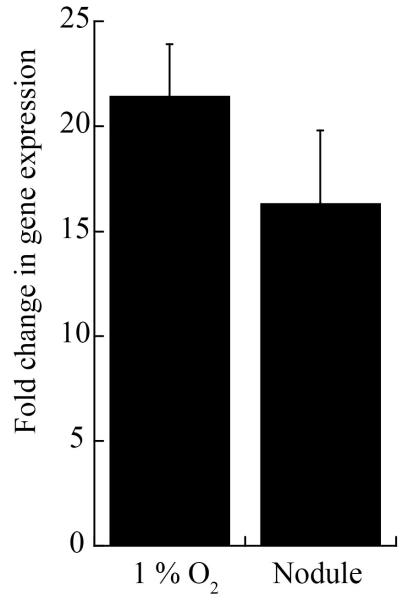
The fact that Nia is encoded by one of the two symbiotic plasmids in S. meliloti suggests a possible role in the symbiosis with legumes. In support of this notion, mutation in the symbiotic regulator FixJ affects Nia expression.36 Real time qPCR studies of Nia induction were carried out using RNA obtained from nodules or from free-living bacteria grown in 1% O2 (Fig. 3). The results show a 20-fold induction in both cases compared to free bacteria living in 20% O2, and hint at a role in symbiotic nitrogen fixation. However, M. truncatula plants inoculated with either wild type or nia strains displayed no change in nodule number, nodulation kinetics or in shoot or root dry weight (data not shown). This could be due to some other gene complementing nia function or to a plant-specific effect in which a phenotype may be manifested in some hosts but not in others.
Fig. 3.
Nia induction in microaerobic and symbiotic conditions. Gene expression levels were determined by qPCR on RNA extracted from cultures grown in 1 % O2 or from M. truncatula root nodules. Expression levels were standardized to the expression levels of 16S rRNA and referred to the expression in bacteria grown in TY medium in aerobic conditions (aprox. 20 % O2). Data are the mean ± SE of three independent experiments.
Nia is a putative Ni2+/Fe2+ transporter
Three Nia constructs were expressed in E. coli and purified: Nia (the full-length protein), ΔHr-Nia (a truncated protein lacking the C-terminal Hr domain), and Nia-Hr (the C-terminal Hr domain) (Fig. S1). The isolated transmembrane constructs showed no detectable levels of Fe2+/3+, Ni2+, Co2+, Cu+/2+ and Zn2+, whereas the purified Nia-Hr contained 0-1.5 Fe/monomer. Incubation with excess Fe2+ under anaerobic conditions resulted in 5 Fe/monomer for Nia, 3-4 for ΔHr-Nia, and 2 for Nia-Hr (Table 2). These values suggest that the transmembrane region of Nia is capable of binding between 2 and 3 iron ions. Given that the highest metal stoichiometry observed in the P1B class is 2 metal ions/transmembrane sites, it is more likely that 2 iron ions bind to the Nia transmembrane sites. Any excess iron may be bound to the (His)6 tag in Nia and ΔHr-Nia. Interestingly, stoichiometric loading of the transmembrane regions could only be achieved if a 10-fold molar excess was added to the mix. This observation suggests a low iron affinity, which would be consistent with the relatively high iron concentrations present in the nodule, where the expression of Nia is the highest (Fig. 3). Furthermore, attempts at iron loading under ambient aerobic conditions were unsuccessful, with no iron observed, which would be consistent with iron loading in the microaerobic conditions of the nodule.
Nickel binding was also tested (Table 2). Strep-tagged Nia-Hr binds one Ni/monomer, and a similar stoichiometry was observed for Nia. ΔHr-Nia binds less than 50% of the Ni2+ bound to the other two proteins, which is likely the result of lacking the C-terminal Hr domain. Thus, the transmembrane region cannot be readily loaded with Ni2+, and the observed binding may be attributable to the (His)6 tag in Nia and ΔHr-Nia. It is also possible that ATP is required for high affinity Ni2+ binding.
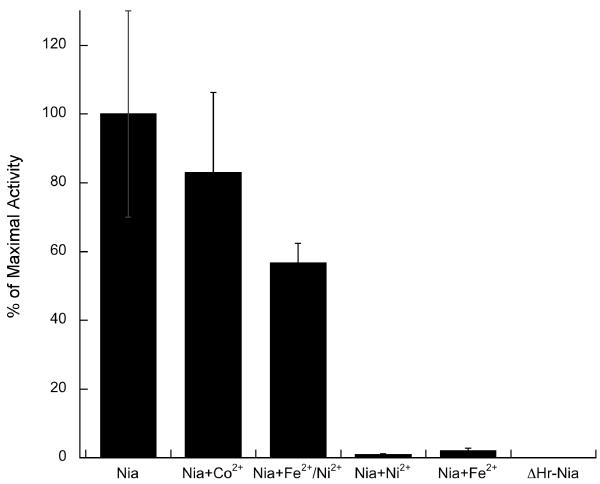
Determinations of metal-dependent ATPase activity were unsuccessful, perhaps due to a very slow turnover of this enzyme or the need for unknown cofactors in the media. Alternatively, the interaction of metals with the active enzyme can be assessed by measuring pNPPase activity. This activity is observed for P-type ATPases in the E2 form and is independent of phosphoprotein formation (Fig. S2). However, metal binding drives the transition of the enzyme to the E1 form, and the effect is manifested in the inhibition of pNPPase activity in the presence of a metal substrate.37-39 Thus, this assay is a useful indicator of metal specificity. The pNPPase activity of freshly purified Nia is ~4 nmol Pi mg−1 min−1, indicating that the enzyme is properly folded (Fig. 4).33 However, deletion of the C-terminal Hr domain abolishes the activity, suggesting that this domain may interact with ATP-binding domain and affect hydrolysis. Alternatively, its removal might adversely affect the enzyme structure. The effects of Fe2+ and Ni2+ on pNPPase were then measured. Consistent with Fe2+ and Ni2+ acting as substrates of Nia, both metals inhibited pNPPase activity (Fig. 4). This inhibition appears to be specific, and not associated with the metals binding to the (His)6 tag, since Co2+ has no effect.
Fig. 4.
Effect of metal ions on pNPPase activity. Maximal activity for Nia is 1.74 nmol Pi mg−1 min−1. Data are the mean ± SE of three independent experiments.
Very little is known about iron and nickel homeostasis in S. meliloti. In free-living conditions, it has been suggested that iron, and perhaps nickel, are acquired via siderophores, such as rhizobactins,40 or by other complexes such as heme or citrate.41,42 At least some of these iron complexes cross the outer membrane through TonB-dependent transporters43 Some of the ABC, Nramp or ZIP transporters encoded in the S. meliloti genome could be involved in iron translocation across the inner membrane, although no experimental evidence is available. Nickel transport could proceed similarly, although specific nickel importers such as NiCoT or HupE homologues, have been not identified in S. meliloti genome in contrast to other rhizobia.44,45 One possible explanation for the lack of importers is that nickel demands in S. meliloti are probably reduced in comparison to other rhizobia since it does not synthesize hydrogenase, and consequently does not need this element as a cofactor. Interestingly, most of the genes involved in iron uptake in free-living conditions are down-regulated by nodulation,46 indicating that iron levels within the nodule are much higher than in free-living conditions and that iron is incorporated through nodule-specific mechanisms. This local accumulation of relatively high levels of iron has been recently illustrated by synchrotron-based X-ray fluorescence47 and explains why detoxifying mechanisms are necessary to deal with the unusual accumulation of this metal.
Nia C-terminal Hr domain
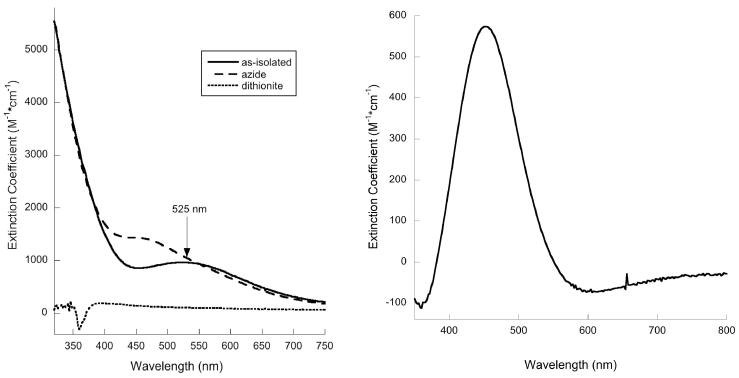
Purified and iron-loaded forms of Nia-Hr are purple in color. This domain has spectroscopic characteristics similar to those of the previously studied P1B-5-ATPase Hr domain from A. cellulolyticus.18 The optical spectrum of iron-loaded Nia-Hr exhibits a ligand-to-metal charge transfer (LMCT) band at ~525 nm (ε ~ 1000 M−1cm−1) (Fig. 5A). This feature is consistent with the oxy form of Hr, which contains two Fe3+ ions and a terminally bound hydroperoxide ligand.28,48,49 Addition of azide results in the appearance of new feature at ~450 nm (Fig. 5B). The azide adduct is an analogue state for the met form of Hr proteins in which the terminal hydroxyl group of the met form is replaced by azide. The 450 nm feature is consistent with the azide adduct features at 453 nm observed for A. cellulolyticus P1B-5-Hr,18 Methylococcus capsulatus (Bath) Hr,49 and DcrH-Hr.28 Addition of dithionite bleaches the color and abolishes the peak at 525 nm, but upon exposure for longer than 30 min to air, the color and oxy spectral feature are restored. The color and spectral feature persist for up to two weeks, indicating an unusually stable oxy state with a very long half life for autooxidation. Generally, prokaryotic Hr proteins exhibit rapid autooxidation, with t1/2 < 1 min reported for both A. cellulolyticus P1B-5-Hr18 and DcrH-Hr,28 and t1/2 ~ 50 min for M. capsulatus (Bath) Hr.49 Nia-Hr appears to behave more like eukaryotic Hrs with t1/2 ~ 20 h.50
Fig. 5.
Optical spectra of Nia-Hr. (A) Spectra of as-isolated, azide-treated, and dithionite-reduced samples in 25 mM Tris pH 7.5, 100 mM NaCl. The dithionite-reduced data were background-corrected to account for the high absorbance of dithionite. (B) Difference spectrum of as-isolated Nia-Hr subtracted from azide-treated Nia-Hr.
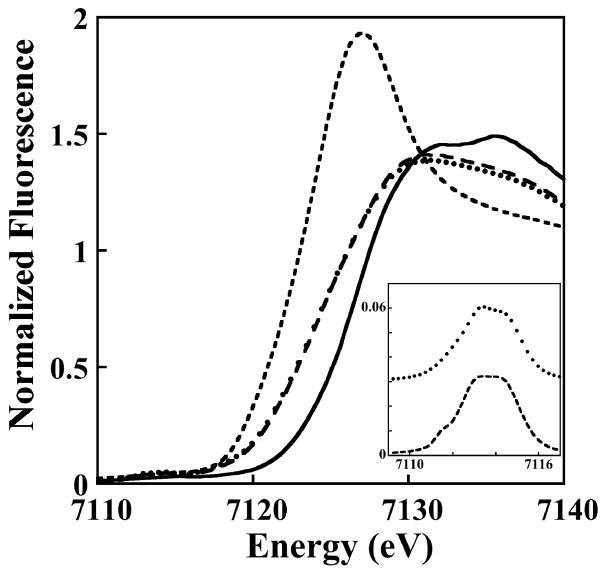
Metal binding to Nia-Hr was also investigated. Nia-Hr is capable of binding one Ni/monomer and 2 Fe/monomer. A previous study of Hr domains of P1B-5-ATPases indicates that the domain is able to adventitiously bind a variety of metals, but only Fe is bound in a stoichiometric and biologically relevant manner.18 Accordingly, we examined the Fe-bound form of Nia-Hr by XAS. These data are consistent with the presence of a diiron center in iron loaded and azide treated Nia-Hr. The X-ray absorption near edge spectra (XANES) for the iron loaded and azide treated samples are similar and indicate a stable mixture of Fe2+ and Fe3+ (Fig. 6). First inflection point energies obtained from the protein XANES spectra (7124.2 eV for iron loaded and 7124.4 eV for azide-treated) when compared with controls indicate 59% (± 5%) Fe2+ and 41% (±5%) Fe3+ for the native sample and 54% (±5%) Fe2+ and 46% (±5%) Fe3+ for the azide treated sample. The small pre-edge 1s to 3d spectral feature in both samples is consistent with high-spin iron species with octahedral metal-ligand coordination geometries.
Fig. 6.
Normalized XANES spectra of iron-loaded (long dashed line) and azide-treated Nia-Hr (dotted line), compared with the model control spectra for FeSO4 (short dashed line) and Fe2(SO4)3 (solid line). Inset: Expansion of the background subtracted 1s→3d region of the XANES spectra with the azide-treated sample’s pre-edge spectrum (dotted line) offset by 0.03 units for clarity.
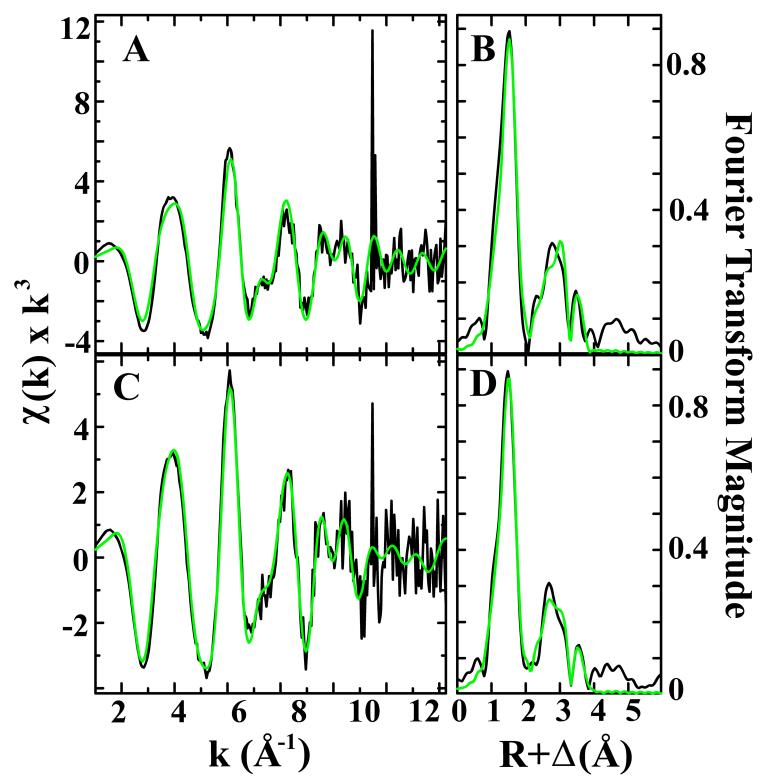
The EXAFS data analysis for both protein samples confirms that the best-fit simulation arises from fitting multiple independent ligand environments to each sample (Fig. 7, Table 3). The nearest neighbor metal-ligand scattering environment is constructed of two O/N ligand environments at 1.95 Å and 2.07 Å for both samples. Bond lengths obtained from simulating the nearest-neighbor ligand environments in both spectra are consistent with five to six coordinate Fe-ligand coordination environments; the low apparent coordination number values from the best-fit simulations are indicative of a semi-disordered ligand environment in the first coordination sphere in both cases. The O/N ligands observed at 1.95 Å would be consistent with iron bound to mono or bidentate carboxylate ligands.51 The longer O/N bond length at 2.07 Å would be consistent with Fe-imidazole nitrogen scattering from histidine residues or with coordinated water.
Fig. 7.
EXAFS and Fourier transforms of Nia-Hr samples. The EXAFS spectra are shown in black for the iron-loaded (A) and azide-treated (C), with corresponding Fourier transforms in (B) and (D) respectively. Simulations for EXAFS and FT data are shown in green.
A predominant Fe-Fe signal is observed in both samples at 3.43-3.44 Å. This Fe-Fe distance is slightly longer than the 3.39 Å distance observed for both the met DcrH-Hr and for P1B-5-ATPase Hr domain from Acidothermus cellulolyticus, and is substantially longer than the 3.25 Å distance typical of invertebrate Hr.18,19,52 The increase in Fe-Fe distance is consistent with the presence of a partly reduced diiron center.20,53 The similar Fe-Fe distances for the azide-treated and iron-loaded samples suggest that the binding of azide does not affect the dinuclear iron core structure in any appreciable manner. Long-range carbon scattering environments at ca. 3.1 and 4.01 Å (4.07 Å for azide-treated) are observed in both samples. Carbon scattering at these bond distances is typical of carboxylate carbon or imidazole carbon/nitrogen scattering from Asp, Glu and His residues coordinated to the metal.54
A variety of biological functions have been associated with Hr domains. Marine invertebrates use Hr proteins to reversibly bind O2 for transport and storage.55 Prokaryotic Hr proteins seem to have a more diverse set of functions, including O2 transport for M. capsulatus (Bath) Hr,49,56 O2 sensing for chemotaxis by DcrH-Hr,28, and iron binding for storage and transport for uncharacterized Hr-like proteins.57 The previous study of A. cellulolyticus P1B-5-Hr suggested that the domain might sense O2 and regulate metal transport. However, the pronounced difference in stabilities of the oxy states of A. cellulolyticus P1B-5-Hr and Nia-Hr suggests that the domain does not function as an O2 sensor for all P1B-5-ATPases. Instead, the Hr-like domain could serve as an iron sensor, analogous to soluble metal binding domains in other P1B-ATPases that play regulatory roles via metal-dependent protein-protein interactions. For example, in the P1B-1-ATPases, Cu+ binding to the N-terminal metal binding domain precludes its interaction with the ATPBD.58,59 Iron binding by Hr domains may similarly affect interdomain interactions. It may be that the massive metal delivery to a maturing nodule47 causes local accumulation of iron and other elements that would have to be detoxified. Nia, which is expressed in the nodule, could detect toxic iron levels with its Hr domain, and activate its efflux.
Iron binding and/or O2 sensing by the Hr domain may also regulate the transport of another metal ion, mostly likely Ni. Previous studies of the Hr domain from a P1B-5-ATPase from A. cellulolyticus suggest interplay between P1B-5-ATPases and nickel-containing superoxide dismutase and NiFe hydrogenase.18 In this case, O2 sensing by the Hr domain could regulate the Ni transport function of the P1B-5-ATPase, thus maintaining appropriate Ni2+ concentrations.
Conclusions
Rhizobia in general and S. meliloti in particular have to thrive in a number of completely different environments, from aerobic soils where local distributions of transition metals can change significantly to a nodular microaerobic environment in which high levels of free-radical forming metals can be present. As a result, versatile transport systems must be in place to ensure cell viability. Nia appears to function in Fe2+ and Ni2+ detoxification both in free-living and in symbiotic conditions. Within the nodule, Nia function likely ensures that the massive iron transport to the developing symbiosome does not reach toxic levels killing the bacteria. This is unlike most bacteria that typically face the opposite problem, either too little iron in the environment, or a competition with the host organism for dwindling concentrations of this element. Its C-terminal Hr domain is likely critical and may act as a sensor responsible for detecting deleterious iron concentrations. Future work will be directed to studying the interaction of the C-terminal Hr domain with the other domains of Nia.
Supplementary Material
Acknowledgements
This work was supported by NIH grants (GM58518 to A. C. R. and DK068139 to T. L. S.) and an Agriculture and Food Research Initiative Competitive Grant, from the USDA-National Institute of Food and Agriculture (Grant 2010-65108-20606 to J.M.A.). E. L. Z. was supported in part by NIH grant 5T32 GM008382. M.G.-G. was supported by the Ramón y Cajal contract RYC-2010-06363. We are grateful to Mr. Michael Gauvin for his technical assistance and Dr. Teresita Padilla-Benavidez (WPI)) for helpful discussions. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL). SSRL is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the NIH, National Center for Research Resources, Biomedical Technology Program.
Footnotes
Electronic supplementary information (ESI) available: Figs. S1, S2, Tables S1, S2, S3
Notes and References
- 1.Palmgren MG, Nissen P. Annu. Rev. Biophys. 2011;40:243–266. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- 2.Argüello JM, Eren E, M González-Guerrero. Biometals. 2007;20:233–248. doi: 10.1007/s10534-006-9055-6. [DOI] [PubMed] [Google Scholar]
- 3.Fan B, Rosen BP. J. Biol. Chem. 2002;277:46987–46992. doi: 10.1074/jbc.M208490200. [DOI] [PubMed] [Google Scholar]
- 4.Lutsenko S, Gupta A, Burkhead JL, Zuzel V. Arch. Biochem. Biophys. 2008;476:22–32. doi: 10.1016/j.abb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argüello JM. J. Membr. Biol. 2003;195:93–108. doi: 10.1007/s00232-003-2048-2. [DOI] [PubMed] [Google Scholar]
- 6.Dutta SJ, Liu J, Hou Z, Mitra B. Biochemistry. 2006;45:5923–5931. doi: 10.1021/bi0523456. [DOI] [PubMed] [Google Scholar]
- 7.Forbes JR, Cox DW. Am. J. Hum. Genet. 1998;63:1663–1674. doi: 10.1086/302163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowe J, Vieyra A, Catty P, Guillain F, Mintz E, Cuillel M. J. Biol. Chem. 2004;279:25986–25994. doi: 10.1074/jbc.M308736200. [DOI] [PubMed] [Google Scholar]
- 9.González-Guerrero M, Eren E, Rawat S, Stemmler TL, Arguello JM. J. Biol. Chem. 2008;283:29753–29759. doi: 10.1074/jbc.M803248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandal AK, Cheung WD, Argüello JM. J. Biol. Chem. 2002;277:7201–7208. doi: 10.1074/jbc.M109964200. [DOI] [PubMed] [Google Scholar]
- 11.Sharma R, Rensing C, Rosen BP, Mitra B. J. Biol. Chem. 2000;275:3873–3878. doi: 10.1074/jbc.275.6.3873. [DOI] [PubMed] [Google Scholar]
- 12.Rensing C, Mitra B, Rosen BP. Proc. Natl. Acad. Sci. USA. 1997;94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rensing C, Sun Y, Mitra B, Rosen BP. J. Biol. Chem. 1998;273:32614–32617. doi: 10.1074/jbc.273.49.32614. [DOI] [PubMed] [Google Scholar]
- 14.Eren E, Argüello JM. Plant Physiol. 2004;136:3712–3723. doi: 10.1104/pp.104.046292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mana-Capelli S, Mandal AK, Argüello JM. J. Biol. Chem. 2003;278:40534–40541. doi: 10.1074/jbc.M306907200. [DOI] [PubMed] [Google Scholar]
- 16.Zielazinski EL, Cutsail GE, Hoffman BM, Stemmler TL, Rosenzweig AC. Biochemistry. 2012;51:7891–7900. doi: 10.1021/bi3006708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raimunda D, Long JE, Sassetti CM, Argüello JM. Mol. Microbiol. 2012;84:1139–1149. doi: 10.1111/j.1365-2958.2012.08082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Traverso ME, Subramanian P, Davydov R, Hoffman BM, Stemmler TL, Rosenzweig AC. Biochemistry. 2010;49:7060–7068. doi: 10.1021/bi100866b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenkamp RE. Chem. Rev. 1994;94:715–726. [Google Scholar]
- 20.Isaza CE, Silaghi-Dumitrescu R, Iyer RB, Kurtz DM, Chan MK. Biochemistry. 2006;45:9023–9031. doi: 10.1021/bi0607812. [DOI] [PubMed] [Google Scholar]
- 21.van Rhijn P, Vanderleyden J. Microbiol. Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beringer JE. J. Gen. Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 23.Lagares A, Caetanoanolles G, Niehaus K, Lorenzen J, H.D. L, Puhler A, Favelukes G. J. Bacteriol. 1002;174:5941–5952. doi: 10.1128/jb.174.18.5941-5952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradford MM. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 25.González-Guerrero M, Raimunda D, Cheng X, Argüello JM. Mol. Microbiol. 2010;78:1246–1258. doi: 10.1111/j.1365-2958.2010.07402.x. [DOI] [PubMed] [Google Scholar]
- 26.Livak K, Schmittgen T. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 28.Xiong J, Kurtz DM, Ai J, Sanders-Loehr J. Biochemistry. 2000;39:5117–5125. doi: 10.1021/bi992796o. [DOI] [PubMed] [Google Scholar]
- 29.Cook JD, Bencze KZ, Jankovic AD, Crater AK, Busch CN, Bradley PB, Stemmler AJ, Spaller MR, Stemmler TL. Biochemistry. 2006;45:7767–7777. doi: 10.1021/bi060424r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.George GNG, S. J., Pickering IJ. 2001 [Google Scholar]
- 31.Rehr JJ, Ankudinov AL. J. Synchrotron Radiat. 2001;8:61–65. doi: 10.1107/s0909049500016423. [DOI] [PubMed] [Google Scholar]
- 32.Bencze KZ, Kondapalli KC, Stemmler TL. In: Applications of Physical Methods to Inorganic and Bioinorganic Chemistry. Scott RA, Lukehart CM, editors. John Wiley & Sons, Ltd.; Chichester, UK: 2007. pp. 513–528. [Google Scholar]
- 33.Yang Y, Mandal AK, Bredeston LM, González-Flecha FL, Argüello JM. Biochim. Biophys. Acta. 2007;1768:495–501. doi: 10.1016/j.bbamem.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 34.Campos M, Berberián G, Beaugé L. Biochim. Biophys. Acta, Biomembr. 1988;938:7–16. doi: 10.1016/0005-2736(88)90116-2. [DOI] [PubMed] [Google Scholar]
- 35.Ray TK, Nandi J. Biochem. J. 1986;233:231–238. doi: 10.1042/bj2330231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bobik C, Meilhoc E, Batut J. J. Bacteriol. 2006;188:4890–4902. doi: 10.1128/JB.00251-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrahan PJ, Pouchan MI, Rega AF. J. Physiol. 1969;202:305–327. doi: 10.1113/jphysiol.1969.sp008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glynn IM. In: The enzymes of biological membranes. Matonosi AM, editor. Vol. 3. Plenum Press; New York: 1985. pp. 35–114. [Google Scholar]
- 39.Glynn IM, Karlish SJD. Annu. Rev. Physiol. 1975;37:13–55. doi: 10.1146/annurev.ph.37.030175.000305. [DOI] [PubMed] [Google Scholar]
- 40.O’Brian MR, Fabiano E. In: Iron Uptake and Homeostasis in Microorganisms. Cornelis P, Andrews SC, editors. Caister Academic Press; Wymondham: 2010. pp. 37–63. [Google Scholar]
- 41.Fabiano E, Gill PR, Noya F, Bagnasco P, delaFuente L, Arias A. Symbiosis. 1995;19:197–211. [Google Scholar]
- 42.Guerinot ML, Meidl EJ, Plessner O. J. Bacteriol. 1990;172:3298–3303. doi: 10.1128/jb.172.6.3298-3303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch D, O’Brien J, Welch T, Clarke P, Cuiv PO, Crosa JH, O’Connell M. J. Bacteriol. 2001;183:2576–2585. doi: 10.1128/JB.183.8.2576-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brito B, Prieto RI, Cabrera E, Mandrand-Berthelot MA, Imperial J, Ruiz-Argüeso T, Palacios JM. J. Bacteriol. 2010;192:925–935. doi: 10.1128/JB.01045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu CL, Javedan S, Moshiri F, Maier RJ. Proc. Natl. Acad. Sci. USA. 1994;91:5099–5103. doi: 10.1073/pnas.91.11.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becker A, Bergès H, Krol E, Bruand C, Rüberg S, Capela D, Lauber E, Meilhoc E, Ampe F, de Bruijn FJ, Fourment J, Francez-Charlot A, Kahn D, Küster H, Liebe C, Pühler A, Weidner S, Batut J. MPMI. 2004;17:292–303. doi: 10.1094/MPMI.2004.17.3.292. [DOI] [PubMed] [Google Scholar]
- 47.Rodríguez-Haas B, Finney L, Vogt S, González-Melendi P, Imperial J, González-Guerrero M. Metallomics. 2013;5:1247–1253. doi: 10.1039/c3mt00060e. [DOI] [PubMed] [Google Scholar]
- 48.Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee S-K, Lehnert N, Neese F, Skulan AJ, Yang Y-S, Zhou J. Chem. Rev. 1999;100:235–350. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 49.Kao W-C, Wang VCC, Huang Y-C, Yu SSF, Chang T-C, Chan SI. J. Inorg. Biochem. 2008;102:1607–1614. doi: 10.1016/j.jinorgbio.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 50.Xiong J, Phillips RS, Kurtz DM, Jin S, Ai J, Sanders-Loehr J. Biochemistry. 2000;39:8526–8536. doi: 10.1021/bi9929397. [DOI] [PubMed] [Google Scholar]
- 51.Hikichi S, Ogihara T, Fujisawa K, Kitajima N, Akita M, M.-O. Y. Inorg. Chem. 1997;36:4539–4547. doi: 10.1021/ic960903m. [DOI] [PubMed] [Google Scholar]
- 52.Holmes MA, Stenkamp RE. J. Mol. Biol. 1991;220:723–737. doi: 10.1016/0022-2836(91)90113-k. [DOI] [PubMed] [Google Scholar]
- 53.Holmes MA, Le Trong I, Turley S, Sieker LC, Stenkamp RE. J. Mol. Biol. 1991;218:583–593. doi: 10.1016/0022-2836(91)90703-9. [DOI] [PubMed] [Google Scholar]
- 54.Stemmler TL, Sossong TM, Goldstein JI, Ash DE, Elgren TE, Kurtz DM, Penner-Hahn JE. Biochemistry. 1997;36:9847–9858. doi: 10.1021/bi9702795. [DOI] [PubMed] [Google Scholar]
- 55.Klotz IM, Kurtz DM. Acc. Chem. Res. 1984;17:16–22. [Google Scholar]
- 56.Karlsen OA, Ramsevik L, Bruseth LJ, Larsen Ø, Brenner A, Berven FS, Jensen HB, Lillehaug JR. FEBS J. 2005;272:2428–2440. doi: 10.1111/j.1742-4658.2005.04663.x. [DOI] [PubMed] [Google Scholar]
- 57.French CE, Bell JML, Ward FB. FEMS Microbiol. Lett. 2008;279:131–145. doi: 10.1111/j.1574-6968.2007.01011.x. [DOI] [PubMed] [Google Scholar]
- 58.González-Guerrero M, Hong D, Argüello JM. J. Biol. Chem. 2009;284:20804–20811. doi: 10.1074/jbc.M109.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsivkovskii R, MacArthur BC, Lutsenko S. J. Biol. Chem. 2001;276:2234–2242. doi: 10.1074/jbc.M003238200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.