Abstract
The incidence of tuberculosis (TB) and especially multidrug-resistant TB (MDR) continues to increase alarmingly worldwide, and reliable and fast diagnosis of MDR is essential for the adequate treatment of patients. In contrast to the standard culture methods, nucleid acid amplification tests (NAATs) provide information about presence of Mycobacterium tuberculosis complex (MTBC) DNA and a potential resistance pattern within hours. We analyzed specimens of 110 patients from Nigeria comparing culture-based drug susceptibility testing (DST) to NAAT assays detecting isoniazid (INH), rifampicin (RMP) (GenoType MTBDRplus), and ethambutol (EMB) (GenoType MTBDRsl) resistance. Compared to DST, the GenoType MTBDRplus and MTBDRsl showed a specificity of 100% (86.3–100) and a sensitivity of 86% (42.1–99.6%) for detection of INH and a specificity of 100% (86.3–100) and a sensitivity of 83% (35.9–99.6%) for detection of RMP, and a sensitivity 100% (47.8–100%) for EMB resistance. However, in two strains, the NAAT assays provided false susceptible results as the mutations causing resistance were in genomic regions not covered by the probes of the GenoType MTBDRplus assay. We show that, in combination to DST, application of the GenoType MTBDRplus and GenoType MTBDRsl assays might be a useful additional tool to allow a rapid and safe diagnosis of MDR and extensively drug-resistant (XDR) MTBC.
Keywords: drug susceptibility testing, GenoType, Mycobacterium tuberculosis complex, Nigeria
Introduction
The incidence of tuberculosis (TB) and especially multidrug-resistant TB (MDR) continues to increase alarmingly worldwide. Nigeria has a population of about 150 million people and ranks among the world’s high-burden countries. The World Health Organization (WHO) report 2009 stated an incidence of TB at 311/100,000, a prevalence of 772,000 cases, and the rate of new sputum smear positive cases was approximately 131/100,000 per year. Studies on drug susceptibility profiles of Mycobacterium tuberculosis (MTB) in Nigeria are scarcely reported, and the prevalence of MDR strains is numeralized between 2% and 53% in new cases of TB [1, 2]. Treatment on MDR TB involves second line drugs which are costly, scarce, and toxic and require longer duration of therapy [2]. Therefore, a rapid diagnosis of MDR TB is essential to medicate patients adequate to the drug resistance pattern. In 2006, the outbreak of rapidly fatal TB among hospitalized HIV patients in South Africa [3] led to the definition of extensively drug-resistant tuberculosis (XDR TB) as TB resistant to fluoroquinolones (FQs) and an injectable second line drug (amikacin, kanamycin, or capreomycin) [3, 4].
The GenoType MTBDRplus assay (Hain) allows identification of isoniazid (INH) and rifampicin (RMP) resistant TB stains directly from clinical isolates. This assay detects a broad variety of rpoB gene mutations and is described to identify more than 95% of all RMP resistant strains [5, 6]. The mutations causing isoniazid (INH) resistance are located in several genes and regions like in 50%–95% in the codon 315 of the katG gene [5–8], in 20%–35% in the inhA regulatory region [5, 6, 8, 9] and in 10%–15% in the ahpC–oxyR intergenic region [5, 6, 9, 10], often in conjunction with katG mutations outside of the codon 315 [11].
In order to rapidly detect second line drug resistance, the GenoType MTBDRsl test has been developed. This assay detects mutations in the gyrA, rrs, and embB genes and, therefore, resistance to fluoroquinolone (FQ), amikacin (AM)/capreomycin (CM), and ethambutol (EMB). The sensitivity for detection of FQ resistance is described as about 75%–90% with specificity of 100%, for aminoglycoside resistance sensitivity, and specificity was 100% [4]; other studies report on sensitivity of 83.3% for AM and 86.8% for CM with a specificity of 100% for detection of AM and 99.1% of CM resistance [12]. For detection of EMB resistance, the sensitivity is indicated with 59%–64% and specificity 100% [4, 12]. Within the present study, we analyzed 110 specimens from Nigerian patients using the MTBDRplus and MTBDRsl tests. The major scientific objective was to determine the accuracy of these tests for direct detection of MTB and for determination of resistance pattern of MTB directly from the patients’ samples.
Materials and methods
Clinical specimens
A total of 110 sputum specimens sent to our laboratory from a tertiary hospital in Hospital Nigeria were processed by conventional N-acetyl-L cystein-NaOH method (final NaOH concentration 1%). After decontamination, the concentrated sediment was suspended in 1.0 to 1.5 ml sterile phosphate. After inoculation of solid and liquid media for the growth detection and the GenoType MTBDRplus assay (Hain), the leftover sediment was stored at −20 °C.
DST
DST (drug susceptibility testing) with INH, RMP, EMB, STR, and PZA was performed by the BACTEC MGIT 960 method (Becton Dickinson, Heidelberg, Germany). Tests were performed with the standard critical concentrations of INH (0.1 µg/ml), RMP (1.0 µg/ml), EMB (5.0 µg/ml), STR (1.0 µg/ml), and PZA (100µg/ml).
Identification of the MTBC strains from clinical specimens
For identification and differentiation of the MTBC strains isolated from the grown cultures, we used the genotype MTBC assay as recommended by the manufacturer (Hain Lifescience, Nehren, Germany).
GenoType MTBDRplus assay
From all specimens, the strip assay was performed as recommended by the manufacturer (Hain Lifescience, Nehren, Germany).
GenoType MTBDRsl assay
From the leftover sediment of the specimens that turned out in the DST to harbor RMP and INH resistant strains, the strip assay was performed as recommended by the manufacturer (Hain Lifescience, Nehren, Germany).
Sequencing
Sequencing of the key regions, involved in the development of resistance, was performed by Hain Lifescience (Nehren, Germany) as published previously [5].
Statistical analysis
Sensitivity and specificity were calculated from cross tabulation, comparing the strip assays (MTBDRplus and MTBDRsl) (Hain Lifescience, Nehren, Germany) with gold standard culture techniques.
Results
Host characteristics
Of the 110 specimens, in 40, we were able to either cultivate MTBC or to detect MTBC complex DNA. Of these patients, 18 were female and 22 male, the mean age of the female patients was 37.27 years (14–82 years), and the mean age of the male patients was 33.28 years (18–82 years). Six females and four males were tested HIV positive, and seven females and ten males had a history of TB treatment or were undergoing TB treatment (Table 1).
Table 1.
Patient characteristics
| Sex |
||
|---|---|---|
| Female | Male | |
| Number of patients | 18 | 22 |
| Age (years) | 37.27 (14–82) | 33.28 (18–82) |
| HIV positive | 6 | 4 |
| HIV negative | 12 | 18 |
| History of TB treatment | 7 | 10 |
Frequency of MTBC detected by culture or GenoType MTBDRplus assay
We analyzed the frequency of MTBC in the specimens using two different methods. The specimens were inoculated in liquid and on three solid media and incubated at 37 °C for at least 7 weeks. The solid media were checked weekly for growth of mycobacteria; growth in the liquid media was monitored by the BACTEC MGIT 960 method.
In cultures from patients without history of MTBC treatment, we detected, in 19 (20.3%) samples, growth of M. tuberculosis complex; 74 samples (79.56%) were negative for growth of M. tuberculosis up to 10 weeks of incubation. Using the GenoType MTBDRplus assay, 22 samples (23.6%) were tested positive for presence of M. tuberculosis complex DNA and 71 (76.3%) were tested negative. Four specimens showed positive detection of M. tuberculosis complex DNA by the GenoType MTBDRplus assay but negative results of culture; one specimen was culture positive but GenoType MTBDRplus negative. Compared to the results from the culture growth, the GenoType MTBDRplus assay showed specificity for detection of M. tuberculosis complex DNA of 95% (74.0–99.9%) and a sensitivity of 95% (86.7–98.5%) (Table 2A).
Table 2A.
Frequency of MTBC detected by culture or Genotype MTBDRplus strip assay in patients without history of MTBC treatment
| Strip assay MTBDR plus
(HAIN) |
|||
|---|---|---|---|
| + | − | ||
| Culture | + | 18 | 1 |
| − | 4 | 70 | |
In patients undergoing MTBC treatment (n = 16), we detected, in 12 (75%) samples, growth of M. tuberculosis complex; four samples (25%) were negative for growth of M. tuberculosis up to 10 weeks of incubation. Using the GenoType MTBDRplus assay, 14 samples (87.5%) were tested positive for presence of M. tuberculosis complex DNA and 2 (12.5%) were tested negative. From the 16 specimens, four showed positive detection of M. tuberculosis complex DNA by the GenoType MTBDRplus assay but negative results of culture, and two specimens were culture positive but GenoType MTBDRplus negative. Therefore, compared to the results from culture as gold standard technique, the GenoType MTBDRplus assay showed a sensitivity for detection of M. tuberculosis complex of 83% (51.6–97.9%) in patients undergoing treatment of MTBC (Table 2B).
Table 2B.
Frequency of MTBC detected by culture or Genotype MTBDRplus strip assay in patients undergoing MTBC treatment
| Strip assay MTBDR plus
(HAIN) |
|||
|---|---|---|---|
| + | − | ||
| Culture | + | 10 | 2 |
| − | 4 | 0 | |
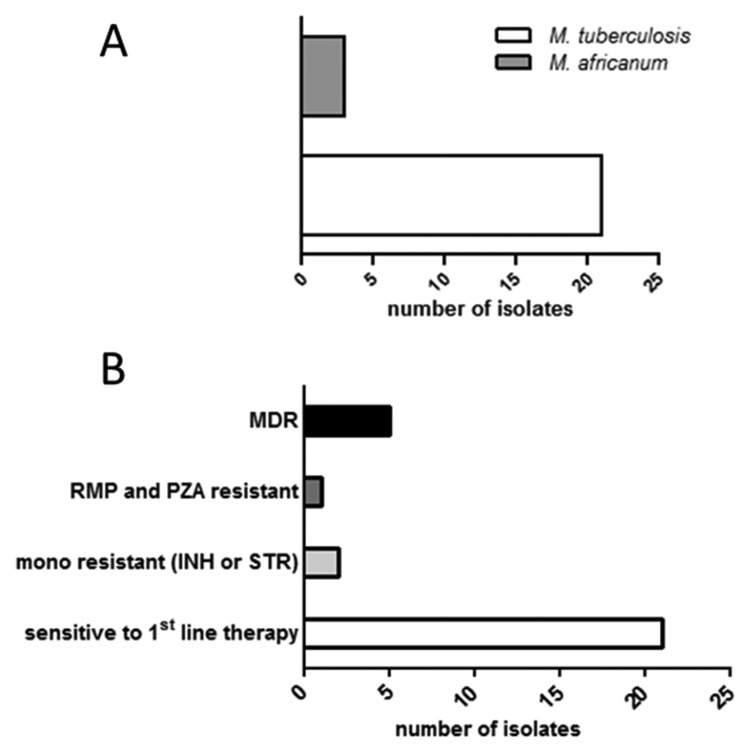
For differentiation and identification of the grown MTBC strains, we used the GenoType MTBC. We identified 21 (67%) M. tuberculosis and 11 (34%) M. africanum strains (Fig. 1A).
Fig. 1.
(A) Differentiation and identification of the MTBC strains obtained from culture. (B) Drug resistance pattern detected by culture based drug susceptibility testing (DST)
Frequency of drug resistance and MDR detected by culture methods
Using the BACTEC MGIT 960 method, we determined the drug resistance and susceptibility pattern of the cultivated MTB strains. Drug susceptibility testing (DST) revealed 21 (65.6%) of INH, RMP, PZA, STR, and EMB sensitive strains. Two strains (6.25%) showed a mono-resistance for either INH or STR. One strain (3.125%) was RMP and PZA resistant. We isolated five MDR MTB stains, of which one strain (3.125%) was resistant to INH, RMP, and STR and four strains (12.5%) were resistant to INH, RMP, STR, EMB, and PZA (Fig. 1B). The MDR strains and the RMP/PZA resistant strain were sent to the National Reference Laboratory in Borstel for verification of the resistance pattern and for additional testing of second line drugs. All six strains were susceptible to protionamide, ofloxacin, PAS, cycloserine/terizidone, amikacin, and capreomycin.
Frequency of drug resistance and MDR detected by usage of the GenoType MTBDRplus and MTBDRsl assays
In order to analyze INH and RMP directly from the sediments of decontaminated sputum samples, we used the GenoType MTBDRplus assay and compared the results with the results from DST, which was used as gold standard technique. Twenty-six specimens (81.25%) were tested INH susceptible, six (18.75%) were tested INH resistant; this corresponds to a specificity of 100% (86.3–100%) and a sensitivity of 86% (42.1–99.6%). Twenty-seven specimens (84.4%) were tested RMP susceptible, five (15.6%) were tested RMP resistant, corresponding to a specificity of 100% (86.3–100%) and a sensitivity of 83% (35.9–99.6%) (Table 3). In case of INH and/or RMP resistance, we additionally performed the GenoType MTBDRsl assay, which detects for fluoroquinolone (FQ), amikacin/capreomycin (AM/CM), and EMB resistance. In five from five tested samples, EMB resistance was detected (sensitivity 100%) (47.8–100%), whereas no FQ or AM/CM resistance was detectable (specificity 100%) (54–100%) (Table 3). This is in line with the DST pattern from the liquid cultures, performed by the German National Reference Centre for Mycobacteria (Borstel, Germany).
Table 3.
Sensitivity and specificity of the strip assay GenoType MTBDR plus assay and the GenoType MTBDR sl assay in comparison to gold standard method culture
| MTBDR plus |
MTBDR sl |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| INH |
RIF |
ETB |
Chinolone |
Aminoglycoside/ capreomycin |
||||||||
| sen | res | sen | res | sen | res | sen | res | sen | res | |||
| Culture | Resistance | sen | 25 (32) | 0 (32) | 26 (32) | 0 (32) | 0 | 0 | 6 (6) | 0 | 6 (6) | 0 |
| res | 1 (32) | 6 (32) | 1 (32) | 5 (32) | 5 (5) | 0 | 0 | 0 | 0 | 0 | ||
Resistance to RMP and INH in M. tuberculosis is most often attributed to mutations in the rpoB, katG, and inhA genes. The GenoType MTBDRplus assay detects INH and RMP resistance-associated mutations in rpoB, katG, and inhA genes. The GenoType MTBDRsl assay detects mutations gyrA, rrs, and embB genes leading to resistance to FQ, AM/CM, and EMB. We found different RMP, INH, and EMB pattern; in case of INH resistance, most of the assays showed katG mutation specific bands with probe MUT 1, whereas in RMP and EMB resistance, pattern was diverse (Table 4).
Table 4.
Detection of INH and RMP resistance-associated mutations in rpoB, katG, and inhA genes by the strip assay GenoType MTBDR plus assay and mutations in the gyrA, rrs, and embB genes leading to resistance to FQ, AM/CM, and EMB by the GenoType MTBDR sl assay
| No. (%) of strains |
MTBDR plus assay |
MTBDR sl assay |
Result | ||
|---|---|---|---|---|---|
| RMP pattern (rpoB) | INH pattern (katG) | INH pattern (inhA) | EMB pattern (embB) | ||
| 2 (25) | ∆WT 7, Mut2A | ∆WT, Mut1 | WT | ∆WT, Mut1A | MDR |
| 1 (12,5) | ∆WT 7 | ∆WT, Mut1 | WT | ∆WT, Mut1A | MDR |
| 1 (12,5) | ∆WT 4, Mut1 | ∆WT, Mut1 | WT | ∆WT, Mut1B | MDR |
| 1 (12,5) | ∆WT 8, Mut3 | ∆WT, Mut1 | WT | ∆WT, Mut1A | MDR |
| 2 (25) | WT | ∆WT, Mut1 | WT | WT | INH resistance |
| 1 (12,5) | WT | WT | ∆WT1, Mut1 | WT | INH resistance |
In sample no. 9, the GenoType MTBDRplus resulted in RMP susceptibility, whereas the DTS showed an RMP resistance of the strain. In sample no. 16, the GenoType MTBDRplus resulted in INH susceptibility, but the DTS showed an INH resistance of the strain. In both strains, sequencing of the rpoB gene and, respectively, the inhA promoter region and katG gene did not show any resistance-associated mutations. This might lead to the conclusion that mutations in regions not covered by the probes of the GenoType MTBDRplus assay might cause the resistant phenotype.
Discussion
Within the present study, we analyzed 110 respiratory specimens from Nigerian patients for infection with MTB and analyzed the MTB drug resistance pattern. Only few publications that deal with the susceptibility pattern of bacteria of the M. tuberculosis complex in Nigeria are available. In our study, 68% of the isolated stains were pan-susceptible to all five drugs (INH, RMP, STR, PZA, and EMB) tested in the DST. This is in line with other studies from Nigeria, reporting on 59–62% pan-susceptible isolates [1, 13]. However, data collected in a tertiary hospital in Nigeria report on pan-susceptibility in only 46% of the isolated TB strains [2]. We found 6.25% of INH or STR mono-resistant strains. Other publications report on a streptomycin resistance rate of 11% but did not find INH mono-resistant strains [2]. The low number of isolates ranging from 32 (this study) to 50 [1, 2] might account for that effect. However, we identified 16% of the isolates as MDR TB stains; these strains were at least resistant to INH, RMP, STR, and EMB, and one was additionally resistant to PZA. The MTB prevalence described in the literature ranges between 4% and 31% [1, 2]. It is suggested that the incidence of MDR tuberculosis is associated to the type of hospital. Tertiary hospitals show a higher rate of MDR strains as compared to other hospitals, as patients treated in these hospitals might represent a selection of referred difficult cases that may have failed or defaulted from previous treatment [1, 2]. In our study, the specimens were obtained from patients treated in a tertiary hospital. However, for a better understanding of the epidemiological situation of MDR and XDR tuberculosis in Nigeria further studies are necessary, including large patient cohorts from different parts of the country.
Interestingly, our data revealed a sensitivity and specificity of 95% of the GenoType MTBDRplus assay on detection of M. tuberculosis complex DNA in the specimens of patients without history of TB treatment. Other commercial nucleid acid amplification tests (NAATs) for diagnosis of tuberculosis directly from respiratory specimens report on sensitivity between 85% and 92% and specificity between 96% and 97% [14, 15]. Therefore, the GenoType MTBDRplus assay seems to reliably detect M. tuberculosis complex DNA and INH and RMP resistance. Certainly, NAATs should be applied only in combination with gold standard culture methods.
As a rapid diagnosis of MDR or even XDR tuberculosis is of a high importance for the patient outcome and of a high epidemiological importance, we evaluated the GenoType MTBDRplus and GenoType MTBDRsl assays on the patient’s specimens. These tests allow information about the MTB resistance pattern within 1 day. The time to positivity of MTB cultures can take up to 7 weeks, and the conventional DST testing takes additional 2 weeks after cultivation of the bacterium. We found a sensitivity of 86% and a specificity of 100% for INH resistance and a sensitivity of 83% and specificity of 100% for RMP resistance. Other studies report on sensitivity for INH resistance of 90% and 98% for RMP resistance [5]. We had two strains that were tested resistant to INH or, respectively, RMP in the DST and susceptible in the GenoType MTBDRplus. In both strains, sequencing of the rpoB gene and respectively the inhA promoter region and katG gene did not show any resistance associated mutations. This might lead to the conclusion that mutations in regions not covered by the probes of the GenoType MTBDRplus assay might cause the resistant phenotype. Analysis of EMB, AM/CM, and FQ resistance using the GenoType MTBDRsl corresponded to the results obtained by DST; however, the number of isolates was low, and we did not detect any resistance to FQ or AM/CM. As mutations in loci other than those covered by the GenoType MTBDRplus and GenoType MTBDRsl assays might lead to false sensitive results, the NAAT-based methods need to be confirmed by conventional DST, based on culture techniques. Therefore, the stripe assays GenoType MTBDRplus and GenoType MTBDRsl cannot replace gold standard culture methods but represent an additional tool for the first identification of drug resistant M. tuberculosis complex strains, resulting in a fast, adapted and more sufficient therapy.
Taken together, our data represent an important addition to the rare epidemiological data concerning resistance pattern of MTB in Nigeria and showed that the application of the GenoType MTBDRplus and GenoType MTBDRsl assays might be a useful additional tool to allow a rapid and safe diagnosis of MDR and XDR tuberculosis.
Contributor Information
Michael Felkel, 1Institute of Tropical Medicine, University Hospital of Tübingen, Tübingen, Germany.
Robert Exner, 1Institute of Tropical Medicine, University Hospital of Tübingen, Tübingen, Germany.
Regina Schleucher, 1Institute of Tropical Medicine, University Hospital of Tübingen, Tübingen, Germany.
Helga Lay, 2Institute of Medical Microbiology and Hygiene, University Hospital of Tübingen, Tübingen, Germany.
Ingo B. Autenrieth, 2Institute of Medical Microbiology and Hygiene, University Hospital of Tübingen, Tübingen, Germany.
Volkhard A. J. Kempf, 3Institute of Medical Microbiology and Hygiene, Johann-Wolfgang Goethe University, Frankfurt, Germany.
Julia-Stefanie Frick, 2Institute of Medical Microbiology and Hygiene, University Hospital of Tübingen, Tübingen, Germany.
References
- 1.Ani AE, Idoko J, Dalyop YB, Pitmang SL. Drug resistance profile of Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients in Jos, Nigeria. Trans R Soc Trop Med Hyg. 2009 Jan;103(1):67–71. doi: 10.1016/j.trstmh.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Kehinde AO, Obaseki FA, Ishola OC, Ibrahim KD. Multidrug resistance to Mycobacterium tuberculosis in a tertiary hospital. J Natl Med Assoc. 2007 Oct;99(10):1185–1189. [PMC free article] [PubMed] [Google Scholar]
- 3.Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, Zeller K, Andrews J, Friedland G. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006 Nov 4;368(9547):1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- 4.Kiet VS, Lan NT, An DD, Dung NH, Hoa DV, van Vinh Chau N, Chinh NT, Farrar J, Caws M. Evaluation of the MTBDRsl test for detection of second-line-drug resistance in Mycobacterium tuberculosis. J Clin Microbiol. 2010 Aug;48(8):2934–2939. doi: 10.1128/JCM.00201-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hillemann D, Rüsch-Gerdes S, Richter E. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol. 2007 Aug;45(8):2635–2640. doi: 10.1128/JCM.00521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Telenti A, Honoré N, Bernasconi C, March J, Ortega A, Heym B, Takiff HE, Cole ST. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J Clin Microbiol. 1997 Mar;35(3):719–723. doi: 10.1128/jcm.35.3.719-723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mokrousov I, Narvskaya O, Otten T, Limeschenko E, Steklova L, Vyshnevskiy B. High prevalence of KatG Ser315Thr substitution among isoniazid-resistant Mycobacterium tuberculosis clinical isolates from northwestern Russia, 1996 to 2001. Antimicrob Agents Chemother. 2002 May;46(5):1417–1424. doi: 10.1128/AAC.46.5.1417-1424.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musser JM, Kapur V, Williams DL, Kreiswirth BN, van Soolingen D, van Embden JD. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J Infect Dis. 1996 Jan;173(1):196–202. doi: 10.1093/infdis/173.1.196. [DOI] [PubMed] [Google Scholar]
- 9.Piatek AS, Telenti A, Murray MR, El-Hajj H, Jacobs Jr WR, Kramer FR, Alland D. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob Agents Chemother. 2000 Jan;44(1):103–110. doi: 10.1128/aac.44.1.103-110.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelley CL, Rouse DA, Morris SL. Analysis of ahpC gene mutations in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997 Sep;41(9):2057–2058. doi: 10.1128/aac.41.9.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sreevatsan S, Pan X, Zhang Y, Deretic V, Musser JM. Analysis of the oxyR-ahpC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob Agents Chemother. 1997 Mar;41(3):600–606. doi: 10.1128/aac.41.3.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hillemann D, Rüsch-Gerdes S, Richter E. Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol. 2009 Jun;47(6):1767–1772. doi: 10.1128/JCM.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ani A, Bruvik T, Okoh Y, Agaba P, Agbaji O, Idoko J, Dahle UR. Genetic diversity of Mycobacterium tuberculosis Complex in Jos, Nigeria. BMC Infect Dis. 2010 Jun 26;10:189. doi: 10.1186/1471-2334-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett A, Magee JG, Freeman R. An evaluation of the BD ProbeTec ET system for the direct detection of Mycobacterium tuberculosis in respiratory samples. J Med Microbiol. 2002 Oct;51(10):895–898. doi: 10.1099/0022-1317-51-10-895. [DOI] [PubMed] [Google Scholar]
- 15.Ling DI, Flores LL, Riley LW, Pai M. Commercial nucleic-acid amplification tests for diagnosis of pulmonary tuberculosis in respiratory specimens: meta-analysis and meta-regression. PLoS One. 2008 Feb 6;3(2):e1536. doi: 10.1371/journal.pone.0001536. [DOI] [PMC free article] [PubMed] [Google Scholar]