Abstract
Background and Aims
Riparian systems are prone to invasion by alien plant species. The spread of invasive riparian plants may be facilitated by hydrochory, the transport of seeds by water, but while ecological studies have highlighted the possible role of upstream source populations in the establishment and persistence of stands of invasive riparian plant species, population genetic studies have as yet not fully addressed the potential role of hydrochoric dispersal in such systems.
Methods
A population genetics approach based on a replicated bifurcate sampling design is used to test hypotheses consistent with patterns of unidirectional, linear gene flow expected under hydrochoric dispersal of the invasive riparian plant Impatiens glandulifera in two contrasting river systems.
Key results
A significant increase in levels of genetic diversity downstream was observed, consistent with the accumulation of propagules from upstream source populations, and strong evidence was found for organization of this diversity between different tributaries, reflecting the dendritic organization of the river systems studied.
Conclusions
These findings indicate that hydrochory, rather than anthropogenic dispersal, is primarily responsible for the spread of I. glandulifera in these river systems, and this is relevant to potential approaches to the control of invasive riparian plant species.
Keywords: Dispersal, gene flow, Himalayan balsam, hydrochory, Impatiens glandulifera, invasive species, propagule pressure, riparian
INTRODUCTION
The introduction of non-native species into new environments represents one of the major threats to ecosystems and biodiversity worldwide and is now ranked second only to habitat loss as a potential cause of ecological catastrophe (Gurevitch and Padilla, 2004). Indeed, a review of recent animal extinctions found that invasive species were responsible for >50 % of cases in which the cause of extinction was known (Clavero and García-Berthou, 2005). Plant invasions can also be extremely detrimental and are usually associated with deliberate anthropogenic introductions, including exotic ornamentals and feral crop species (Pimentel et al., 2001; Weber, 2003). Problems associated with the introduction of exotic plant species into new habitats include hybridization with native species, impacts on species–species interactions and alteration of soil chemistry (Sakai et al., 2001; Tickner et al., 2001; Henderson et al., 2006; Pearson, 2009), but the key detrimental effect is usually the rapid formation of largely monospecific stands that reduce biodiversity in impacted habitats (Cronk and Fuller, 2001; van der Wal et al., 2008; Hejda et al., 2009). Invasive plant species can often outcompete native species for a wide range of resources, including light, nutrients and pollinators, and such ‘biotic homogenization’ has been implicated as a potential causal factor in future mass extinction events (McKinney and Lockwood, 1999).
Riparian systems are of particular ecological interest as they encompass both terrestrial and freshwater habitats where biodiversity is maximal and important ecological, hydrological and geomorphological processes occur (Tickner et al., 2001). They are especially prone to invasion by exotic plant species as their dynamic physical nature regularly provides suitable habitats for colonizing species due to the periodic deposition of nutrient-rich sediments and the creation of large areas with sufficient space, light and moisture for the successful germination of seedlings (Hupp and Osterkamp, 1996; Myers and Bazely, 2003). In addition to the impacts on native biodiversity mentioned above, invasive plants can also alter the geomorphological and hydrological aspects of riparian ecosystems (Tickner et al., 2001). The establishment of dense populations of non-native plants can significantly lower water-table levels with serious consequences not only for the existing flora but also for entire mesocosms (Loope et al., 1988; DiTomaso, 1998). Newly established populations of invasive plant species may also interfere with sediment distribution, leading to aggradation, which in turn can radically alter the geomorphology of river systems and may increase the risk of periodic flooding (Graf, 1982; Birkeland, 1996).
Propagule pressure (in terms of seeds, fruits or vegetative fragments) has been shown to be a major factor determining the successful establishment and spread of invasive species (Lockwood et al., 2005; Colautti et al., 2006; Martínez-Ghersa and Ghersa, 2006; Eschtruth and Battles, 2009; Simberloff, 2009). This is particularly important in riparian systems, as propagules may be transported longitudinally downstream via hydrochory, the dispersal of seeds via water, potentially facilitating the spread of invasive riparian plants (Burkart, 2001; Nakayama et al., 2007; Tassin et al., 2007). Thus, the unidirectional, linear connectivity between potentially suitable habitats may compound the potential for invasion due to increased propagule pressure. Ecological studies have highlighted the possible role of upstream source populations in the establishment and persistence of stands of invasive plant species in riparian habitats (e.g. Dawson and Holland, 1999; Samuel and Kowarik, 2010). The occurrence of such patterns of dispersal has implications for control strategies: when downstream dispersal is a factor, eradication programmes may be most efficient when they specifically target upstream populations, thus reducing propagule pressure.
This study aims to test the hypothesis that hydrochory, or water-mediated seed dispersal, is a key factor in the spread of the invasive riparian plant species Impatiens glandulifera (Himalayan balsam). The species was introduced into Britain and Ireland as an ornamental in the 19th century and has subsequently become a naturalized invader. It is an annual herb which can outcompete even hardy, well-established, native perennial species in riparian habitats (Beerling and Perrins, 1993). Flowers are self-compatible and protandrous, and are primarily pollinated by Diptera and Hymenoptera (Lopezaraiza-Mikel et al., 2007). Individual plants produce around 500–5000 seeds and short-distance dispersal (<5 m) occurs through explosive dehiscence, whereas long-distance dispersal is believed to occur either anthropogenically or via hydrochory. To date, only a single population genetic study of I. glandulifera has been published, and this did not explicitly test hypotheses on hydrochoric dispersal (Walker et al., 2009). The two main hypotheses concerning the role of hydrochory to be tested here are based on the patterns of gene flow expected under a model of linear, unidirectional gene flow. First, there is expected to be an increase in levels of genetic diversity downstream due to the accumulation of propagules from upstream source populations. Secondly, if hydrochory, rather than overland transport (anthropogenic or by animals), is the major factor in the dispersal of seeds, populations on different tributaries not linked by water flow will be more genetically differentiated than populations from different areas of the river which are linked by the flow of the river (i.e. above and below a confluence in a bifurcate or dendritic network), which in turn will be more genetically differentiated than populations from the same tributary. The first hypothesis has been tested previously in several studies (e.g. Ritland, 1989; Gornall et al., 1998; Russell et al., 1999; Lundqvist and Andersson, 2001; Tero et al., 2003; Liu et al., 2006; Markwith and Scanlon, 2007; Pollux et al., 2009; Honnay et al., 2010; Schleuning et al., 2011), but no overall consensus has emerged. The second hypothesis, however, has not been explicitly tested, as studies have not used a bifurcate experimental design, and it has been suggested that the dendritic organization of river systems should be taken into account to gain further insights into the role of hydrochory in the dispersal of riparian plants (Pollux et al., 2009).
MATERIALS AND METHODS
Experimental design, sampling and DNA extraction
To test the predictions of the hydrochory hypothesis, it was necessary to sample Impatiens grandulifera over entire river drainage basins (i.e. with river lengths of at least 25 km and with a bifurcate organization). Replication involved studying populations in two contrasting river basins in Britain and Ireland (Fig. 1). The Western Cleddau in Pembrokeshire, Wales, was chosen based on data from the Joint Nature Conservation Committee (www.jncc.gov.uk). The system has been designated as a Special Area of Conservation (SAC), and is characterized by Callitricho–Batrachion Type 4 (CB4) vegetation communities, where I. glandulifera is a frequent alien (Hatton-Ellis and Grieve, 2003). The second river system selected was the Tempo/Colebrooke river system in Co. Fermanagh, Northern Ireland, and was chosen based on river vegetation survey records from the Quercus Centre for Biodiversity and Conservation Biology (www.quercus.ac.uk). The system is part of the Upper Lough Erne river basin, a predominantly limestone catchment surrounded by large, semi-natural alluvial forests. The exact age of the study populations is unknown, but the species was not recorded in either area prior to 1940 (Beerling and Perrins, 1993). Species occurrence data for both river systems was obtained from the National Biodiversity Network Gateway (www.nbn.org.uk) and surveys and sampling of both systems were carried out during the summer of 2006. Nine and ten sites representing populations from two upstream tributaries and populations from below the point of the confluence of those tributaries were sampled from the Western Cleddau and Tempo/Colebrooke systems, respectively (Table 1). Geographical distances between sites were calculated using the ArcView GIS system. Between 15 and 35 individuals per site were sampled and DNA was extracted from individual leaves using the Qiagen DNeasy Plant Mini system.
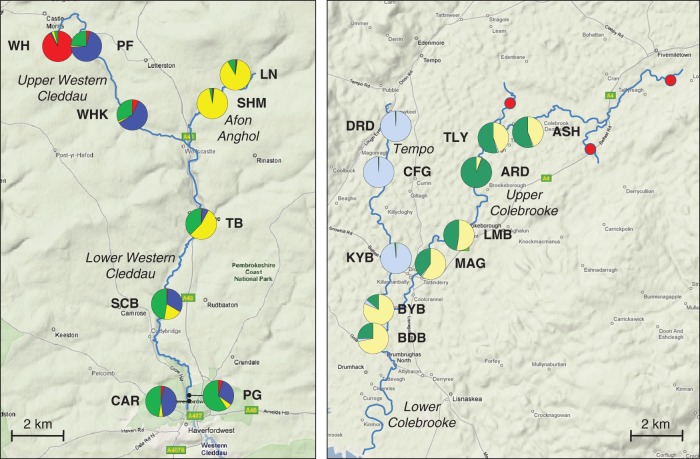
Fig. 1.
Maps showing locations of populations sampled in this study. Left, Western Cleddau; right, Tempo/Colebrooke. Population codes are given in Table 1. Pie charts indicate probability of assignment of individuals to one of K = 4 (Western Cleddau) or K = 3 (Tempo/Colebrooke) genetic clusters based on the Structure analysis (see Materials and methods). Red circles in the Tempo/Colebrooke system indicate areas surveyed but where no I. glandulifera was found. For clarity, only tributaries sampled or surveyed are shown.
Table 1.
Locations of sampled sites and sample numbers
| System | Tributary | Population | Grid ref. | Code | n | HO | HE | AR | FIS |
|---|---|---|---|---|---|---|---|---|---|
| Western Cleddau | Upper Western Cleddau | Withy Hill | SM904297 | WH | 26 | 0·366 | 0·408 | 1·851 | –0·291 |
| Priskilly Forest | SM919299 | PF | 21 | 0·130 | 0·122 | 1·461 | –0·062 | ||
| Welsh Hook | SM932278 | WHK | 20 | 0·140 | 0·164 | 1·636 | 0·149* | ||
| Afon Anghol | Little Newcastle | SM979289 | LN | 19 | 0·202 | 0·149 | 1·547 | –0·377 | |
| Sealyham Bridge | SM967279 | SHM | 24 | 0·175 | 0·118 | 1·358 | –0·500 | ||
| Lower Western Cleddau | Treffgarne Bridge | SM959230 | TB | 21 | 0·195 | 0·175 | 1·556 | –0·116 | |
| St. Catherine's Bridge | SM945198 | SCB | 18 | 0·152 | 0·206 | 1·578 | 0·267* | ||
| Haverfordwest playground | SM953161 | PG | 17 | 0·194 | 0·221 | 2·027 | 0·127** | ||
| Haverfordwest car park | SM953164 | CAR | 15 | 0·223 | 0·234 | 1·993 | 0·045 | ||
| Tempo/Colebrooke | Tempo | Drumderg | H341446 | DRD | 30 | 0·262 | 0·217 | 1·572 | –0·211 |
| Cornafannoge | H335426 | CFG | 30 | 0·205 | 0·194 | 1·500 | –0·053 | ||
| Killynure Bridge | H339388 | KYB | 21 | 0·222 | 0·222 | 1·600 | –0·001 | ||
| Ballyvelin Bridge | H334372 | BYB | 30 | 0·174 | 0·292 | 2·170 | 0·410*** | ||
| Upper Colebrooke | Ashbrooke | H392442 | ASH | 30 | 0·266 | 0·282 | 2·115 | 0·057** | |
| Tullyreagh Bridge | H379442 | TLY | 30 | 0·273 | 0·303 | 2·149 | 0·100*** | ||
| Ardunshin | H372426 | ARD | 35 | 0·231 | 0·231 | 1·885 | 0·003 | ||
| Littlemount Bridge | H370405 | LMB | 31 | 0·245 | 0·257 | 2·121 | 0·045 | ||
| Maguiresbridge | H350387 | MAG | 30 | 0·282 | 0·328 | 2·327 | 0·145*** | ||
| Lower Colebrooke | Ballindarragh Bridge | H331360 | BDB | 30 | 0·257 | 0·326 | 2·526 | 0·215*** |
n, number of individuals sampled; HO, expected heterozygosity; HE, expected heterozygosity; AR, allelic richness; FIS, inbreeding coefficient. *P < 0·05; **P < 0·01; ***P < 0·001.
Microsatellite genotyping
Individuals were genotyped at ten microsatellite loci, representing all the published markers available for I. glandulifera (Provan et al., 2007; Walker et al., 2009) with the exception of locus A2 from Walker et al. (2009), which regularly failed to amplify in several populations, probably due to the presence of null alleles. Loci IGNSSR103, IGNSSR105, IGNSSR106, IGNSSR 210 and IGNSSR240 were genotyped following the procedures described by Provan et al. (2007). For the remaining five loci, forward primers were modified by the addition of a 19-bp M13 tail (5′-CACGACGTTGTAAAACGAC-3′) and reverse primers were modified by the addition of a 7-bp tail (5′-GTGTCTT-3′). PCR was carried out in a total volume of 10 µL containing 100 ng genomic DNA, 10 pmol PET-, 6-FAM- or HEX-labelled M13 primer, 1 pmol M13-tailed forward primer, 10 pmol reverse primer, 1× PCR reaction buffer, 200 µm each dNTP, 2·5 mm MgCl2 and 0·25 U GoTaq Flexi DNA polymerase (Promega, Sunnyvale, CA). PCR cycling conditions followed those given by Provan et al. (2007) and Walker et al. (2009). Genotyping was carried out on an AB3730xl capillary genotyping system. Allele sizes were scored using LIZ-500 size standards and were checked by comparison with previously sized control samples.
Data analysis
GenePop (V3·4; Raymond and Rousset, 1995) was used to test for linkage disequilibrium between nuclear loci. Levels of genetic diversity measured as allelic richness (AR) and observed and expected heterozygosity (HO and HE) were calculated using the Fstat software package (V2·9.3·2; Goudet, 2001) and the Arlequin software package (V3·01; Excoffier et al., 2005), respectively. Inbreeding coefficients (FIS) were also calculated using the Fstat software package. To test the hypothesis that genetic diversity increases downstream, a linear regression analysis was carried out for both allelic richness and expected heterozygosity against the distance downstream from the furthest upstream population sampled in each system using the MedCalc software package (V12·2.1; www.medcalc.org, Ostend, Belgium). For both river systems, tributaries plus all sites downstream of their confluence with the main river were considered separately in each case (Upper Western Cleddau/Lower Western Cleddau; Afon Anghol/Lower Western Cleddau; Tempo/Lower Colebrooke; Upper Colebrooke/Lower Colebrooke).
Levels of population differentiation were calculated in the analysis of molecular variance (AMOVA) framework (Excoffier et al., 1992) as pairwise ΦST values using the Arlequin software package. To test the hypothesis of patterns of differentiation consistent with linear, unidirectional gene flow, populations on different tributaries unlinked by water flow were classified as ‘unlinked’, whereas those from different areas of the river which are linked by the flow of the river (i.e. above and below a confluence) were defined as ‘linked’. Populations from the same tributary were classed as ‘same’. The significance of differences between values for ‘unlinked’, ‘linked’ and ‘same’ groups of populations was determined using the Student–Newman–Keuls post hoc test implemented in the MedCalc software package. Genetic structuring was also assessed at the individual level using a Bayesian procedure implemented in the Structure software package (V2·3.3; Pritchard et al., 2000). The program was run using no prior knowledge and the admixture ancestry model. For each river system, five independent runs were carried out for each value of K, the number of genetic clusters, up to K = 9 (Western Cleddau) or K = 10 (Tempo/Colebrooke). Each Markov chain Monte Carlo analysis used a burn-in period of 10 000 followed by a further 100 000 iterations. The most likely value for K was estimated using the ΔK statistic of Evanno et al. (2005) implemented in the Structure Harvester software package (V0·6.1; Earl et al., 2012).
RESULTS
Two to eight alleles were detected across all populations studied at each of the ten microsatellite loci analysed and no evidence of linkage disequilibrium was detected. All loci were variable in each river system. Mean within-population values for allelic richness (AR) averaged over all loci ranged from 1·358 (SHM population) to 2·027 (PG population) in the Western Cleddau system, and from 1·500 (CFG population) to 2·525 (BDB population) in the Tempo/Colebrooke system. Mean levels of expected heterozygosity averaged over all loci ranged from 0·118 (SHM population) to 0·408 (WH population) in the Western Cleddau system, and from 0·194 (CFG population) to 0·328 (MAG population) in the Tempo/Colebrooke system. Values of FIS ranged from –0·500 (SHM population) to 0·267 (SCB population) in the Western Cleddau system, and from –0·211 (DRD population) to 0·410 (BYB population) in the Tempo/Colebrooke system (Table 1). The regression analyses indicated a significant downstream increase in allelic richness in both systems and a significant downstream increase in expected heterozygosity in all courses except the Upper Colebrooke/Lower Colebrooke (Table 2).
Table 2.
Regression analyses of levels of genetic diversity vs. distance downstream from uppermost population
| Tributaries |
AR |
HE |
||
|---|---|---|---|---|
| R2 | P | R2 | P | |
| Upper Western Cleddau/Lower Western Cleddau | 0·657 | 0·025* | 0·945 | <0·001*** |
| Afon Anghol/Lower Western Cleddau | 0·739 | 0·014* | 0·909 | 0·002** |
| Tempo/Lower Colebrooke | 0·771 | 0·025* | 0·772 | 0·025* |
| Upper Colebrooke/Lower Colebrooke | 0·583 | 0·039* | 0·252 | n.s. |
AR, allelic richness; HE, expected heterozygosity. *P < 0·05; **P < 0·01; ***P < 0·001.
Population-pairwise ΦST values (see Supplementary Data Table S1) in the Western Cleddau system ranged from zero (PF-WHK) to 0·560 (PF-SHM). In the Tempo/Colebrooke system, values ranged from zero (DRD-CFG and TLY-MAG) to 0·536 (DRD-ARD). In both systems, the Student–Newman–Keuls post hoc test indicated significant (P < 0·05) differences between population-pairwise ΦST values for ‘unlinked’, ‘linked’ and ‘same’ groups in the Western Cleddau system. In the Tempo/Colebrooke system, the mean population-pairwise ΦST between ‘unlinked’ tributaries was significantly higher (P < 0·05) than that for both the ‘linked’ and the ‘same’ groups. Although the mean population-pairwise ΦST between ‘linked’ tributaries was higher than that between populations on the same tributary, this difference was not significant (Fig. 2). This organization of genetic variation between tributaries was further highlighted by the Structure analysis, which identified K = 4 and K = 3 as the most likely numbers of genetic clusters for the Western Cleddau and Tempo/Colebrooke systems, respectively (Fig. 1). In the Western Cleddau, individuals from the WH population, a population isolated from the main river system for >20 years (see Discussion), were almost exclusively assigned to a single cluster, depicted in red, which was present at low frequency elsewhere in the system. The remaining two populations from the Upper Western Cleddau (PF and WHK) were associated with the two clusters indicated in blue and light green. Individuals from both populations on the Afon Anghol tributary (LN and SHM) were generally assigned to a fourth genetic cluster, depicted in yellow. Populations downstream of the confluence of the two tributaries were associated to a greater or lesser degree with combinations of the yellow, blue and light green clusters. In the Tempo/Colebrooke system, individuals from three of the four Tempo populations (DRD, CFG and KYB) were almost exclusively assigned to a single genetic cluster depicted in light blue, which was almost completely absent elsewhere in the system. Individuals from the remaining population on this tributary (BYB) and those from the populations on the Upper Colebrooke (ASH, TLY, ARD, LMB and MAG) and the sole population below the confluence (BDB) were primarily assigned to one of two clusters, depicted in light yellow and dark green.
Fig. 2.
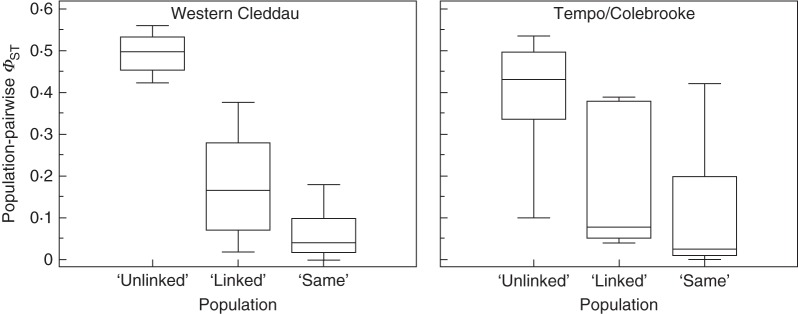
Box-whisker plots of mean population pairwise ΦST values between ‘unlinked’, ‘linked’ and ‘same’ populations.
DISCUSSION
Although our study revealed low levels of genetic variation, which is characteristic of the founder effects often associated with invasions, they confirm our hypotheses concerning increasing genetic variation downstream and structuring of genetic diversity between tributaries, which are indicative of hydrochoric seed dispersal. Water-mediated dispersal and the subsequent establishment of new individuals and populations of riparian plant species is the end result of a combination of factors including fertilization, seed production, transport and germination. Population genetic approaches can provide an overview of the integrated end result of all these processes and thus offer valuable insights into the spread, persistence and management of invasive riparian plants. The observed patterns of genetic variation within and between the populations analysed in the present study suggest that hydrochory, rather than anthropogenic dispersal, is primarily responsible for the spread of I. glandulifera in the Western Cleddau and Tempo/Colebrooke river systems. Hydrogeomorphology of rivers was also found to influence patterns of dispersal. The implications of these findings are relevant to concepts of dispersal in riparian plants in general, and to possible modes of spread and potential approaches to the control of invasive riparian plant species.
Evidence for hydrochoric dispersal
Previous molecular genetic studies of the potential role of hydrochoric dispersal in riparian plants have been somewhat equivocal, with some (Gornall et al., 1998; Lundqvist and Andersson, 2001; Chen et al., 2009; Pollux et al., 2009; Schleuning et al., 2011) reporting the expected increase in genetic diversity downstream and others (Russell et al., 1999; Markwith and Scanlon, 2007; Prentis and Mather, 2007; Honnay et al., 2010) finding no clear evidence for hydrochory. Here, the significant downstream increase in genetic diversity coupled with the structuring of this diversity between tributaries indicates a major role for hydrochory in the dispersal of I. glandulifera seeds. Although the biparentally inherited nuclear markers used to estimate levels of dispersal would also reflect the effects of pollen flow, the effects of downstream seed movement are still evident. The seeds of I. glandulifera are negatively buoyant, but they may still be dispersed by currents of fast-flowing water (Beerling and Perrins, 1993). Furthermore, laboratory and field observations have shown that intact seed pods of I. glandulifera can float for several days and that ‘sprung’ pods sometimes retain seeds and thus might also provide a vector for downstream seed dispersal (H. M. Love et al., unpubl. res.). It has been suggested that this dependence on water courses for long-distance dispersal has most likely limited I. glandulifera primarily to riparian habitats in its invasive range (Pyšek and Prach, 1993) and a logistic regression analysis at both national and regional levels in England found a correlation between the presence of the species and the abundance of rivers (Collingham et al., 2000).
Further evidence for hydrochoric dispersal can be seen in the organization of genetic variation by tributaries. The pairwise ΦST values and the results of the Structure analysis show that gene flow tends to occur primarily between populations within tributaries and, to a lesser degree, between populations on linked tributaries, with little dispersal between unlinked tributaries. Although geographically proximal populations are expected to be genetically similar under a simple, unrestricted two-dimensional model of gene flow, the KYB (Tempo) and MAG (Upper Colebrooke) populations, which are <1 km apart but on two unlinked tributaries, exhibit a pairwise ΦST value of 0·393 (P < 0·0001), whereas the KYB and DRD populations on the same tributary (Tempo) are separated by 8 km but exhibit a pairwise ΦST value of only 0·002 (n.s.; P = 0·315). More random patterns of gene flow such as anthropogenic transport of seeds, i.e. not restricted to unidirectional dispersal between populations linked by watercourses, would be unlikely to give rise to such a pattern of structuring. The only previous study on the population genetics of I. glandulifera did not explicitly aim to test the role of hydrochory in the dispersal of the species but, nevertheless, noted that the average population-pairwise FST within catchments in north-eastern England was significantly lower than the average value between catchments (Walker et al., 2009). Pollination by bees is important for I. glandulifera (Thijs et al., 2012), but because bees are central-place foragers, and must return to their nest after feeding, movement between forage resources is constrained and the resulting pollen-mediated gene flow is normally over short distances (R. J. Paxton, Martin-Luther-Universität, Germany, pers. comm.).
The role of hydrochory in shaping patterns of genetic diversity in I. glandulifera is particularly apparent in the Structure analysis of the Western Cleddau system. Populations from the two upstream tributaries are associated with four genetic clusters, which are rarely shared across the two tributaries. Populations below the confluence of the two, however, show evidence of admixture of three of these clusters, reflecting gene flow from the two separate upstream tributaries, and it has been shown that CB4 river systems, like the Western Cleddau (see Methods), are often characterized by recolonization from upstream source populations (Hatton-Ellis and Grieve, 2003). The WH population at the upstream extent of the Upper Western Cleddau consists almost exclusively of individuals associated with the remaining genetic cluster, which is only represented at extremely low frequencies across the remainder of the system. Interviews with local residents and gardeners confirmed that the WH population was planted next to a garden pond which had been dammed off from the main river 22 years earlier. Thus, the genetic distinctness of the WH population reflects its lack of any aquatic connection to the remaining populations, further indicating the importance of hydrochory in gene flow and dispersal in I. glandulifera.
The assignment of individuals from the BYB population on the Tempo river to genetic clusters associated with the Colebrooke system highlights the importance of the hydrogeomorphology of individual river systems and tributaries in hydrochoric dispersal. This population is situated near the confluence of the Tempo and the much larger Colebrooke river, but the distance from the point of confluence (approx. 100 m) means that backing up of water from the larger tributary is unlikely to account for the observed pattern of gene flow that contradicts the expected linear, unidirectional movement of propagules. The region is prone to flooding, however, and this is most likely responsible for the observed gene flow between the two tributaries at this point. Previous genetic (Jacquemyn et al., 2006) and ecological (Boedeltje et al., 2004; Gurnell et al., 2008; Moggridge and Gurnell, 2009) studies have indicated the importance of periodic flooding events in the dispersal of riparian plants, including invasive species (Truscott et al., 2006). The low levels of assignment of individuals to the light blue cluster in the downstream BDB population indicates that dispersal is also limited from the Tempo into the much larger Colebrook watercourse, further confirming the role of hydrological factors.
Although the results of this study strongly support hydrochoric downstream dispersal of I. glandulifera, it should be borne in mind that there are several other potential, non-exclusive processes that could also shape the observed pattern of genetic diversity in the river systems studied. First, lower upstream genetic diversity could result from founder effects as a result of human-mediated dispersal, as human population densities tend to be higher in the lower reaches of river systems. This, however, would not necessarily lead to the observed structuring of diversity between tributaries, as human-mediated dispersal would be equally likely between tributaries. Secondly, the distribution of genetic diversity might reflect a series of garden escapes, although this would not explain the increase in genetic diversity downstream. Finally, increased habitat heterogeneity downstream might lead to relaxed selection pressures, thus supporting a wider range of genotypes (Ward and Stanford, 1995). Despite these possibilities, hydrochoric dispersal would appear to be by far the most parsimonious explanation for the observed distribution of genetic diversity in populations of I. glandulifera in both river systems.
In the majority of angiosperm species, including I. glandulifera (van Went, 1984), the plastid genome is transmitted uniparentally via the female parent and thus molecular markers specific to the plastid genome can provide valuable insights into seed-mediated gene flow, including hydrochory (McCauley, 1995). In the present study, we hoped to be able to dissect the effects of pollen and seed flow by comparing patterns of genetic variation in nuclear and plastid markers. Eight chloroplast microsatellite markers were developed and utilized for this study, but failed to display any polymorphism (data not shown). Given that such markers represent the most variable regions of the plastid genome (reviewed by Provan et al., 2001; Ebert and Peakall, 2009), this lack of cytoplasmic variation, coupled with the low levels of diversity found at the nuclear loci analysed in the present study, suggests that I. glandulifera populations in Britain and Ireland have a narrow genetic base and is consistent with a limited number of introductions. Furthermore, it suggests that large-scale sequencing of the plastid genome of I. glandulifera for single nucleotide polymorphisms (SNPs) may be necessary to develop seed-specific markers for the further study of hydrochory in this species.
Implications for control of invasive riparian plant species
The primary point of control with respect to invasive species is to prevent introduction from the native range, but for already established populations, knowledge of modes and patterns of secondary dispersal will help to prevent the subsequent spread of these species (Kowarik and Samuel, 2008). The unique value of genetic approaches lies in revealing the accumulation over time of the results of multiple complex processes rather intractable to experimental ecological study in natural systems (variation in water flow, interactions between aquatic organisms, etc.). I. glandulifera can be treated with glyphosate, but the widespread use of such herbicides in riparian habitats should be viewed as an extreme measure. It has previously been suggested that targeting upstream populations might reduce the impact of propagule pressure where hydrochory plays a major role in dispersal (Dawson and Holland, 1999; Wadsworth et al., 2000; Clements et al., 2008) and the results of the present study give added weight to these recommendations. In addition, the observed genetic differentiation between tributaries would imply that separate tributaries should have their own eradication programmes, again probably focusing on upstream populations, but consideration should also be given to hydrological factors, i.e. targeting larger tributaries first as they typically represent greater sources for downstream areas, as highlighted by the fact that the predominant genetic cluster in the Tempo is barely represented below its confluence with the much larger Colebrooke. Given that intensity (i.e. the percentage of occupied area) of control is the major determinant of the cost of eradication programmes, and that control intensities of a previous modelling study were far above those typical of real programmes in the UK yet failed to control the spread of I. glandulifera, such a multi-faceted, targeted approach might offer the most rational and cost-effective means of control. This concept might also be extended to the control of other invasive riparian species that display evidence of primarily hydrochoric dispersal. The fact that we found similar results in two contrasting systems means that we can infer that our observations are probably widely applicable.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Jane Preston for information on the Tempo/Colebrooke river system, Andy Davies and Peter McEvoy for GIS assistance, and Robert Beatty for advice on statistical procedures. We are also grateful to two anonymous referees for constructive comments on the original version of the manuscript.
LITERATURE CITED
- Beerling DJ, Perrins JM. Impatiens glandulifera Royle (Impatiens roylei Walp.) Journal of Ecology. 1993;81:367–382. [Google Scholar]
- Birkeland GH. Riparian vegetation and sandbar morphology along the lower Little Colorado River, Arizona. Physical Geography. 1996;17:534–553. [Google Scholar]
- Boedeltje G, Bakker JP, Ten Brinke A, van Groenendaal JM, Soesbergen M. Dispersal phenology of hydrochorous plants in relation to discharge, seed release time and buoyancy of seeds: the flood pulse concept supported. Journal of Ecology. 2004;92:786–796. [Google Scholar]
- Burkart M. River corridor plants (Stromtalpflanzen) in Central European lowland: a review of a poorly understood plant distribution pattern. Global Ecology and Biogeography. 2001;10:449–468. [Google Scholar]
- Chen YY, Yang W, Li W, Li ZZ, Huang HW. High allozyme diversity and unidirectional linear migration patterns within a population of tetraploid Isoetes sinensis, a rare and endangered pteridophyte. Aquatic Botany. 2009;90:52–58. [Google Scholar]
- Clavero M, García-Berthou E. Invasive species are a leading cause of animal extinctions. Trends in Ecology and Evolution. 2005;20:110. doi: 10.1016/j.tree.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Clements DR, Feenstra KR, Jones K, Stainforth R. The biology of invasive alien plants in Canada. 9. Impatiens glandulifera Royle. Canadian Journal of Plant Sciences. 2008;88:403–417. [Google Scholar]
- Cloutier D, Póvoa JSR, Procopio LC, et al. Chloroplast DNA variation of Carapa guianensis in the Amazon basin. Silvae Genetica. 2005;54:270–274. [Google Scholar]
- Colautti RI, Grigorovich IA, MacIsaac HJ. Propagule pressure: a null model for biological invasions. Biological Invasions. 2006;8:1023–1037. [Google Scholar]
- Collingham YC, Wadsworth RA, Huntley B, Hulme PE. Predicting the spatial distribution of non-indigenous riparian weeds: issues of spatial scale and extent. Journal of Applied Ecology. 2000;37(S1):13–27. [Google Scholar]
- Cronk QCB, Fuller JL. Plant invaders: the threat to natural ecosystems. London: Earthscan Publications; 2001. [Google Scholar]
- Dawson FH, Holland D. The distribution in bankside habitats of three alien invasive plants in the UK in relation to the development of control strategies. Hydrobiologia. 1999;415:193–201. [Google Scholar]
- DiTomaso JM. Impact, biology and ecology of saltcedar (Tamarix spp.) in the southwest United States. Weed Technology. 1998;12:326–336. [Google Scholar]
- Earl DA, vonHoldt BM. Structure Harvester: a website and program for visualizing Structure output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Ebert D, Peakall R. Chloroplast simple sequence repeats (cpSSRs): technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Molecular Ecology Resources. 2009;9:673–690. doi: 10.1111/j.1755-0998.2008.02319.x. [DOI] [PubMed] [Google Scholar]
- Eschtruth AK, Battles JJ. Assessing the relative importance of disturbance, herbivory, diversity and propagule pressure in exotic plant invasions. Ecological Monographs. 2009;79:265–280. [Google Scholar]
- Evanno G, Regnaud S, Goudet J. Detecting the number of clusters of individuals using the software Structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes – application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval LG, Schneider S. Arlequin, Version 3·0: an integrated software package for population genetic data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Gornall RJ, Hollingsworth PM, Preston CD. Evidence for spatial structure and directional gene flow in a population of an aquatic plant Potamogeton coloratus. Heredity. 1998;80:414–421. [Google Scholar]
- Goudet J. Fstat, version 2·9.3, A program to estimate and test gene diversities and fixation indices. 2001 http://www2.unil.ch/popgen/softwares/fstat.htm . [Google Scholar]
- Graf WL. Tamarisk and river channel management. Environmental Management. 1982;6:283–296. [Google Scholar]
- Gurevitch J, Padilla DK. Are invasive species a major cause of extinctions? Trends in Ecology and Evolution. 2004;19:470–474. doi: 10.1016/j.tree.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Gurnell A, Thompson K, Goodson J, Moggridge H. Propagule deposition along rivers: linking hydrology and ecology. Journal of Ecology. 2008;96:553–565. [Google Scholar]
- Hatton-Ellis TW, Grieve N. Peterborough: English Nature; 2003. Ecology of watercourses characterized by Ranunculion fluitantis and Callitricho-Batrachion vegetation. Conserving Natura 2000 Rivers Ecology Series Number 11. [Google Scholar]
- Hejda M, Pyšek P, Jarošik V. Impact of invasive plants on the species richness, diversity and composition of invaded communities. Journal of Ecology. 2009;97:393–403. [Google Scholar]
- Henderson S, Dawson TP, Whittaker RJ. Progress in invasive plants research. Progress in Physical Geography. 2006;30:25–46. [Google Scholar]
- Honnay O, Jacquemyn H, Nackaerts K, Breyne P, van Looy K. Patterns of population genetic diversity in riparian and aquatic plant species along rivers. Journal of Biogeography. 2010;37:1730–1739. [Google Scholar]
- Hupp CR, Osterkamp WR. Riparian vegetation and fluvial geomorphic processes. Geomorphology. 1996;14:277–295. [Google Scholar]
- Jacquemyn H, Honnay O, van Looy K, Breyne P. Spatiotemporal structure of genetic variation of a spreading plant population on dynamic riverbanks along the Meuse River. Heredity. 2006;96:471–478. doi: 10.1038/sj.hdy.6800825. [DOI] [PubMed] [Google Scholar]
- Kowarik I, Samuel I. Water dispersal as an additional pathway to invasions by the primarily wind-dispersed tree Ailanthus altissima. Plant Ecology. 2008;198:241–252. [Google Scholar]
- Liu Y, Wang Y, Huang H. High interpopulation genetic differentiation and unidirectional linear migration patterns in Myricaria laxiflora (Tamaricaceae), an endemic riparian plant in the Three Gorges Valley of the Yangtze River. American Journal of Botany. 2006;93:206–215. doi: 10.3732/ajb.93.2.206. [DOI] [PubMed] [Google Scholar]
- Lockwood JL, Cassey P, Blackburn T. The role of propagule pressure in explaining species invasions. Trends in Ecology and Evolution. 2005;20:223–228. doi: 10.1016/j.tree.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Loope LL, Sanchez PG, Tarr PW, Loope WL, Anderson RL. Biological invasions of arid land nature reserves. Biological Conservation. 1988;44:95–118. [Google Scholar]
- Lopezaraiza-Mikel ME, Hayes RB, Whalley MR, Memmott J. The impact of an alien plant on a native plant-pollinator network: an experimental approach. Ecology Letters. 2007;10:539–550. doi: 10.1111/j.1461-0248.2007.01055.x. [DOI] [PubMed] [Google Scholar]
- Lundqvist E, Andersson E. Genetic diversity in populations of plants with different breeding and dispersal strategies in a free-flowing boreal river system. Hereditas. 2001;135:75–83. doi: 10.1111/j.1601-5223.2001.00075.x. [DOI] [PubMed] [Google Scholar]
- McCauley DE. The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trends in Ecology and Evolution. 1995;10:198–202. doi: 10.1016/s0169-5347(00)89052-7. [DOI] [PubMed] [Google Scholar]
- McKinney ML, Lockwood JL. Biotic homogenization: a few winners replacing many losers in the next mass extinction. Trends in Ecology and Evolution. 1999;14:450–453. doi: 10.1016/s0169-5347(99)01679-1. [DOI] [PubMed] [Google Scholar]
- Markwith SH, Scanlon MJ. Multiscale analysis of Hymenocallis coronaria (Amaryllidaceae) genetic diversity, genetic structure and gene movement under the influence of unidirectional stream flow. American Journal of Botany. 2007;94:151–160. doi: 10.3732/ajb.94.2.151. [DOI] [PubMed] [Google Scholar]
- Martínez-Ghersa MA, Ghersa CM. The relationship of propagule pressure to invasion potential in plants. Euphytica. 2006;148:87–96. [Google Scholar]
- Moggridge HL, Gurnell AM. Hydrological controls on the transport and deposition of plant propagules within riparian zones. River Research and Applications. 2009;26:512–527. [Google Scholar]
- Myers JH, Bazely D. Ecology and control of introduced plants. Cambridge: Cambridge University Press; 2003. [Google Scholar]
- Nakayama N, Nishihiro J, Kayaba Y, Muranaka T, Washitani I. Seed deposition of Eragrostis curvula, an invasive alien plant on a river floodplain. Ecological Research. 2007;22:696–701. [Google Scholar]
- Pearson DE. Invasive plant architecture alters trophic interactions by changing predator abundance and behavior. Oecologia. 2009;159:549–558. doi: 10.1007/s00442-008-1241-5. [DOI] [PubMed] [Google Scholar]
- Pimentel D, McNair S, Janecka J, et al. Economic and environmental threats of alien plant, animal, and microbe invasions. Agriculture, Ecosystems and Environment. 2001;84:1–20. [Google Scholar]
- Pollux BJA, Luteijn A, van Groenendael JM, Ouborg NJ. Gene flow and genetic structure of the aquatic macrophyte Sparganium emersum in a linear unidirectional river. Freshwater Biology. 2009;54:64–76. [Google Scholar]
- Prentis PJ, Mather PB. Micro-geographic landscape features demarcate seedling genetic structure in the stream lily, Helmholtzia glaberrima. Aquatic Botany. 2007;87:111–115. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provan J, Powell W, Hollingsworth PM. Chloroplast microsatellites: new tools for studies in plant ecology and systematics. Trends in Ecology and Evolution. 2001;16:142–147. doi: 10.1016/s0169-5347(00)02097-8. [DOI] [PubMed] [Google Scholar]
- Provan J, Love HM, Maggs CA. Development of microsatellites for the invasive riparian plant Impatiens glandulifera (Himalayan balsam) using intersimple sequence repeat cloning. Molecular Ecology Notes. 2007;7:451–453. [Google Scholar]
- Pyšek P, Prach K. Plant invasions and the role of the riparian habitats: a comparison of four species. Journal of Biogeography. 1993;20:413–420. [Google Scholar]
- Raymond M, Rousset F. GenePop (version 1·2): population genetic software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Ritland K. Genetic differentiation, diversity and inbreeding in the mountain monkeyflower (Mimulus caespitosus) of the Washington cascades. Canadian Journal of Botany. 1989;67:2017–2024. [Google Scholar]
- Russell JR, Weber JC, Booth A, Powell W, Sotelo-Montes C, Dawson IK. Genetic variation of Calycophyllum spruceanum in the Peruvian Amazon Basin, revealed by amplified fragment length polymorphism (AFLP) analysis. Molecular Ecology. 1999;8:199–204. doi: 10.1046/j.1365-294x.1999.00589.x. [DOI] [PubMed] [Google Scholar]
- Sakai AK, Allendorf FW, Holt JS, et al. The population biology of invasive species. Annual Review of Ecology and Systematics. 2001;32:305–332. [Google Scholar]
- Samuel I, Kowarik I. Urban rivers as dispersal corridors for primarily wind-dispersed invasive tree species. Landscape and Urban Planning. 2010;94:244–249. [Google Scholar]
- Schleuning M, Becker T, Vadillo GP, Hahn T, Matthies D, Durka W. River dynamics shape clonal diversity and genetic structure of an Amazonian understorey herb. Journal of Ecology. 2011;99:373–382. [Google Scholar]
- Simberloff D. The role of propagule pressure in biological invasions. Annual Review of Ecology, Evolution and Systematics. 2009;40:81–102. [Google Scholar]
- Tassin J, Riviere JN, Clergeau P. Reproductive versus vegetative recruitment of the invasive tree Schinus terebenthifolius: implications for restoration on Reunion Island. Restoration Ecology. 2007;15:412–419. [Google Scholar]
- Tero N, Aspi J, Siikamäki P, Jäkäläniemi A, Tuomi J. Genetic structure and gene flow in a metapopulation of an endangered plant species Silene tatarica. Molecular Ecology. 2003;12:2073–2085. doi: 10.1046/j.1365-294x.2003.01898.x. [DOI] [PubMed] [Google Scholar]
- Thijs KW, Brys R, Verboven HAF, Hermy M. The influence of an invasive plant species on the pollination success and reproductive output of three riparian plant species. Biological Invasions. 2012;14:355–365. [Google Scholar]
- Tickner DP, Angold PG, Gurnell AM, Mountford JO. Riparian plant invasions: hydrogeomorphological control and ecological impacts. Progress in Physical Geography. 2001;25:22–52. [Google Scholar]
- Truscott A-M, Soulsby C, Palmer SCF, Newell L, Hulme PE. The dispersal characteristics of the invasive plant Mimulus guttatus and the ecological significance of increased occurrence of high-flow events. Journal of Ecology. 2006;94:1080–1091. [Google Scholar]
- van der Wal R, Truscott AM, Pearce ISK, Cole L, Harris MP, Wanless S. Multiple anthropogenic changes cause biodiversity loss through plant invasion. Global Change Biology. 2008;14:1428–1436. [Google Scholar]
- van Went JL. Unequal distribution of plastids during vegetative cell formation in Impatiens. Theoretical and Applied Genetics. 1984;68:305–310. doi: 10.1007/BF00267882. [DOI] [PubMed] [Google Scholar]
- Wadsworth RA, Collingham YC, Willis SG, Huntley B, Hulme PE. Simulating the spread and management of alien riparian weeds: are they out of control? Journal of Applied Ecology. 2000;37(S1):28–38. [Google Scholar]
- Walker NF, Hulme PE, Hoelzel AR. Population genetics of an invasive riparian species, Impatiens glandulifera. Plant Ecology. 2009;203:243–252. [Google Scholar]
- Ward JV, Stanford JA. The serial discontinuity concept: extending the model to floodplain rivers. Regulated Rivers: Research & Management. 1995;10:159–168. [Google Scholar]
- Weber E. Invasive plant species of the World: a reference guide to environmental weeds. Wallingford, UK: CABI Publishing; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.