Abstract
Background
Auxin is a versatile plant hormone with important roles in many essential physiological processes. In recent years, significant progress has been made towards understanding the roles of this hormone in plant growth and development. Recent evidence also points to a less well-known but equally important role for auxin as a mediator of environmental adaptation in plants.
Scope
This review briefly discusses recent findings on how plants utilize auxin signalling and transport to modify their root system architecture when responding to diverse biotic and abiotic rhizosphere signals, including macro- and micro-nutrient starvation, cold and water stress, soil acidity, pathogenic and beneficial microbes, nematodes and neighbouring plants. Stress-responsive transcription factors and microRNAs that modulate auxin- and environment-mediated root development are also briefly highlighted.
Conclusions
The auxin pathway constitutes an essential component of the plant's biotic and abiotic stress tolerance mechanisms. Further understanding of the specific roles that auxin plays in environmental adaptation can ultimately lead to the development of crops better adapted to stressful environments.
Keywords: Abiotic stress, arabidopsis, auxin, biotic stress, hormone crosstalk, lateral root development, plant hormones
INTRODUCTION
Plants are extremely flexible organisms adaptable to a range of diverse environments. Their intrinsic ability to simultaneously inhabit both above- and below-ground domains makes them unique among most other living organisms, which occupy a single habitat at a given time. In response to diverse environmental signals, plants modify their development through the perception and integration of exogenous signals into the signalling pathways of plant hormones. Auxin is one of the most versatile plant hormones and plays essential roles in growth and development. The revelation of the existence of an auxin biosynthesis, signalling and transport apparatus in single-celled green algae is a clear indication that auxin has played an important evolutionary role during the adaptation of plants to diverse land environments (De Smet et al., 2010). In recent years, significant progress has been made towards understanding how this hormone regulates plant growth and development. However, less is known about the roles of auxin as a regulator of biotic and abiotic stress responses. In this review, after a brief account of auxin biosynthesis, signalling and transport, interesting new insights into the role of auxin as an integrator of environmental signals are highlighted.
AUXIN BIOSYNTHESIS, SIGNALLING AND TRANSPORT
Auxin biosynthesis, signalling and transport processes, which are particularly relevant to the findings discussed in this article, have been extensively reviewed elsewhere (Vanneste and Friml, 2009; Tromas and Perrot-Rechenmann, 2010; Zhao, 2010; Finet and Jaillais, 2012; Mano and Nemoto, 2012; Rosquete et al., 2012; Swarup and Péret, 2012). Therefore, only a brief account of these processes will be presented here.
Auxin biosynthesis
As reviewed extensively elsewhere (Zhao, 2010; Mano and Nemoto, 2012; Rosquete et al., 2012; Sauer et al., 2013), auxin [indole-3-acetic acid (IAA)], is produced in meristematic tissues through tryptophan-dependent and -independent biosynthetic pathways. So far, three tryptophan-dependent auxin biosynthetic pathways, named after the intermediate compounds generated in each pathway, have been identified. These include the IPA (indole-3-pyruvic acid) pathway, the IAM (indole-3-acetamide) pathway, the TAM (tryptamine) pathway and the IAOx (indole-3-acetaldoxime) pathway. To maintain optimal concentrations and ratios of IAA and IAA derivatives in plant tissues, auxin homeostasis is regulated by processes such as degradation, conjugation to amino acids and transport. The apparent complexity and redundancy of auxin biosynthesis, signalling and transport seem to indicate the absolute requirement of this hormone for multiple plant processes.
Auxin signalling
In recent years, tremendous progress has been made towards dissecting the signalling pathway of auxin (reviewed by Vanneste and Friml, 2009; Tromas and Perrot-Rechenmann, 2010; Swarup and Péret, 2012). Briefly, at low auxin concentrations, auxin responses are suppressed by AUX/IAA (AUXIN/INDOLE-3-ACETIC ACID) proteins. AUX/IAA proteins repress AUXIN RESPONSE FACTORS (ARFs), a class of transcription factors that regulate auxin-responsive gene expression. Auxin perception by TIR1 (TRANSPORT INHIBITOR RESPONSE 1) and related AUXIN F-BOX (AFB) proteins AFB1, AFB2 and AFB3 leads to the degradation of AUX/IAAs by the 26S proteasome and the subsequent release of ARFs from suppression. By binding to the auxin-responsive element (ARE) commonly found in the promoters of auxin-responsive genes, ARFs regulate (activate or repress) auxin-responsive gene expression, leading to a variety of auxin-mediated phenotypic alterations.
Auxin transport
Plant processes involved in auxin transport have also been extensively reviewed (Vanneste and Friml, 2009; Spalding, 2013). Briefly, auxin synthesized in aerial tissues (e.g. apical meristems) is transported locally and systemically throughout the plant. The cell-to-cell active movement of auxin is known as polar auxin transport (PAT), as opposed to the direct and rapid transport of auxin from shoots to roots through the phloem. Two general classes of transporters involved in PAT are auxin influx carriers such as AUX1 (AUXIN RESISTANT 1) and LAX1 (like aux1),which pump auxin into the cell, and auxin efflux carriers such as the PIN (PIN-FORMED) and ATP-binding cassette type B (ABCB) families [also known as the multidrug resistant proteins or P-glycoproteins (MDR-PGPs)], which pump auxin out of the cell. The specific localization of PIN proteins within the cell influences the direction of auxin transport (see below). More recently another class of auxin carriers, called PILS (PIN-LIKES), has been identified based on their structural similarity to the PIN family (Barbez et al., 2012).
Lateral root development
One of the plant processes regulated by auxin is lateral root development, which has been extensively reviewed elsewhere (Osmont et al., 2007; Nibau et al., 2008; Péret et al., 2009; Overvoorde et al., 2010; Smith and De Smet, 2012; Petricka et al., 2012) and will not be discussed here in detail. Briefly, upon germination, young seedlings contain only primary roots formed directly from the radicle present in the embryo. Lateral root primordia originate from pericycle cells located in front of the xylem of primary roots. Additional lateral roots reiteratively generated during root growth play essential roles in the water and nutrient uptake required to sustain proper plant growth and development. Auxin biosynthesis, signalling and transport are required for lateral root formation since auxin mutants show reduced or defective lateral root production, and exogenous treatment of pericycle cells with auxin promotes lateral root formation. Over recent years, research into lateral root development has introduced a new paradigm whereby the role of auxin as a key regulator of plant root architecture in response to environmental stimuli has been studied (Malamy, 2005). In this review I will briefly highlight some of the recent studies in this exciting research area.
AUXIN- AND NUTRIENT-DEPENDENT ALTERATIONS IN ROOT ARCHITECTURE
Given the importance of macro- and micro-nutrient elements for plant growth and development, it is not surprising that plant roots have developed unique capabilities to sense and respond to nutrients available in soil. Emerging evidence implicates auxin as one of the main players involved in this essential adaptive response. Since different plant species may respond differently to the lack or excess of a particular nutrient (Niu et al., 2012), for consistency this review will discuss mainly examples from the dicot model plant arabidopsis (Arabidopsis thaliana).
Nitrogen
Nitrogen (N) is an essential element taken up mostly in the form of nitrate (NO3−) from the soil solution. It has long been known that nitrogen deficiency leads to directional root growth towards nitrogen-rich regions of the soil (Zhang and Forde, 2000). Until recently how this nutritional signal is integrated into plant root development had remained elusive. So far, a number of plasma membrane-located transporters involved in nitrate uptake have been identified. One of such transporters, the arabidopsis NITRATE TRANSPORTER1·1 (NRT1·1) protein (also known as CHL1), has dual functionality as a high-activity nitrate influx carrier and nitrate sensor (Ho et al., 2009). At high nitrate concentrations, the chl1/nrt1·1 mutant shows decreased lateral root proliferation relative to wild-type, suggesting that the nitrate-sensing mechanism is compromised in this mutant (Remans et al., 2006). Remarkably, recent research has provided a mechanistic new insight into the role of NRT1·1 in lateral root development. It appears that under low NO3− concentrations NRT1·1 promotes basipetal auxin transport (from the lateral root tip shootward) to inhibit auxin accumulation in lateral root initials. At high NO3− levels, NRT1·1-dependent auxin transport out of the lateral roots is inhibited, leading to the accumulation of auxin in lateral root initials and promotion of lateral root growth (Krouk et al., 2010; Gojon et al., 2011; Bouguyon et al., 2012). Therefore, it appears that auxin transport and nitrate uptake and sensing mechanisms are linked so that rapid alterations in root architecture can be achieved during adaptive responses to this essential nutrient.
Potassium
Similar to the dual roles of NRT1·1, the arabidopsis TRH1 (TINY ROOT HAIR 1) gene, which encodes a KT/KUP/HAK family protein, has been proposed to play a role in both auxin and potassium (K+) transport (Vicente-Agullo et al., 2004). TRH1 is expressed in root-cap cells known to be involved in gravity perception and auxin redistribution (Vicente-Agullo et al., 2004). The arabidopsis trh1 mutant shows defects in root hair development and gravitropism and a reduced ability to transport K+ (Rigas et al., 2001). These phenotypic defects could be restored by an exogenous auxin supply, suggesting that auxin is involved in TRH1-mediated root development (Vicente-Agullo et al., 2004). Indeed, recent research has shown that TRH1 regulates auxin transport by influencing the localization of the auxin efflux protein PIN1 (Rigas et al., 2013), although, as highlighted by Dolan (2013), additional research is required to further dissect the possible roles of TRH1 in auxin and K+ transport. Recent transcriptome analyses of rice roots during K+ deficiency have also identified a large number of auxin-related genes (Ma et al., 2012), further suggesting that auxin regulates root responses to this nutrient.
Phosphorus
Plants also alter their root development in response to low phosphate (Pi) (López-Bucio et al., 2002, 2005; Nacry et al., 2005; Svistoonoff et al., 2007; Niu et al., 2012). Such alterations include the reduction of primary root growth but promotion of lateral root development to facilitate exploration of the rhizosphere for new nutrient sources. The response to Pi deficiency of the auxin signalling mutant axr1 is similar to that of wild-type plants (Williamson et al., 2001). Also, in contrast to N deficiency, which alters lateral root development through alteration of auxin transport, no increase in free auxin levels or auxin transport was found in the roots of Pi-deprived seedlings (Pérez-Torres et al., 2008). However, pericyle cells in the primary roots of Pi-deprived seedlings show increased sensitivity to exogenous auxin as determined by analysis of expression of the DR5-GUS synthetic auxin reporter in these cells (Pérez-Torres et al., 2008). As mentioned above, lateral root primordium originates from pericycle founder cells and the increased sensitivity of these cells to auxin under low-Pi conditions suggests that Pi starvation primes these cells to produce lateral roots. The sensitivity of lateral root formation to Pi deficiency was reduced in the tir1 mutant and nearly completely lost in the tir1 afb2 afb3 triple auxin receptor mutant. Furthermore, in the absence of Pi, the AUX/IAA protein AXR3/IAA7 shows an increased degradation rate (Pérez-Torres et al., 2008). Together, these findings suggest that functional auxin perception is required for proper root responses to Pi.
Another link between auxin transport and Pi-mediated lateral root development has recently been uncovered through the analysis of the arabidopsis siz1 mutant. SIZ1, which encodes a SUMO E3 ligase, was previously identified as a negative regulator of lateral root development under Pi-deficient conditions (Miura et al., 2005). The siz1 mutant displays extreme sensitivity to Pi deficiency with a reduction in primary root but an increase in lateral root development. This response appears to occur through earlier accumulation of auxin in siz1 roots (Miura et al., 2011). N-1-naphthylphthalamic acid (NPA), a chemical inhibitor of auxin (efflux) activity, reverses the phenotypes caused by the siz1 mutation under Pi-deficient conditions. This suggests that SIZ1 is a negative regulator of auxin transport and lateral root development in plants grown under Pi-deficient conditions (Miura et al., 2011).
Other nutrient elements
Metal ions such as copper (Cu2+), aluminium (Al3+), iron (Fe), boron (B) and cadmium (Cd) also cause drastic alterations in lateral root development, elongation and overall plant root architecture (Lequeux et al., 2010; Aquea et al., 2011; Martín-Rejano et al., 2011; Peto et al., 2011; Giehl et al., 2012; Hu et al., 2013; Yuan et al., 2013). Altered auxin biosynthesis, signalling and/or transport seem to have an underlying effect on these alterations as transcriptional activation of auxin-related genes and increased auxin levels were observed in roots exposed to various concentrations of these metals (Mattiello et al., 2010; Lequeux et al., 2010; Giehl et al., 2012). For instance, genetic analyses show that root growth is inhibited less by Al3+ in aux1 and pin2 mutants than in wild-type plants. Chemical inhibition of auxin transport also reduces the negative effects of Al3+ on root growth (Sun et al., 2010). Similarly, the AUX1 auxin transporter is required for Fe-triggered lateral root elongation (Giehl et al., 2012). Sulphur (S) deficiency also suppresses lateral root development in wild-type arabidopsis, while this response is compromised in the axr1–3 mutant (Dan et al., 2007).
Together, these findings are consistent with the view that auxin signalling and transport play important roles in regulating root responses to soil nutrients.
AUXIN-MEDIATED ALTERATIONS OF ROOT ARCHITECTURE DURING ABIOTIC STRESS ADAPTATION
Low-temperature stress
During gravitropic responses, asymmetrical localization of auxin to one side of the cell through the action of auxin transport proteins, in particular PINs, redirects the root growth towards the centre of gravity (Friml, 2010). Exposure of roots to cold inhibits such gravity responses in arabidopsis (Rahman, 2013). In cold-exposed root cells, trafficking and lateral localization of the auxin efflux proteins PIN2 and PIN3 is inhibited until the seedlings are returned to normal growth temperatures (Shibasaki et al., 2009). Possible roles of auxin as a regulator of cold stress responses of arabidopsis have recently been reviewed (Rahman, 2013) and will not be discussed here in detail.
Water stress
The importance of roots during water stress has been studied in much less detail compared with above-ground parts. Nevertheless, plant roots are capable of sensing and responding to the presence of moisture in soil, a phenomenon known as hydrotropism. The auxin response pathway, but not auxin transport, appears to play a role in hydrotropism (Kaneyasu et al., 2007). The gravitropic and hydrotropic responses are antagonistic, and the overall involvement of auxin in hydrotropism is predicted to be less than that on gravitropism (Cassab et al., 2013). Hydrotropic root responses seem to be mainly regulated by abscisic acid (ABA) signalling, which can overcome the effect of auxin-mediated responses (e.g. gravitropism) when these two forces are at odds to facilitate the extension of roots towards moist regions of the soil profile (Taniguchi et al., 2010). Under moderate water stress conditions, ABA is required for maintaining primary root and root hair growth by modulating auxin transport in both arabidopsis and rice. This response is also accompanied by an enhanced proton secretion process through the action of plasma membrane-located H-ATPases, which are essential for maintaining root elongation (Xu W et al., 2013). Further research is needed to dissect the complex interactions between auxin and other plant hormones that may be at play during water stress- or drought-mediated alterations in root architecture.
Recently, an important role for arabidopsis IAR3 (IAA-Ala Resistant3) as a modulator of root architecture during osmotic stress has been shown (Kinoshita et al., 2012). IAR3 is a hydrolase capable of generating free auxin by hydrolysing an inactive auxin form such as an IAA–amino acid conjugate (e.g. IAA-Ala) (Rampey et al., 2004). It was therefore proposed that, under drought stress, IAR3 generates bioactive auxin which then stimulates lateral root development and contributes to survival under drought stress (Kinoshita et al., 2012).
Genetic screens designed to identify mutants compromised in hydrotropism have also identified MIZU-KUSSEI (MIZ)1, encoding a novel protein in arabidopsis. Roots of miz1 mutants grow vertically towards gravity, but do not show curved growth towards moisture (Kobayashi et al., 2007). MIZ1 over-expression lines show earlier and more pronounced growth curvature towards moisture and reduced primary root elongation, lateral root development and auxin levels (Miyazawa et al., 2012).
Genome-wide expression analyses of plants under water stress often reveal differentially expressed auxin-responsive genes. For instance, genes encoding various members of the ARF transcription factor family are differentially expressed during dehydration stress in soybean roots, leading to the suggestion that these genes may be potential candidates for the generation of soybeans with increased drought tolerance (Ha et al., 2013).
Salt stress
Salinity is an abiotic stress that severely affects plant and root development. A mild salt stress results in a drastic reduction of lateral root elongation but an increase in lateral root numbers, while higher salt levels completely inhibit root elongation (Zolla et al., 2010). Increased lateral root numbers due to salt stress were reduced in auxin signalling mutants axr1, axr4 and tir1, and completely blocked in the auxin influx mutant aux1 (Wang et al., 2009; Zolla et al., 2010).
As stated above, uneven distribution of auxin efflux carriers, in particular PIN2, in the cells of the root elongation zone is known to control gravitropic root responses through the regulation of basipetal auxin transport (reviewed by Vanneste and Friml, 2009). Salt stress, in addition to altering PIN2 cellular localization, inhibits PIN2 expression (Sun et al., 2008). Therefore, like cold stress, salt stress interferes with root gravitropism, which appears to be an adaptive response to reduce the damaging effects of salt stress (Galvan-Ampudia and Testerink, 2011).
The SOS (SALT OVERLY SENSITIVE) pathway characterized by molecular genetic analysis of several mutants (e.g. sos1, sos2 and sos3) is required for salt tolerance in arabidopsis (Ji et al., 2013). The tolerance provided by this pathway seems to occur at least partly through the promotion of lateral root development as transgenic plants over-expressing SOS genes show both increased salt tolerance and lateral root development under salt stress (Yang et al., 2008). Lateral root development in the sos3 mutant shows increased inhibition (salt sensitivity) at low salt concentrations, accompanied by reduced auxin levels in lateral root primordia. The reduced lateral root emergence observed in sos3 appears to be due to the reduced shoot-to-root (acropetal) auxin transport as well as reduced basipetal auxin transport within the roots. The reduced expression of the auxin efflux protein PIN2 in the roots of sos3 plants supports this proposal (Zhao et al., 2011). These findings indicate that auxin signalling and influx are both required for lateral root development under salt stress.
pH
Soil acidity or pH directly and indirectly influences root development. Plant roots respond to changes in soil pH with massive transcriptional alterations in the expression of a large number of auxin-responsive genes, suggesting that pH-mediated changes in root architecture are at least partly mediated by auxin (Lager et al., 2010). Plant roots grown under alkaline conditions (e.g. pH 8) show increased auxin transport activity mediated by PIN2 (Xu W et al., 2012). PIN2-transported auxin is also required for the activation of plasma membrane H+-ATPase-mediated proton secretion from the root tips. This process appears to be essential in acidifying the environment around the roots and maintaining primary root growth (Xu W et al., 2012). Additional studies are certainly required to dissect the effect of pH on root development and the potential roles of auxin signalling and transport in the integration of pH-mediated effects into root architecture.
Waterlogging
Submergence or waterlogging is another abiotic stress factor that negatively affects root development by restricting the O2 supply to the roots. Plants tend to develop adventitious roots when submerged to alleviate the negative effects of this stress on plant growth and development. The accumulation of auxin in the base of the stem promotes adventitious root formation. In the tomato, adventitious root formation requires auxin sensitivity as the formation of these roots is inhibited in submerged tomato plants compromised in auxin sensing (Vidoz et al., 2010). In addition, chemical inhibition of PAT inhibits adventitious root formation, suggesting that auxin transport is required for their formation (Vidoz et al., 2010). Furthermore, recent evidence discussed by Muday et al. (2012) indicates that the stress hormone ethylene, which accumulates in waterlogged plants, can contribute to the regulation of lateral and adventitious root formation in a complex crosstalk with auxin.
Redox status/reactive oxygen
The redox status of cells is altered during biotic and abiotic stress responses and affects auxin signalling and lateral root development. The arabidopsis triple mutant ntra ntrb cad2, which lacks the key components of the thioredoxin and glutaredoxin signalling involved in redox regulation, shows compromised auxin transport (Bashandy et al., 2010). Nitric oxide accumulates in response to auxin treatment during lateral root formation and chemical inhibition of nitric oxide accumulation abolishes lateral root formation. Nitric oxide is also required for normal operation of auxin signalling by promoting the degradation of AUX/IAAs (Terrile et al., 2012). During cadmium stress, rice root growth is regulated by an interplay between reactive oxygen species (ROS) (H202) and auxin signalling, in which increased ROS alters the expression of key auxin signalling components (Zhao et al., 2012). Together, these findings point to an essential link between the signalling pathways of auxin and ROS during the adaptive responses to stress.
The involvement of auxin signalling in salt and oxidative stress tolerance seems to occur, at least in part, through modulation of the cellular redox status. The primary root growth of tir1 afb2 seedlings show reduced sensitivity to salt (Iglesias et al., 2010). Similarly, tir1 afb2 and tir1 afb3 double receptor mutants exhibit a higher percentage of primary root elongation and reduced H202-induced cell death than wild-type roots under oxidative stress. To explain this phenomenon, it was proposed that the mutant seedlings may have reduced levels of endogenous ROS. Indeed, higher levels of anti-oxidant enzymes such as CAT (CATALASE) and APX (ASCORBATE PEROXIDASE) were detected in salt-stressed tir1 abf2 plants. Increased activities of these ROS-degrading enzymes also correlated with transient induction of GST1, encoding glutathione S-transferase 1 (GST1), APX1, encoding a cytosolic ascorbate peroxidase, and ZAT12, encoding a zinc finger transcription factor in response to salt stress (Iglesias et al., 2010). These findings have led to the suggestion that under stress conditions auxin signalling promotes the production of ROS, which potentiates tissue damage. Therefore, an attenuated auxin signalling pathway may be a strategy employed by plants to enhance tolerance to ROS-generating abiotic stresses (Iglesias et al., 2010).
BIOTIC FACTORS AND AUXIN-MEDIATED ALTERATIONS OF PLANT ROOT ARCHITECTURE
In a complex environment like soil, plant roots encounter many living organisms, such as symbiotic/endophytic bacteria and fungi as well as bacterial and fungal pathogens, nematodes, insects and even parasitic plants. In the following sections, recent findings on how diverse biotic signals alter root development by directly or indirectly modifying auxin signalling and transport will be briefly reviewed.
Beneficial microbes
Many plant species establish symbiotic relationships with fungi such as arbuscular mycorrhiza (AM). AM enter plant roots through lateral roots and manipulates the plant's root system architecture, at least partly through the host auxin signalling pathway (Hanlon and Coenen, 2011; Sukumar et al., 2013). Arabidopsis is not susceptible to infection by AM and therefore the interaction between AM and plant roots has been mostly studied in species other than arabidopsis. Nevertheless, arabidopsis roots can recognize the signals generated by AM. For instance, the mycorrhizal fungus Laccaria bicolor can induce lateral root development after indirect contact with arabidopsis roots (Felten et al., 2009). Chemical or genetic inhibition of PAT through NPA treatment or the use of the pin2 mutant leads to a dramatic reduction in L. bicolor-mediated lateral root induction during this interaction. Similar results were also reported for the interaction between arabidopsis roots and the mycorrhizal fungi known as truffles (Tuber borchii and T. melanosporum) (Splivallo et al., 2009). Furthermore, Trichoderma virens, a plant-beneficial fungus, promotes lateral root growth in arabidopsis through an auxin signalling- and transport-dependent mechanism. The lateral root-promoting ability of T. virens is attenuated in the auxin transport or signalling mutants aux1, big1, eir1/pin2 and axr1 (Contreras-Cornejo et al., 2009). It should be noted that AM can also produce auxin and/or auxin-like compounds that can potentially contribute to the alterations observed in root development. For instance, the growth-promoting effects of the beneficial fungus Piriformospora indica on arabidopsis appears to be mediated through a highly branched root system promoted by fungally produced auxin (Sirrenberg et al., 2007).
Root inoculation of arabidopsis with the rhizobacterium Phyllobacterium brassicaceae leads to a 50 % increase in lateral root growth, while this effect is abolished in aux1 and axr1 mutants, again suggesting that auxin signalling and transport are both required for this effect (Contesto et al., 2010). Similarly, beneficial Pseudomonas, which promotes plant growth, inhibits primary root elongation while promoting lateral root formation in arabidopsis in an auxin-dependent manner, as the auxin receptor mutant tir1 afb2 afb3 mutant shows insensitivity to Pseudomonas-stimulated lateral root formation (Zamioudis et al., 2013). The nitrogen-fixing nodules formed during the interaction of plant roots with the beneficial soil bacterium Frankia are structurally and developmentally related to lateral roots (Pawlowski et al., 2011). The involvement of auxin in the formation of root nodules in legumes by rhizobia has recently been reviewed (Mathesius, 2010) and will not be discussed here.
Pathogenic microbes
Plant roots are constantly exposed to a variety of soil-inhabiting organisms, including pathogenic bacteria and fungi that modify the plant's root architecture in an auxin-dependent manner. The root-infecting pathogenic bacterium Ralstonia solanacearum, for instance, reduces the formation of lateral roots in petunia (Zolobowska and Van Gijsegem, 2006). Similarly, the soil-borne fungal pathogen Fusarium oxysporum infects arabidopsis through lateral root initials (Kidd et al., 2011). Several arabidopsis auxin signalling and transport mutants in which lateral root development is known to be altered show increased resistance to F. oxysporum, indicating the possible involvement of the host's auxin signalling and transport pathways in the infection process (Kidd et al., 2011).
Recognition of the conserved molecules collectively known as microbe-associated molecular patterns (MAMPs) by pattern recognition receptors (PRRs) elicits developmental alterations that may be executed through the auxin pathway. For instance, flg22, a conserved MAMP from the flagellin of the bacterial pathogen Pseudomonas syringae, inhibits root growth in arabidopsis (Gómez-Gómez et al., 1999). This effect is most likely mediated by flg22-responsive microRNA393, a known inhibitor of auxin receptors TIR1, AFB2 and AFB3 (Navarro et al., 2006). As expected, the inhibition of auxin receptor gene expression leads to the stabilization of IAA/AUX repressors, which act as repressors of auxin-responsive genes, including those involved in lateral root development.
Oligogalacturonides, elicitors derived from plant cell wall hydrolysis during parasitism, reduce primary root cell elongation and development but promote lateral root formation in an auxin-dependent manner (Hernández-Mata et al., 2011). Interestingly, in contrast to flg22-mediated effects, the stabilization of IAA/AUX repressors is not required for the antagonistic interaction between oligogalacturonides and auxin signalling (Savatin et al., 2011). It was proposed that the effect of oligogalacturonides on auxin signalling is at least partly due to the promoting effects of oligogalacturonides on flavonoids (Savatin et al., 2011), known inhibitors of auxin transport (Brown et al., 2001). Supporting this view, auxin signalling (e.g. tir1) and transport (e.g. doc1, pgp1, pgp4, pgp19 and tt4) mutants show altered responses to oligogalacturonides (Hernández-Mata et al., 2011).
Nematodes
Nematodes are soil-dwelling organisms that cause significant damage to many crop plants. Root-knot nematodes (Meloidogyne spp.) induce multinucleated giant cells in infected roots while cyst nematodes (e.g. Heterodera sp.) modify root cells to form specialized cells called syncytia. Like lateral roots, these nematode-induced root cells originate from pericyle cells, and their formation requires components of the host's auxin signalling and transport pathways (Grunewald et al., 2009a). Indeed, nematode infectivity is compromised in auxin signalling and transport mutants (Grunewald et al., 2009b). Numbers of the infected beet cyst nematode (Heterodera schachtii) are significantly reduced in the aux1 lax3 double mutant, suggesting that this nematode takes advantage of the host's auxin transport process during the colonization of arabidopsis roots (Lee et al., 2011). Supporting this view, Hs19C07, an effector of H. schachtii, physically interacts with the arabidopsis auxin influx transporter LAX3, a close relative of the better known auxin influx protein AUX1 (Lee et al., 2011). A possible mechanism for the reduced nematode infectivity observed in the mutant would be that LAX3-mediated auxin transport may be required for the activation of the cell wall-loosening enzymes (e.g. expansins) required for both lateral root formation (Swarup et al., 2008) and syncytium development.
Other arabidopsis auxin transport proteins are also known to have roles in nematode susceptibility. Based on the analysis of single and double mutants of the PIN auxin efflux family, it was shown that PIN1 is required for the initiation and PIN3 and PIN4 are required for the expansion of nematode feeding sites in arabidopsis (Grunewald et al., 2009b). Whether nematode effectors are involved in this phenomenon is not known; however, this finding further supports the view that nematodes can exploit the host's auxin transport process for their benefit (Gheysen and Mitchum, 2011; Haegeman et al., 2012).
TRANSCRIPTIONAL AND POSTTRANSCRIPTIONAL REGULATION OF ENVIRONMENT- AND AUXIN-MEDIATED ROOT SYSTEM ARCHITECTURE
Transcription factors
So far, a few transcriptional regulators involved in the environmental regulation of lateral root development have been identified. For instance, the Medicago truncatula HD-Zip I transcription factor HB1 is required for the inhibition of lateral root emergence under salt stress (Ariel et al., 2010). HB1 represses LBD1, which encodes an auxin-responsive lateral organ boundaries (LOB) domain-containing transcription factor (reviewed by Majer and Hochholdinger, 2011) by directly binding to the promoter of this transcription factor (Ariel et al., 2010). Several members of the LBD gene family (e.g. LBD18/ASL20, LBD16/ASL18 and LBD29/ASL16) regulate lateral root formation and the expression of these genes is directly regulated by ARFs (Okushima et al., 2007; Lee et al., 2009). More recently, the involvement of LBD18 in the regulation of EXPANSINA 14 (EXPA14) and EXPA17 genes, which encode the cell wall-loosening expansin enzymes implicated in lateral root formation, has been shown (Lee et al., 2012; Lee and Kim, 2013). Indeed, knocking down EXPA17 expression delays lateral root emergence while EXPA17 over-expression promotes lateral root density in the presence of exogenous auxin (Lee and Kim, 2013). Another member of the LBD gene family, JAGGED LATERAL ORGAN (JLO), is required for all auxin responses in the root (Bureau et al., 2010). Identification of environmental signals that activate root-specific expression of these transcription factors may lead to new insights into the regulation of root architecture.
The arabidopsis transcription factor MYB77 is proposed to be a positive regulator of lateral root development under low IAA or low nutrient levels. MYB77 interacts with ARF7 and promotes auxin-responsive gene expression (Shin et al., 2007). MYB77 expression is negatively regulated by K+ deprivation and the lateral root density of the myb77 mutant is lower than that of wild-type plants under K-deprived conditions. The response of myb77 to N and P remains unchanged, suggesting that MYB77 is required for correct responses to K+.
Over-expression of NAC2, a NAC (NAM-ATAF1/2-CUC2) domain-containing transcription factor whose salt-responsive induction is dependent on the auxin receptor TIR1, promotes lateral root development under salt stress, although the exact mechanism(s) of NAC2-mediated salt tolerance is currently unknown (He et al., 2005). NTM2, another NAC transcription factor of arabidopsis, integrates auxin and salt signals during seed germination (Park et al., 2011) while NAC4, acting downstream from the auxin receptor AFB3, controls nitrate-mediated lateral root development, most likely by directly or indirectly regulating the expression of other transcription factors, such as the zinc finger protein OCS ELEMENT BINDING FACTOR 4 (OBF4) (Vidal et al., 2013). The arabidopsis WRKY75 transcription factor is induced by Pi deprivation. wrky75 knockdown plants were more sensitive to Pi deprivation and exhibited increased lateral root length and numbers under both normal or Pi deprivation conditions. It was proposed that this transcription factor might exert its effect on lateral root development by regulating the genes involved in auxin transport (Devaiah et al., 2007). GbWRKY1, a novel cotton (Gossypium barbadense) transcription factor, also positively regulates Pi deprivation tolerance by altering auxin sensitivity when over-expressed in arabidopsis (Xu L et al., 2012). OsWRKY72, the rice orthologue of WRKY75, has also recently been associated with auxin transport (Yu et al., 2010). Finally, the role of OsARF16, a positive regulator of auxin responses, in primary root, lateral root and root hair development under Pi deficiency, has recently been shown (Shen et al., 2013). OsARF16 and rice PIN genes such as OsPINb, OsPIN4 and OsPIN9 are co-regulated, suggesting that OsARF16-mediated effects occur through altered auxin transport (Shen et al., 2013).
More recently, the involvement of three PLETHORA (PLT3, PLT5 and PLT7) transcription factors, which seem to act downstream from ARF7 and ARF19, in lateral root development was shown (Hofhuis et al., 2013). It is unknown, however, whether these transcription factors regulate environmentally related root responses.
MicroRNAs
Over the years, a number of microRNAs that target plant genes involved in the regulation of root architecture have been identified (reviewed by Khan et al., 2011). The mode of action of some of these microRNAs, which can also be responsive to signals such as nutrient deficiency or other stresses, clearly involves auxin. For instance, arabidopsis plants over-expressing miR393, a microRNA that downregulates the expression of the AFB3 auxin receptor, resemble afb3 mutant plants, which show compromised primary root and lateral root growth in response to nitrate (Vidal et al., 2010). Interestingly, drought stress activates miR393. The miR393-dependent degradation of TIR1 and AFB2 transcripts contributes to osmotic stress-mediated inhibition of lateral root growth by attenuating auxin signalling (Chen et al., 2012a). These findings indicate that diverse signals converge on these microRNAs, which in turn regulate the expression of key genes involved in auxin signalling and lateral root development. More recently, IAR3 has been identified as a target of miR167. As discussed above, IAR3 regulates lateral root development and contributes to osmotic stress tolerance in arabidopsis. miR167 also controls the expression of ARF8 and modulates N-responsive lateral root initiation (Kinoshita et al., 2012). Other stress-responsive microRNAs regulating lateral root development by targeting the auxin pathway have been reported. For instance, miR164 affects lateral root development by targeting NAC1, which in turn regulates auxin signalling in arabidopsis (Guo et al., 2005).
CONCLUDING REMARKS: IS AUXIN A COMPONENT OF THE OVERALL BIOTIC AND ABIOTIC STRESS TOLERANCE MECHANISM OF PLANTS?
Better understanding of how plant roots integrate environmental signals can lead to the development of potential remedies to improve crop productivity, such as the use of soil microbes to optimize plant growth in stressful environments (Remans et al., 2012; Schenk et al., 2012). Although this review article has mainly focused on the role of auxin as an integrator of environmental signals in plant root development, emerging evidence also implicates auxin as an integral part of the plant's overall biotic and stress tolerance mechanism (see for instance Zhang et al., 2008, 2012; Kazan and Manners, 2009; Shen et al., 2010; Wang et al., 2010; Stirnberg et al., 2012). For instance, the arabidopsis activation-tagged yuc7-1D mutant, with constitutively elevated auxin levels, shows increased expression of the stress-associated genes RD29A (RESPONSIVE TO DESSICATION 29A) and COR15A (COLD-REGULATED 15A) and increased drought tolerance (Lee et al., 2012). A similar role in drought tolerance has been shown for another arabidopsis YUCCA gene implicated in auxin production when expressed in potato (Solanum tuberosum) (Kim et al., 2012). In rice, a novel YUCCA protein, CONSTITUTIVELY WILTED1, is involved in maintaining water homeostasis and an appropriate root-to-shoot ratio (Woo et al., 2007). A role in drought tolerance has also recently been shown for the putative auxin efflux carrier OsPIN3t in rice (Zhang et al., 2012).
In their natural environments, plants also encounter strong competition from nearby plants. The auxin pathway has long been known to be an essential part of the plant's shade-avoidance mechanism, an evolutionarily important response triggered by reduced red:far red light ratios under light-limiting conditions (Morelli and Ruberti, 2000; Keuskampa et al., 2010). New evidence also suggests extensive communication between roots of different plants in the rhizosphere (Cahill et al., 2010; Chen et al., 2012b; Faget et al., 2013; Falik et al., 2013). Although possible mechanisms involved in this phenomenon are not yet clear, root tips seem to function as sensors for detecting the presence of other roots from nearby plants (Fang et al., 2013). Given the prominent role of auxin in root adaptive responses, it would not be surprising if auxin also plays a role in regulating root–root communication. In fact, auxin is known to play a role in root–shoot communication, a process that can be critical for survival during environmental adaptation (Kabouw et al., 2012).
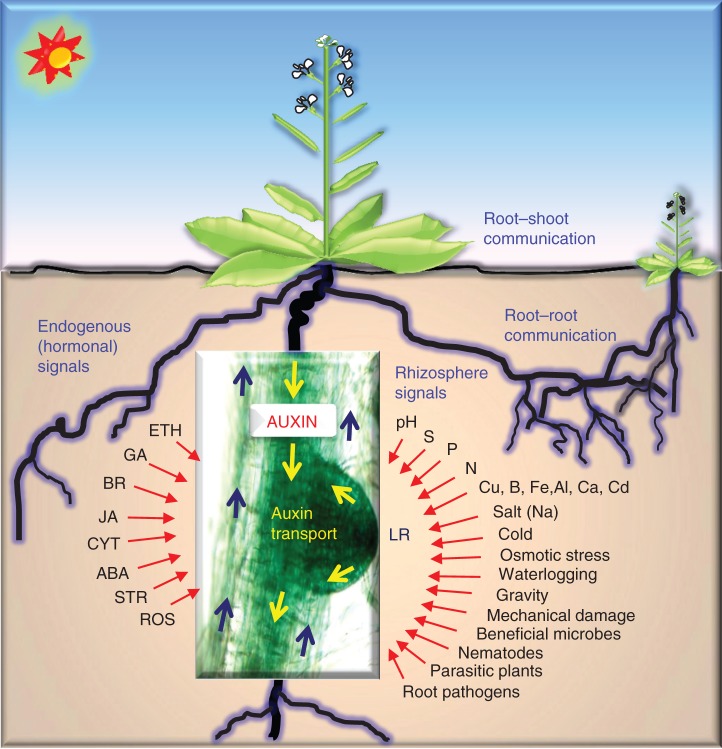
The extensive interplay reported between auxin and other plant hormones might be at least partly responsible for some of the stress-related developmental alterations in auxin biosynthesis, signalling or transport mutants (Ivanchenko et al., 2008; Teale et al., 2008; Seo and Park, 2009; Fukaki and Tasaka, 2009; Gou et al., 2010; Blomster et al., 2011; Lewis et al., 2011; Gutierrez et al., 2012; Bielach et al., 2012; Durbak et al., 2012; Muday et al., 2012; Shani et al., 2013; Löfke et al., 2013; Rahman, 2013). In addition, as reviewed here, auxin also crosstalks with other stress-responsive signalling pathways, such as those of Ca2+ and ROS, produced in the plant during adaptive responses to various biotic and abiotic stresses. Despite all these complexities, different stresses appear to induce an overlapping set of plant responses, called the ‘stress induced morphogenic response’ (SIMR) (Potters et al., 2007, 2009), which helps the plant to reallocate available resources between defence and development (Fig. 1). An improved understanding of the role of auxin during plants’ adaptation to environmental signals will help in the design of future strategies aimed at improving biotic and abiotic stress tolerance in crop plants.
Fig. 1.
Auxin plays an essential role in the integration of diverse biotic and abiotic environmental signals to plant root development. Auxin signalling interacts with the signalling pathways of all other known plant hormones. Auxin is also proposed to have roles in regulating the communication both within the same plant (root–shoot communication) and between different plants (root–root communication). The central image is a β-glucuronidase (GUS)-stained section of a primary root with emerging lateral root from a DR5-GUS-expressing arabidopsis seedling (Ulmasov et al., 1997) and the intensity of GUS staining correlates with increased auxin activity. The arrows indicate an effect but do not necessarily suggest a positive interaction. ABA, abscisic acid; LR, lateral root; ETH, ethylene; GA, gibberellin; BR, brassinosteroid; STR, strictogalactones; JA, jasmonate; CYT, cytokinins; ROS, reactive oxygen species. See the text for additional details.
ACKNOWLEDGEMENTS
This review is dedicated to the memory of Mitsuyo Ito. I apologize to colleagues whose relevant work could not be included due to space restrictions and thank Dr Rebecca Lyons for critical reading of the paper and helpful suggestions. I also thank two anonymous reviewers for their useful comments on the manuscript.
LITERATURE CITED
- Aquea F, Federici F, Moscoso C, et al. A molecular framework for the inhibition of arabidopsis root growth in response to boron toxicity. Plant Cell and Environment. 2011;35:719–734. doi: 10.1111/j.1365-3040.2011.02446.x. [DOI] [PubMed] [Google Scholar]
- Ariel F, Diet A, Verdenaud M, et al. Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell. 2010;22:2171–2183. doi: 10.1105/tpc.110.074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E, Kubeš M, Rolčík J, et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature. 2012;485:119–122. doi: 10.1038/nature11001. [DOI] [PubMed] [Google Scholar]
- Bashandy T, Guilleminot J, Vernoux T, et al. Interplay between the NADP-linked thioredoxin and glutathione systems in arabidopsis auxin signaling. Plant Cell. 2010;22:376–391. doi: 10.1105/tpc.109.071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielach A, Duclercq J, Marhavý P, Benková E. Genetic approach towards the identification of auxin-cytokinin crosstalk components involved in root development. Philosophical Transactions of the Royal Society of London. B, Biological Sciences. 2012;367:1469–1478. doi: 10.1098/rstb.2011.0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomster T, Salojärvi J, Sipari N, et al. Apoplastic reactive oxygen species transiently decrease auxin signaling and cause stress-induced morphogenic response in arabidopsis. Plant Physiology. 2011;157:1866–1883. doi: 10.1104/pp.111.181883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, et al. Flavonoids act as negative regulators of auxin transport in vivo in arabidopsis. Plant Physiology. 2001;126:524–535. doi: 10.1104/pp.126.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau M, Rast MI, Illmer J, Simon R. JAGGED LATERAL ORGAN (JLO) controls auxin dependent patterning during development of the arabidopsis embryo and root. Plant Molecular Biology. 2010;74:479–491. doi: 10.1007/s11103-010-9688-2. [DOI] [PubMed] [Google Scholar]
- Bouguyon E, Gojon A, Nacry P. Nitrate sensing and signaling in plants. Seminars in Cellular and Developmental Biology. 2012;23:648–654. doi: 10.1016/j.semcdb.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Cahill JF, Jr, McNickle GG, Haag JJ, Lamb EG, Nyanumba SM, St Clair CC. Plants integrate information about nutrients and neighbors. Science. 2010;328:1657. doi: 10.1126/science.1189736. [DOI] [PubMed] [Google Scholar]
- Cassab GI, Eapen D, Campos ME. Root hydrotropism: an update. American Journal of Botany. 2013;100(14–24) doi: 10.3732/ajb.1200306. [DOI] [PubMed] [Google Scholar]
- Chen H, Li Z, Xiong L. A plant microRNA regulates the adaptation of roots to drought stress. FEBS Letters. 2012a;586:1742–1747. doi: 10.1016/j.febslet.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Chen BJW, During HJ, Anten NPR. Detect thy neighbor: identity recognition at the root level in plants. Plant Science. 2012b;195:157–167. doi: 10.1016/j.plantsci.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Contesto C, Milesi S, Mantelin S, et al. The auxin-signaling pathway is required for the lateral root response of arabidopsis to the rhizobacterium Phyllobacterium brassicacearum. Planta. 2010;232:1455–1470. doi: 10.1007/s00425-010-1264-0. [DOI] [PubMed] [Google Scholar]
- Contreras-Cornejo HA, Macias-Rodriguez L, Cortes-Penagos C, López-Bucio J. Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in arabidopsis. Plant Physiology. 2009;149:1579–1592. doi: 10.1104/pp.108.130369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan H, Yang G, Zheng ZL. A negative regulatory role for auxin in sulphate deficiency response in Arabidopsis thaliana. Plant Molecular Biology. 2007;63:221–235. doi: 10.1007/s11103-006-9084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Voss U, Lau S, et al. Unraveling the evolution of auxin signaling. Plant Physiology. 2010;155:209–221. doi: 10.1104/pp.110.168161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in arabidopsis. Plant Physiology. 2007;143:1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L. Pointing PINs in the right directions: a potassium transporter is required for the polar localization of auxin efflux carriers. New Phytologist. 2013;197:1027–1028. doi: 10.1111/nph.12151. [DOI] [PubMed] [Google Scholar]
- Durbak A, Yao H, McSteen P. Hormone signaling in plant development. Current Opinion in Plant Biology. 2012;15:92–96. doi: 10.1016/j.pbi.2011.12.004. [DOI] [PubMed] [Google Scholar]
- Faget M, Nagel KA, Walter A, et al. Root-root interactions – extending our perspective to be more inclusive of the range of theories in ecology and agriculture using in-vivo analyses. Annals of Botany. 2013;112:253–266. doi: 10.1093/aob/mcs296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falik O, Mordoch Y, Ben-Natan D, et al. Plant responsiveness to root–root communication of stress cues. Annals of Botany. 2013;110:271–280. doi: 10.1093/aob/mcs045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Clark RT, Zheng Y, et al. Genotypic recognition and spatial responses by rice roots. Proceedings of the National Academy of Sciences of the USA. 2013;110:2670–2675. doi: 10.1073/pnas.1222821110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felten J, Kohler A, Morin E, et al. The ectomycorrhizal fungus Laccaria bicolour stimulates lateral root formation in poplar and arabidopsis through auxin transport and signaling. Plant Physiology. 2009;151:1991–2005. doi: 10.1104/pp.109.147231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finet C, Jaillais Y. Auxology: when auxin meets plant evo-devo. Developmental Biology. 2012;369:19–31. doi: 10.1016/j.ydbio.2012.05.039. [DOI] [PubMed] [Google Scholar]
- Friml J. Subcellular trafficking of PIN auxin efflux carriers in auxin transport. European Journal of Cell Biology. 2010;89:231–235. doi: 10.1016/j.ejcb.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tasaka M. Hormone interactions during lateral root formation. Plant Molecular Biology. 2009;69:437–449. doi: 10.1007/s11103-008-9417-2. [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia CS, Testerink C. Salt stress signals shape the plant root. Current Opinions in Plant Biology. 2011;14:296–302. doi: 10.1016/j.pbi.2011.03.019. [DOI] [PubMed] [Google Scholar]
- Gheysen G, Mitchum MG. How nematodes manipulate plant development pathways for infection. Current Opinions in Plant Biology. 2011;14:415–421. doi: 10.1016/j.pbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Giehl RF, Lima JE, von Wirén N. Localized iron supply triggers lateral root elongation in arabidopsis by altering the AUX1-mediated auxin distribution. Plant Cell. 2012;24:33–49. doi: 10.1105/tpc.111.092973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojon A, Krouk G, Perrine-Walker F, Laugier E. Nitrate transceptor(s) in plants. Journal of Experimental Botany. 2011;62:2299–2308. doi: 10.1093/jxb/erq419. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant Journal. 1999;18:277–284. doi: 10.1046/j.1365-313x.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- Gou J, Strauss SH, Tsai CJ, et al. Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell. 2010;22:623–639. doi: 10.1105/tpc.109.073239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, van Noorden G, Isterdael GV, Beeckman T, Gheysen G, Mathesius U. Manipulation of auxin transport in plant roots during Rhizobium symbiosis and nematode parasitism. Plant Cell. 2009a;21:2553–2562. doi: 10.1105/tpc.109.069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, Cannoot B, Friml J, Gheysen G. Parasitic nematodes modulate PIN mediated auxin transport to facilitate infection. PLoS Pathogens. 2009b;5 doi: 10.1371/journal.ppat.1000266. e10000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Mongelard G, Floková K, et al. Auxin controls arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell. 2012;24:2515–2517. doi: 10.1105/tpc.112.099119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CV, Le DT, Nishiyama R, Watanabe Y, et al. The auxin response factor transcription factor family in soybean: genome-wide identification and expression analyses during development and water stress. DNA Research. 2013 doi: 10.1093/dnares/dst027. doi:10.1093/dnares/dst027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman A, Mantelin S, Jones JT, Gheysen G. Functional roles of effectors of plant-parasitic nematodes. Gene. 2012;492:19–31. doi: 10.1016/j.gene.2011.10.040. [DOI] [PubMed] [Google Scholar]
- Hanlon MT, Coenen C. Genetic evidence for auxin involvement in arbuscular mycorrhiza initiation. New Phytologist. 2011;189:701–709. doi: 10.1111/j.1469-8137.2010.03567.x. [DOI] [PubMed] [Google Scholar]
- He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant Journal. 2005;44:903–916. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- Hernández-Mata G, Mellado-Rojas ME, Richards-Lewis A, López-Bucio J, Beltran-Pena E, Soriano-Bello EL. Plant immunity induced by oligogalacturonides alters root growth in a process involving flavonoid accumulation in Arabidopsis thaliana. Journal of Plant Growth Regulation. 2011;29:441–454. [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Hofhuis H, Laskowski M, Du Y, et al. Phyllotaxis and rhizotaxis in arabidopsis are modified by three PLETHORA transcription factors. Current Biology. 2013;23:956–962. doi: 10.1016/j.cub.2013.04.048. [DOI] [PubMed] [Google Scholar]
- Hu YF, Zhou G, Na XF, et al. Cadmium interferes with maintenance of auxin homeostasis in arabidopsis seedlings. Journal of Plant Physiology. 2013;170:965–975. doi: 10.1016/j.jplph.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Iglesias MJ, Terrile MC, Bartoli CG, D'Ippólito S, Casalongué CA. Auxin signaling participates in the adaptative response against oxidative stress and salinity by interacting with redox metabolism in arabidopsis. Plant Molecular Biology. 2010;74:215–222. doi: 10.1007/s11103-010-9667-7. [DOI] [PubMed] [Google Scholar]
- Ivanchenko MG, Muday GK, Dubrovsky JG. Ethylene-auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant Journal. 2008;55:335–347. doi: 10.1111/j.1365-313X.2008.03528.x. [DOI] [PubMed] [Google Scholar]
- Ji H, Pardo JM, Batelli G, Van Oosten MJ, Bressan RA, Li X. The salt overly sensitive (SOS) pathway: established and emerging roles. Molecular Plant. 2013;6:275–286. doi: 10.1093/mp/sst017. [DOI] [PubMed] [Google Scholar]
- Kabouw P, van Dam NM, van der Putten WH, Biere A. How genetic modification of roots affects rhizosphere processes and plant performance. Journal of Experimental Botany. 2012;63:3475–3483. doi: 10.1093/jxb/err399. [DOI] [PubMed] [Google Scholar]
- Kaneyasu T, Kobayashi A, Nakayama M, Fujii N, Takahashi H, Miyazawa Y. Auxin response, but not its polar transport, plays a role in hydrotropism of arabidopsis roots. Journal of Experimental Botany. 2007;58:1143–1150. doi: 10.1093/jxb/erl274. [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. Linking development to defense: auxin in plant-pathogen interactions. Trends in Plant Science. 2009;14:373–382. doi: 10.1016/j.tplants.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Khan GA, Declerck M, Sorin C, Hartmann C, Crespi M, Lelandais-Brière C. MicroRNAs as regulators of root development and architecture. Plant Molecular Biology. 2011;77:47–58. doi: 10.1007/s11103-011-9793-x. [DOI] [PubMed] [Google Scholar]
- Kidd BN, Kadoo NY, Dombrecht B, et al. Auxin signaling and transport promote susceptibility to the root-infecting fungal pathogen Fusarium oxysporum in arabidopsis. Molecular Plant-Microbe Interactions. 2011;24:733–748. doi: 10.1094/MPMI-08-10-0194. [DOI] [PubMed] [Google Scholar]
- Kim JI, Baek D, Park HC, et al. Overexpression of arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Molecular Plant. 2012;6:337–349. doi: 10.1093/mp/sss100. [DOI] [PubMed] [Google Scholar]
- Kinoshita N, Wang H, Kasahara H, et al. A-Ala Resistant3, an evolutionarily conserved target of miR167, mediates arabidopsis root architecture changes during high osmotic stress. Plant Cell. 2012;24:3590–3602. doi: 10.1105/tpc.112.097006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Takahashi A, Kakimoto Y, et al. A gene essential for hydrotropism in roots. Proceedings of the National Academy of Sciences of the USA. 2007;104:4724–4729. doi: 10.1073/pnas.0609929104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskampa DH, Pollmannb S, Voeseneka LACJ, Peetersa AJM, Pierik R. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proceedings of the National Academy of Sciences of the USA. 2010;107:22740–22744. doi: 10.1073/pnas.1013457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, et al. Nitrate-regulated auxin transport by NRT1·1 defines a mechanism for nutrient sensing in plants. Developmental Cell. 2010;18:927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Lager I, Andréasson O, Dunbar TL, et al. Changes in external pH rapidly alter plant gene expression and modulate auxin and elicitor responses. Plant Cell and Environment. 2010;33:1513–1528. doi: 10.1111/j.1365-3040.2010.02161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Chronis D, Kenning C, et al. The novel cyst nematode effector protein 19C07 interacts with the arabidopsis auxin influx transporter LAX3 to control feeding site development. Plant Physiology. 2011;155:866–880. doi: 10.1104/pp.110.167197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim J. EXPANSINA17 upregulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant and Cell Physiology. 2013 doi: 10.1093/pcp/pct105. doi:10.1093/pcp/pct105. [DOI] [PubMed] [Google Scholar]
- Lee HW, Kim NY, Lee DJ, Kim J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in arabidopsis. Plant Physiology. 2009;151:1377–1389. doi: 10.1104/pp.109.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim MJ, Kim NY, Lee SH, Kim J. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant Journal. 2013;73:212–224. doi: 10.1111/tpj.12013. [DOI] [PubMed] [Google Scholar]
- Lee M, Jung JH, Han DY, Seo PJ, Park WJ, Park CM. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in arabidopsis. Planta. 2012;235:923–938. doi: 10.1007/s00425-011-1552-3. [DOI] [PubMed] [Google Scholar]
- Lequeux H, Hermans C, Lutts S, Verbruggen N. Response to copper excess in Arabidopsis thaliana: impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiology and Biochemistry. 2010;48:673–82. doi: 10.1016/j.plaphy.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Lewis DR, Negi S, Sukumar P, Muday GK. Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development. 2011;138:3485–3495. doi: 10.1242/dev.065102. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. Phosphate availability alters architecture and causes changes in hormone sensitivity in the arabidopsis root system. Plant Physiology. 2002;129:244–256. doi: 10.1104/pp.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, et al. An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiology. 2005;137:681–691. doi: 10.1104/pp.104.049577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfke C, Zwiewka M, Heilmann I, Van Montagu MC, Teichmann T, Friml J. Asymmetric gibberellin signaling regulates vacuolar trafficking of PIN auxin transporters during root gravitropism. Proceedings of the National Academy of Sciences USA. 2013;110:3627–3632. doi: 10.1073/pnas.1300107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma TL, Wu WH, Wang Y. Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biology. 2012;12:161. doi: 10.1186/1471-2229-12-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends in Plant Science. 2011;16:47–52. doi: 10.1016/j.tplants.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Malamy JE. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell and Environment. 2005;28:67–77. doi: 10.1111/j.1365-3040.2005.01306.x. [DOI] [PubMed] [Google Scholar]
- Mano Y, Nemoto K. The pathway of auxin biosynthesis in plants. Journal of Experimental Botany. 2012;63:2853–2872. doi: 10.1093/jxb/ers091. [DOI] [PubMed] [Google Scholar]
- Martín-Rejano EM, Camacho-Cristóbal JJ, Herrera-Rodríguez MB, et al. Auxin and ethylene are involved in the responses of root system architecture to low boron supply in arabidopsis seedlings. Physiologia Plantarum. 2011;142:170–178. doi: 10.1111/j.1399-3054.2011.01459.x. [DOI] [PubMed] [Google Scholar]
- Mathesius U. The role of auxin in root-symbiont and root-pathogen interactions – from development to defense. In: Lüttge UE, Beyschlag W, Büdel B., Francis D, editors. Progress in botany. Volume 71. Berlin: Springer; 2010. pp. 185–210. [Google Scholar]
- Mattiello L, Kirst M, da Silva FR, Jorge RA, Menossi M. Transcriptional profile of maize roots under acid soil growth. BMC Plant Biology. 2010;10:196. doi: 10.1186/1471-2229-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, et al. The arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proceedings of the National Academy of Sciences of the USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Gong Q, et al. SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation. Plant Physiology. 2011;155:1000–1012. doi: 10.1104/pp.110.165191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa Y, Moriwaki T, Uchida M, Kobayashi A, Fujii N, Takahashi H. Overexpression of MIZU-KUSSEI1 enhances root hydrotropic response by retaining cell viability under hydrostimulated condition in Arabidopsis thaliana. Plant and Cell Physiology. 2012;53:1926–1933. doi: 10.1093/pcp/pcs129. [DOI] [PubMed] [Google Scholar]
- Morelli G, Ruberti I. Shade avoidance responses. Driving auxin along lateral routes. Plant Physiology. 2000;122:621–626. doi: 10.1104/pp.122.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday GK, Rahman A, Binder BM. Auxin and ethylene: collaborators or competitors? Trends in Plant Science. 2012;17:181–195. doi: 10.1016/j.tplants.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Nacry P, Canivenc G, Muller B, et al. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in arabidopsis. Plant Physiology. 2005;138:2061–2074. doi: 10.1104/pp.105.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Nibau C, Gibbs DJ, Coates JC. Branching out in new directions: the control of root architecture by lateral root formation. New Phytologist. 2008;179:595–614. doi: 10.1111/j.1469-8137.2008.02472.x. [DOI] [PubMed] [Google Scholar]
- Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS. Responses of root architecture development to low phosphorus availability: a review. Annals of Botany. 2012;112:391–408. doi: 10.1093/aob/mcs285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmont K, Sibout R, Hardtke C. Hidden branches: developments in root system architecture. Annual Reviews of Plant Biology. 2007;58:93–113. doi: 10.1146/annurev.arplant.58.032806.104006. [DOI] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T. Auxin control of root development. Cold Spring Harbour Perspectives in Biology 2. 2010 doi: 10.1101/cshperspect.a001537. a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim YS, Kim SG, Jung JH, Woo JC, Park CM. Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in arabidopsis. Plant Physiology. 2011;156:537–549. doi: 10.1104/pp.111.177071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski K, Bogusz D, Ribeiro A, Berry A. Progress on research on actinorhizal plants. Functional Plant Biology. 2011;38:633–638. doi: 10.1071/FP11066. [DOI] [PubMed] [Google Scholar]
- Péret B, Larrieu A, Bennett MJ. Lateral root emergence: a difficult birth. Journal of Experimental Botany. 2009;60:3637–3643. doi: 10.1093/jxb/erp232. [DOI] [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, et al. Phosphate availability alters lateral root development in arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell. 2008;20:3258–3272. doi: 10.1105/tpc.108.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto A, Lehotai N, Lozano-Juste J, et al. Involvement of nitric oxide and auxin in signal transduction of copper-induced morphological responses in arabidopsis seedlings. Annals of Botany. 2011;108:449–457. doi: 10.1093/aob/mcr176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN. Control of arabidopsis root development. Annual Review of Plant Biology. 2012;63:563–590. doi: 10.1146/annurev-arplant-042811-105501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potters G, Pasternak TP, Guisez Y, Palme KJ., Jansen MAK. Stress-induced morphogenic responses: growing out of trouble? Trends in Plant Science. 2007;12:98–105. doi: 10.1016/j.tplants.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Potters G, Pasternak TP, Guisez Y, Jansen MA. Different stresses, similar morphogenic responses: integrating a plethora of pathways. Plant Cell and Environment. 2009;32:158–169. doi: 10.1111/j.1365-3040.2008.01908.x. [DOI] [PubMed] [Google Scholar]
- Rahman A. Auxin: a regulator of cold stress response. Physiology Plantarum. 2013;147:28–35. doi: 10.1111/j.1399-3054.2012.01617.x. [DOI] [PubMed] [Google Scholar]
- Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during arabidopsis germination. Plant Physiology. 2004;135:978–988. doi: 10.1104/pp.104.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, et al. The arabidopsis NRT1·1 transporter participates in the signaling pathway triggering root colonization of nitrate rich patches. Proceedings of the National Academy of Sciences of the USA. 2006;103:19206–19211. doi: 10.1073/pnas.0605275103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Thijs S, Truyens S, et al. Understanding the development of roots exposed to contaminants and the potential of plant-associated bacteria for optimization of growth. Annals of Botany. 2012;110:239–252. doi: 10.1093/aob/mcs105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S, Debrosses G, Haralampidis K, et al. TRH1 encodes a potassium transporter required for tip growth in arabidopsis root hairs. Plant Cell. 2001;13:139–151. doi: 10.1105/tpc.13.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S, Ditengou FA, Ljung K, et al. Root gravitropism and root hair development constitute coupled developmental responses regulated by auxin homeostasis in the arabidopsis root apex. New Phytologist. 2013;197:1130–1141. doi: 10.1111/nph.12092. [DOI] [PubMed] [Google Scholar]
- Rosquete MR, Barbez E, Kleine-Vehn J. Cellular auxin homeostasis: gatekeeping is housekeeping. Molecular Plant. 2012;5:772–786. doi: 10.1093/mp/ssr109. [DOI] [PubMed] [Google Scholar]
- Sauer M, Robert S, Kleine-Vehn J. Auxin: simply complicated. Journal of Experimental Botany. 2013;64:2565–2577. doi: 10.1093/jxb/ert139. [DOI] [PubMed] [Google Scholar]
- Savatin DV, Ferrari S, Sicilia F, De Lorenzo G. Oligogalacturonide-auxin antagonism does not require posttranscriptional gene silencing or stabilization of auxin response repressors in arabidopsis. Plant Physiology. 2011;157:1163–1174. doi: 10.1104/pp.111.184663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Carvalhais LC, Kazan K. Unraveling plant-microbe interactions: can multi-species transcriptomics help? Trends in Biotechnology. 2012;30:177–184. doi: 10.1016/j.tibtech.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park CM. Auxin homeostasis during lateral root development under drought condition. Plant Signaling and Behavior. 2009;4:1002–1004. doi: 10.4161/psb.4.10.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E, Weinstain R, Zhanga Y, et al. Gibberellins accumulate in the elongating endodermal cells of arabidopsis root. Proceedings of the National Academy of Sciences of the USA. 2013;110:4834–4839. doi: 10.1073/pnas.1300436110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Bai Y, Wang S, et al. Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS Journal. 2010;277:2954–2969. doi: 10.1111/j.1742-4658.2010.07706.x. [DOI] [PubMed] [Google Scholar]
- Shen C, Wang S, Zhang S, et al. OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.) Plant Cell and Environment. 2013;36:607–620. doi: 10.1111/pce.12001. [DOI] [PubMed] [Google Scholar]
- Shibasaki K, Uemura M, Tsurumi S, Rahman A. Auxin response in arabidopsis under cold stress: underlying molecular mechanisms. Plant Cell. 2009;21:3823–3838. doi: 10.1105/tpc.109.069906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, et al. The arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg A, Göbel C, Grond S, et al. Piriformospora indica affects plant growth by auxin production. Physiologia Plantarum. 2007;131:581–589. doi: 10.1111/j.1399-3054.2007.00983.x. [DOI] [PubMed] [Google Scholar]
- Smith S, De Smet I. Root system architecture: insights from arabidopsis and cereal crops. Philosophical Transactions of the Royal Society of London. B, Biological Sciences. 2012;367:1441–1452. doi: 10.1098/rstb.2011.0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding EP. Diverting the downhill flow of auxin to steer growth during tropisms. American Journal of Botany. 2013;100:203–214. doi: 10.3732/ajb.1200420. [DOI] [PubMed] [Google Scholar]
- Splivallo R, Fischer U, Gobel C, Feussner I, Karlovsky P. Truffles regulate plant root morphogenesis via the production of auxin and ethylene. Plant Physiology. 2009;150:2018–2029. doi: 10.1104/pp.109.141325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Zhao S, Williamson L, Ward S, Leyser O. FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant Journal. 2012;71:907–920. doi: 10.1111/j.1365-313X.2012.05038.x. [DOI] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, et al. Root tip contact with low-phosphate media reprograms plant root architecture. Nature Genetics. 2007;39:792–796. doi: 10.1038/ng2041. [DOI] [PubMed] [Google Scholar]
- Sukumar P, Legué V, Vayssières A, Martin F, Tuskan GA, Kalluri UC. Involvement of auxin pathways in modulating root architecture during beneficial plant-microorganism interactions. Plant Cell and Environment. 2013;36:909–919. doi: 10.1111/pce.12036. [DOI] [PubMed] [Google Scholar]
- Sun F, Zhang W, Hu H, et al. Salt modulates gravity signaling pathway to regulate growth direction of primary roots in arabidopsis. Plant Physiology. 2008;146:178–188. doi: 10.1104/pp.107.109413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Tian QY, Chen J, Zhang WH. Aluminium-induced inhibition of root elongation in arabidopsis is mediated by ethylene and auxin. Journal of Experimental Botany. 2010;61:347–356. doi: 10.1093/jxb/erp306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nature Cell Biology. 2008;10:946–954. doi: 10.1038/ncb1754. [DOI] [PubMed] [Google Scholar]
- Swarup R, Péret B. AUX/LAX family of auxin influx carriers—an overview. Frontiers in Plant Science. 2012;3:225. doi: 10.3389/fpls.2012.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi YY, Taniguchi M, Tsuge T, Oka A, Aoyama T. Involvement of Arabidopsis thaliana phospholipase Dzeta2 in root hydrotropism through the suppression of root gravitropism. Planta. 2010;231:491–497. doi: 10.1007/s00425-009-1052-x. [DOI] [PubMed] [Google Scholar]
- Teale WD, Ditengou FA, Dovzhenko AD, et al. Auxin as a model for the integration of hormonal signal processing and transduction. Molecular Plant. 2008;1:229–237. doi: 10.1093/mp/ssn006. [DOI] [PubMed] [Google Scholar]
- Terrile MC, París R, Calderón-Villalobos LI, et al. Nitric oxide influences auxin signaling through S-nitrosylation of the arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant Journal. 2012;70:492–500. doi: 10.1111/j.1365-313X.2011.04885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A, Perrot-Rechenmann C. Recent progress in auxin biology. Comptes Rendus Biologies. 2010;333:297–306. doi: 10.1016/j.crvi.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Vicente-Agullo F, Rigas S, Desbrosses G, Dolan L, Hatzopoulos P, Grabov A. Potassium carrier TRH1 is required for auxin transport in arabidopsis roots. Plant Journal. 2004;40:523–535. doi: 10.1111/j.1365-313X.2004.02230.x. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, et al. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the USA. 2010;107:4477–4482. doi: 10.1073/pnas.0909571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Riveras E, Contreras-López O, Gutiérrez RA. Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proceedings of the National Academy of Sciences of the USA. 2013;110:12840–12845. doi: 10.1073/pnas.1310937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidoz ML, Loreti E, Mensuali A, Alpi A, Perata P. Hormonal interplay during adventitious root formation in flooded tomato plants. Plant Journal. 2010;63:551–562. doi: 10.1111/j.1365-313X.2010.04262.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Bai Y, Shen C, et al. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Functional and Integrative Genomics. 2010;10:533–546. doi: 10.1007/s10142-010-0174-3. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li K, Li X. Auxin redistribution modulates plastic development of root system architecture under salt stress in Arabidopsis thaliana. Journal of Plant Physiology. 2009;166:1637–1645. doi: 10.1016/j.jplph.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiology. 2001;126:875–882. doi: 10.1104/pp.126.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YM, Park HJ, Su'udi M, Yang JI, et al. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Molecular Biology. 2007;65:125–136. doi: 10.1007/s11103-007-9203-6. [DOI] [PubMed] [Google Scholar]
- Yang Q, Chen Z-Z, Zhou X-F, et al. Overexpression of SOS (salt overly sensitive) genes increases salt tolerance in transgenic arabidopsis. Molecular Plant. 2008;2:22–31. doi: 10.1093/mp/ssn058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Ligang C, Liping Z, Diqiu Y. Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of arabidopsis. Journal of Bioscience. 2010;35:459–471. doi: 10.1007/s12038-010-0051-1. [DOI] [PubMed] [Google Scholar]
- Yuan H-M, Xu H-H, Liu W-C Ying-Tang L. Copper regulates primary root elongation through PIN1-mediated auxin redistribution. Plant and Cell Physiology. 2013;54:766–778. doi: 10.1093/pcp/pct030. [DOI] [PubMed] [Google Scholar]
- Xu L, Jin L, Long L, et al. Overexpression of GbWRKY1 positively regulates the Pi starvation response by alteration of auxin sensitivity in arabidopsis. Plant Cell Reports. 2012;31:2177–2188. doi: 10.1007/s00299-012-1328-7. [DOI] [PubMed] [Google Scholar]
- Xu W, Jia L, Baluska F, et al. PIN2 is required for the adaptation of arabidopsis roots to alkaline stress by modulating proton secretion. Journal of Experimental Botany. 2012;63:6105–6114. doi: 10.1093/jxb/ers259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Jia L, Shi W, et al. Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. New Phytologist. 2013;197:139–150. doi: 10.1111/nph.12004. [DOI] [PubMed] [Google Scholar]
- Zamioudis C, Mastranesti P, Dhonukshe P, Blilou I, Pieterse CM. Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiology. 2013;162:304–318. doi: 10.1104/pp.112.212597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG. Regulation of arabidopsis root development by nitrate availability. Journal of Experimental Botany. 2000;51:51–59. [PubMed] [Google Scholar]
- Zhang J, Peng Y, Guo Z. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Research. 2008;18:508–521. doi: 10.1038/cr.2007.104. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li J, Zhang W, et al. The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant Journal. 2012;72:805–816. doi: 10.1111/j.1365-313X.2012.05121.x. [DOI] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis and its role in plant development. Annual Reviews of Plant Biology. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao FY, Han MM, Zhang SY, et al. Hydrogen peroxide-mediated growth of the root system occurs via auxin signaling modification and variations in the expression of cell-cycle genes in rice seedlings exposed to cadmium stress. Journal of Integrative Plant Biology. 2012;54:991–1006. doi: 10.1111/j.1744-7909.2012.01170.x. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wang T, Zhang W, Li X. SOS3 mediates lateral root development under low salt stress through regulation of auxin redistribution and maxima in arabidopsis. New Phytologist. 2011;189:1122–1134. doi: 10.1111/j.1469-8137.2010.03545.x. [DOI] [PubMed] [Google Scholar]
- Zolla G, Heimer YM, Barak S. Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. Journal of Experimental Botany. 2010;61:211–224. doi: 10.1093/jxb/erp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolobowska L, Van Gijsegem F. Induction of lateral root structure formation on petunia roots: a novel effect of GMI1000 Ralstonia solanacearum infection impaired in Hrp mutants. Molecular Plant-Microbe Interactions. 2006;19:597–606. doi: 10.1094/MPMI-19-0597. [DOI] [PubMed] [Google Scholar]