Abstract
Background and Aims
Subtribe Centaureinae appears to be an excellent model group in which to analyse satellite DNA and assess the influence that the biology and/or the evolution of different lineages have had on the evolution of this class of repetitive DNA. Phylogenetic analyses of Centaureinae support two main phases of radiation, leading to two major groups of genera of different ages. Furthermore, different modes of evolution are observed in different lineages, reflected by morphology and DNA sequences.
Methods
The sequences of 502 repeat units of the HinfI satellite DNA family from 38 species belonging to ten genera of Centaureinae were isolated and compared. A phylogenetic reconstruction was carried out by maximum likelihood and Bayesian inference.
Key Results
Up to eight different HinfI subfamilies were found, based on the presence of a set of diagnostic positions given by a specific mutation shared by all the sequences of one group. Subfamilies V–VIII were mostly found in older genera (first phase of radiation in the subtribe, late Oligocene–Miocene), although some copies of these types of repeats were also found in some species of the derived genera. Subfamilies I–IV spread mostly in species of the derived clade (second phase of radiation, Pliocene to Pleistocene), although repeats of these subfamilies exist in older species. Phylogenetic trees did not group the repeats by taxonomic affinity, but sequences were grouped by subfamily provenance. Concerted evolution was observed in HinfI subfamilies spread in older genera, whereas no genetic differentiation was found between species, and several subfamilies even coexist within the same species, in recently radiated groups or in groups with a history of recurrent hybridization of lineages.
Conclusions
The results suggest that the eight HinfI subfamilies were present in the common ancestor of Centaureinae and that each spread differentially in different genera during the two main phases of radiation following the library model of satellite DNA evolution. Additionally, differential speciation pathways gave rise to differential patterns of sequence evolution in different lineages. Thus, the evolutionary history of each group of Centaureinae is reflected in HinfI satellite DNA evolution. The data reinforce the value of satellite DNA sequences as markers of evolutionary processes.
Keywords: HinfI satellite DNA, concerted evolution, molecular drive, library hypothesis, Centaureinae, radiation
INTRODUCTION
There are four tribes in subfamily Carduoideae (Asteraceae), the most abundant and widespread being tribe Cardueae. This tribe represents 90 % of the species diversity of the group, with 2400 species belonging to 72 genera (Susanna and Garcia-Jacas, 2009). Current classifications of the tribe accept five subtribes: Cardopatiinae, Carlininae, Echinopsinae, Carduinae and Centaureinae (Susanna and Garcia-Jacas, 2009). Cardopatiinae, Carlininae and Echinopsinae are early diverging groups with relatively few species, and Carduinae and Centaureinae are more diversified, derived groups (Susanna and Garcia-Jacas, 2009). Asteraceae in general, and subtribe Centaureinae in particular, provide an excellent opportunity for understanding adaptation in the recent radiation of a plant group at a global scale (Panero and Funk, 2008). Subtribe Centaureinae comprises 32 genera with >800 species distributed in the Mediterranean–Oriental–Turkestanian region, with representatives in tropical regions of Africa, North and South America, East Asia and Australia (Susanna and Garcia-Jacas, 2007); the Mediterranean basin is the main centre of diversity of the subtribe (Wagenitz, 1986; Meusel and Jäger, 1992). Phylogenetic analyses of Centaureinae support two main phases of radiation, leading to two major groups of genera of different ages, one which includes the early diverging genera of the subtribe and the other the derived genera (Garcia-Jacas et al., 2001; Hellwig, 2004). The first phase may date to the late Oligocene and Miocene, whereas the major modern or derived clades appear to have differentiated around the transition from the Pliocene to the Pleistocene (Hellwig, 2004). The derived clade, or second radiation, encompasses most of the species currently classified in the genus Centaurea and the Carthamus complex. Centaurea comprises three subgenera: Acrocentron, Centaurea (formerly the Jacea group) and Cyanus (Susanna and Garcia-Jacas, 2009). The Carthamus complex includes the genera Carduncellus, Carthamus, Femeniasia and Phonus (Garcia-Jacas et al., 2001). The rest of the genera constitute the early diverging groups (first radiation): Cheirolophus, Crupina, Klasea, Psephellus, Rhaponticoides (formerly Centaurea sensu stricto), Rhaponticum, Serratula and Volutaria, among others (Garcia-Jacas et al., 2001). Both groups (early diverging and derived) have recent members in the Mediterranean, but only the derived younger genera are species rich (Hellwig, 2004). Different modes of evolution are observed in different lineages, reflected by morphology and DNA sequences. Hellwig (2004) described the ecogeographical history of Centaureinae (Asteraceae) in the Mediterranean against the background of the geological history of the region. According to Hellwig (2004), representatives of the different clades often grew sympatrically but took different evolutionary routes. The wide range of ecological niches and the fine mosaic-like habitats of different qualities are the basis for the coexistence of many species which have evolved in the Centaureinae (Hellwig, 2004). Expansion and reduction of suitable habitats led to the evolution of groups consisting of many now largely allopatric, vicariant species. Although slow geographic speciation, often preceded by clinal morphological variation and extended reticulation, predominates in several groups of perennial species, other groups, containing mostly annuals, produced many reproductively isolated, but sometimes sympatric species (Hellwig, 2004). Taking these factors into account, subtribe Centaureinae appear to be an excellent model group in which to analyse satellite DNA and the influence that the biology and/or the evolution of its different lineages have had on the evolution of this class of repetitive DNA.
Satellite DNA sequences are families of short, highly repetitive sequences found in heterochromatin mainly in pericentromeric and sub-telomeric regions of eukaryote chromosomes (López-Flores and Garrido-Ramos, 2012; Plohl et al., 2012). Monomer sequences of a satellite DNA evolve concertedly through a process of molecular drive in which mutations are homogenized in a genome and fixed in a population at a higher rate than that at which they arise. This process results in rapid divergence of satellite sequences in reproductively isolated groups of organisms (Plohl et al., 2012). Nevertheless, the overall variability profile of satellite DNA monomers in a genome is a complex feature that depends on genomic conservation and divergence of satellite DNAs, distribution and homogenization patterns among variants, putative selective constraints imposed on them, reproduction mode and population factors (Plohl et al., 2010, 2012). Therefore, concerted evolution might be slowed down due to satellite DNA location, organization and repeat-copy number (Navajas-Pérez et al., 2005, 2009), functional constraints (Mravinac et al., 2005) or biological factors (Luchetti et al., 2003, 2006; Robles et al., 2004; Suárez-Santiago et al., 2007a). In the absence of selective and biological constraints, the rate of concerted evolution of a family of satellite DNA sequences should thus depend basically on the divergence time between species (Pérez-Gutiérrez et al., 2012). However, the high rate of sequence changes in the course of concerted evolution is not the only possible cause that may explain diversity among dominant satellite DNAs in related species (Plohl et al., 2012). In addition to the great potential for sequence change, satellite DNAs are permanently altered in copy number by expanding and contracting arrays of satellite monomers. Since several satellite DNA families or subfamily variants of one family may coexist in a genome, the evolution of species-specific satellite DNA composition can be directed by copy number changes within a library of satellite sequences common for a group of species (Plohl et al., 2012). According to the library hypothesis of satellite DNA evolution, the occurrence of a species-specific profile of satellite DNAs results from differential amplifications and/or contractions within a pool of sequences shared by related genomes (Fry and Salser, 1977). The library of satellite DNAs represents a permanent source of sequences that can be independently amplified in each species into a dominant satellite DNA, rapidly changing any profile of genomic satellite DNA (Plohl et al., 2012).
The HinfI satellite DNA family was isolated from the genomes of several species of Centaurea, Phonus and Carthamus (Suárez-Santiago et al., 2007a). In Centaurea, the absence of concerted evolution was visualized by similar levels of intraspecific variation and interspecific divergence, lack of fixed species-diagnostic nucleotide sites and the coexistence of several subfamilies within the same species, all of these features reflecting the reticulate mode of evolution in this genus. Taking this into account and having found that this satellite DNA is conserved throughout the subtribe, we extend our previous study to analyse here the HinfI repeats in the rest of the Centaureinae in order not only to check the different hypotheses on satellite DNA evolution within and between the two main radiation rounds but also to look for the value of these types of sequences as markers of the different evolutionary processes in this highly diversified group.
MATERIALS AND METHODS
We analysed 502 HinfI repeat units isolated from 38 species belonging to ten genera of subtribe Centaureinae: Centaureae, Carthamus, Phonus, Carduncellus, Rhaponticoides, Rhapon-ticum, Klasea, Crupina, Cheirolophus and Volutaria (Table 1). We included 55 representative sequences, previously analysed by us (Suárez-Santiago et al., 2007a), belonging to Centaurea granatensis, C. alba subsp. alba, C. corymbosa, C. boissieri subsp. willkommii, C. delicatula, C. jaennensis and Phonus arborescens. The mean number of selected sequences (representative of each of three HinfI satellite subfamilies characterized) was eight. The rest of the repeat sequences (447) were determined for the first time for this work and belong to additional Centaurea spp. and species of the other nine genera. The mean number of HinfI sequences studied from each new species analysed was 14 and we never analysed fewer than ten repeats per species in these cases (Table 1). For Crupina crupinastrum, we analysed 29 repeats, because we studied two populations of this species, one formerly considered C. matae, a synonym of C. crupinastrum. The EMBL accession numbers for the 55 sequences selected from our previous study (Suárez-Santiago et al., 2007a) are AM712738–AM712751, AM712761–AM712774, AM712803–AM712809, AM712815–AM712821 and AM712828–AM712840. The EMBL accession numbers for the remaining 447 sequences analysed in this study for the first time are HF571538–HF571985.
Table 1.
List of species analysed
| Taxa | Locality/Voucher/Reference* | No. of repeats analysed |
|---|---|---|
| Centaurea L. | ||
| Centaurea subgenus Acrocentron | ||
| C. clementei Boiss. | SP: Cádiz, Grazalema/GDAC 5102 | 11 |
| C. granatensis Boiss. | SP: Granada, Huétor Santillán/GDA 46119† | 9 |
| C. saxifraga Coincy | SP: Granada, Zújar/GDA 499332 | 10 |
| Centaurea subgenus Cyanus | ||
| C. cyanus L. | SP: Ctra. Ávila-Salamanca/GDAC 23553 | 14 |
| Centaurea subgenus Centaurea | ||
| Centaurea section Acrolophus-Phalolepis (Cass.) Dostál | ||
| C. alba L. subsp. alba | SP: Ávila, Sierra del Águila/GDAC 24948† | 7 |
| C. corymbosa Pourret | FR: Narbonne, La Clape/BC† | 7 |
| Centaurea section Willkommia Blanca | ||
| C. boissieri D.C. subsp. willkommii (Schultz Bip. ex Willk.) Dostál | SP: Murcia, Sierra Espuña/GDAC 6649† | 14 |
| C. delicatula Breitw & Podlech | TN: Djebel Chambi/MPU† | 6 |
| C. jaennensis Degen and Debeaux in Degen | SP: Jaén, Pozo Alcón/GDAC 6724† | 8 |
| Carthamus L. | ||
| C. tinctorius L. | SP: Sevilla/GDAC 13709 | 18 |
| C. lanatus L. | SP: Córdoba, Sierra Morena, Los Patalos/GDAC 39253 | 15 |
| Phonus Hill | ||
| P. arborescens (L.) G.López | SP: Granada, Sierra de Lújar/GDA 18987‡ | 19 |
| Carduncellus Adans. | ||
| C. caeruleus (L.) C.Presl | SP: Málaga/GDA 49305 | 12 |
| C. calvus Boiss. & Reut. | MO: Beni Hosmar, Montis Borgues/GDA 3998 | 16 |
| C. cuatrecasasii G.López | SP: Granada, Pedro Martínez/GDA 25041 | 10 |
| C. dianius Webb | SP: Alicante, Mongó/GDAC 4193 | 14 |
| C. hispanicus Boiss. ex DC. | SP: Granada, Sierra Sagra/GDAC 20480 | 12 |
| C. mitissimus (L.) DC. | SP: León, Solana del Puerto de Ventana/GDAC 13047 | 13 |
| Rhaponticoides Vaill. | ||
| R. alpina (L.) M.V.Agab. & Greuter§ | SP: Jaén, Sa de las Villas/GDAC 55133 | 15 |
| R. linaresii (Lázaro Ibiza) M.V.Agab & Greuter§ | SP: Palencia, Dueñas/GDAC 13180 | 13 |
| Rhaponticum Vaill. | ||
| R. acaule (L.) DC. | MO: Región de Monte Arruit/GDA 5265 | 13 |
| R. berardioides Batt. | MAR: Talasentant/GDA 4998 | 13 |
| R. coniferum (L.) DC. | SP: Granada, Pantano de Cubillas/GDA 42407 | 12 |
| R. centauroides (L.) Holub | SP: Lérida, Valle de Arán/GDAC 24874 | 14 |
| R. exaltatum (Willk.) Greuter | SP: Ávila, Hoyocasero/GDAC 34999 | 15 |
| Klasea Cass. | ||
| K. flavescens (L.) Holub subsp. flavescens | SP: Granada, Lanjarón/GDA 54612 | 11 |
| K. flavescens subsp. leucantha (Cav.) Cantó & Rivas Mart. | SP: Granada, Cerro Gordo/GDAC 10730 | 13 |
| Crupina (Pers.) DC. | ||
| C. crupinastrum (Moris) Vis. | SP: Granada, Alhama de Granada/GDA 54194 | 29 |
| C. vulgaris Cass. | SP: Córdoba, Sa Morena/GDAC 42205 | 13 |
| Cheirolophus Cass. | ||
| C. falcisectus Montelongo & Moraleda | SP: Gran Canaria, Jardín Botánico Viera y Clavijo/GDAC 44225 | 16 |
| C. intybaceus (Lam) Dostál | SP: Almería, Níjar/GDA 25217 | 15 |
| C. sempervirens (L.) Pomel | SP: Granada, Sa de Cázulas/GDA 52606 | 12 |
| C. sventenii (A.Santos) G.Kunkel | SP: Gran Canaria, Jardín Botánico Viera y Clavijo/GDAC 44224 | 14 |
| C. teydis (Buch) G.López | SP: Santa Cruz de Tenerife, Circo de Las Cañadas del Teide/GDA 41753 | 15 |
| Volutaria Cass. | ||
| V. crupinoides (Desf.) Maire | MO: Driouch/GDAC 39797 | 12 |
| V. lippii (L.) Maire | SP: Almería/GDA 52154 | 14 |
| V. muricata (L.) Maire | SP: Málaga/GDAC 42882 | 14 |
| V. tubuliflora (Murb.) Sennen | SP: Almería, Tabernas/GDAC 43411 | 14 |
*FR, France. MO, Morocco. SP, Spain. TN, Tunisia. GDA/GDAC, University of Granada Herbarium. BC, Barcelona herbarium. MPU, University of Montpellier Herbarium.
†Sequences previously described in Suárez-Santiago et al. (2007a).
‡Four repeats previously described in Suárez-Santiago et al. (2007a) plus 15 new repeats.
§These species have been considered synonyms.
Total genomic DNA was extracted from dried leaves from the Herbarium of the University of Granada (Spain) using a DNAeasy Plant Mini Kit (Qiagen). To amplify the monomeric units of the HinfI satellite DNA, the primer pairs CenHinf1 and CenHinf2 (5'-GCTTCGTTTTGATAGTTCGTGG-3' and 5'-TAACTTTTGCTACGGGAGTCCG-3') were used. These primers were internal primers, oriented oppositely with respect to each other, and designed from the most conserved region of HinfI satellite DNA sequences (see below). The location of these primers is indicated in Fig. 1. As satellite DNA sequences are tandemly arranged, we expected to obtain complete monomer and multimer sequences flanked by partial monomer sequences that are excluded from the analysis. This procedure enables the ends of the amplified product to be avoided. PCR amplifications were performed in a GeneAmp 2700 Applied Biosystems Thermocycler. Polymerase chain reactions were performed in a volume of 50 µL of mix containing 10 mm 10× PCR buffer, 2·5 mm MgCl2, 200 µm of a mix of each dNTP, 1 µm of each primer, 2·5 U of Taq DNA polymerase (Amersham Biosciences) and 10 ng of DNA. The PCR conditions were 40 cycles of 94 °C for 60 s, 50 °C for 60 s and 72 °C for 60 s. After these 40 cycles, there was a 10 min extension period at 72 °C. Amplification was detected on 1·5 % agarose gels stained with SYBR safe®. The PCR products were excised from the gels and the DNA was purified using the GFX™ PCR DNA and the Gel Band Purification Kit (Amersham Biosciences). The purified PCR products were ligated into the pSC-A-amp/kan vector of the StrataClone PCR Cloning Kit (StrataGene) and cloned in StrataClone SoloPack competent cells (StrataGene) following the manufacturer's recommendations. The transformed colonies were transferred to a fresh plate and in 10 µL of a PCR mixture to verify their transformation. Positive clones were purified using a commercial kit (Dominion, Spain).
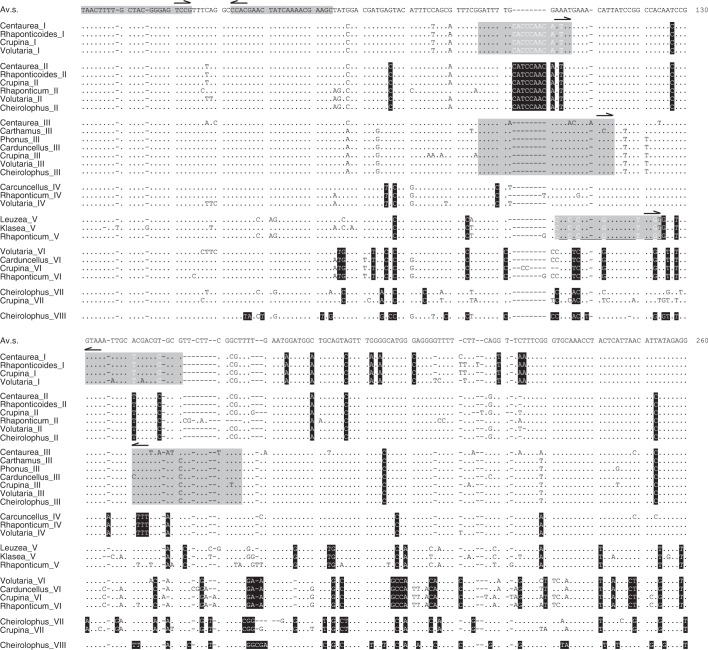
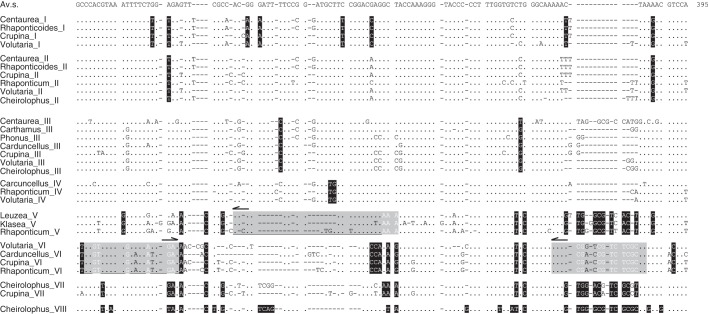
Fig. 1.
Sequence alignment of average sequences of the different HinfI satellite DNA subfamilies from different genera. The first line is an average sequence (Av.s.) for all the HinfI sequences. Only base changes to the average sequence are shown in individual genera and subfamilies. Diagnostic positions for each subfamily are shaded black. The annealing regions of the primer pairs used for the amplification of the repeats are indicated (grey).
Automated sequencing of the plasmid clones was performed in both directions using the generic primers M13F and M13R. Thermal cycling reactions were performed using the ABI Prism® Big Dye™ Terminator Cycle Sequencing Kit (Applied Biosystems). Sequencing was carried out on an ABI PRISM® 3100-Avant Genetic Analyzer (Applied Biosystems).
Multiple sequence alignments were performed using ClustalX (Higgins and Sharp, 1988). Maximum likelihood (ML) and Bayesian analysis were used to infer phylogeny. For selection of the DNA substitution model, we used MEGA v.5 (Tamura et al., 2011). The model with the lowest AIC (Akaike information criterion) score was chosen (GTR + G). The ML analysis was conducted in MEGA v.5 (Tamura et al., 2011). Bootstrap support values were calculated on 1000 replicates (Felsenstein, 1985). Bayesian posterior probabilities (PPs) were estimated using MrBayes v.3·2 (Ronquist et al., 2012). The extent of rate variation across sites for individual data partitions was estimated by the shape parameter of the gamma distribution(s). Each Bayesian analysis was initiated with random starting trees and was run for 1 million generations with the sampling frequency of trees set at the 500th iteration. For all analyses, the variance of split sequences was 0·01. The potential scale reduction factor (PSRF) was close to 1·0 for all of the substitution model parameters. The fraction of sampled values discarded as burn-in was set at 0·25. Posterior probabilities of 0·95–1·00 were considered statistically significant. Trees were displayed with FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
The distribution of nucleotide variability throughout the monomer sequences was determined using the sliding window command (window length = 1, step size = 1) implemented in the DNA polymorphism and DNA divergence between populations options of DnaSP v.5 (Librado and Rozas, 2009). The conserved and variable regions were considered significant when Pi exceed the average Pi ± 2 × s.d. (standard deviation), as described in Mravinac et al. (2004). In addition, conserved regions along the HinfI repeats were determined by the Conserved DNA Region option (minimum window length = 19, conservation threshold = 0·11) of the DnaSP v.5 program.
Sequence analysis revealed the existence of up to eight different HinfI satellite DNA subfamilies amplified by the primers CenHinf1 and CenHinf2. These subfamilies were differentially represented in different species. In order to re-check the distribution of these subfamilies, four additional primer pairs were designed for the specific amplification of repeats of subfamilies I, III, V and VI, and they were probed in a set of representative species. The sequences of these primers were chosen from specific regions defining the subfamily type, and their location is indicated in Fig. 1. To amplify the monomeric units of subfamily I of the HinfI satellite DNA, the primer pairs CentOne-F and CentOne-R (5'-GATTTTGCACCCAACAATAT-3' and 5'-TCCATCTATTCAAACGCGC-3') were used, for those of subfamily III, the primer pairs CentThree-F and CentThree-R (5'-GATTTTGGAAATTAACAATT-3' and 5'-AGCCGAAGAAGGCAATAACT-3') were used, for those of subfamily V, the primers CentFive-F and CentFive-R (5'-CAACGCAACGTTATCCGTCCGC-3' and 5'-TTTCTCGTCCGGACCGACCGTG-3') were used, and for those of subfamily VI, the primers CentSix-F and CentSix-R (5'-CCGTGTAAAGTATCATGGAA-3' and 5'-TAGCGAGACGGTCGGTTT-3') were used. The presence/absence analysis of each subfamily in each analysed species was achieved by gel electrophoresis on 1·5 % agarose gels stained with SYBR safe®. Amplified products were sequenced to verify their subfamily provenance.
RESULTS
The primer pairs CenHinf1 and CenHinf2 were used for the amplification of HinfI repeats from the genomes of 38 species, the PCR products were cloned and 502 HinfI cloned repeats were sequenced. These repeats were ascribed to eight monomer types or subfamilies. These subfamilies were established according to a set of diagnostic positions given by a specific mutation shared by all the sequences of one group (Fig. 1). They were designated with Roman numerals from I to VIII following the nomenclature previously used in Suárez-Santiago et al. (2007a) for three of them (subfamilies I, II and III). Additionally, the different types of sequences have diagnostic deletions found at different positions in the HinfI repeats. To study diversity distribution along the repeat sequences, a sliding window analysis was performed using a window length of 10 and step size 1 (see Supplementary Data Fig. S1). Windows that exhibit diversity ≤ (average + 2 s.d.) were defined as variable, and those with diversity ≥ (average –2 s.d.) were considered as conserved. The analysis reveals one conserved segment from positions 1 to 50 resulting from the overlapping of the neighbouring windows. The Conserved DNA Region option (minimum window length = 19, conservation threshold = 0·11) of the DnaSP v.5 program identified the same region, from sites 1 to 48, as a significant conserved region. This region coincides with that from which the primers were designed (Fig. 1). The rest of the repeat sequences was variable and included the diagnostic positions defining each subfamily.
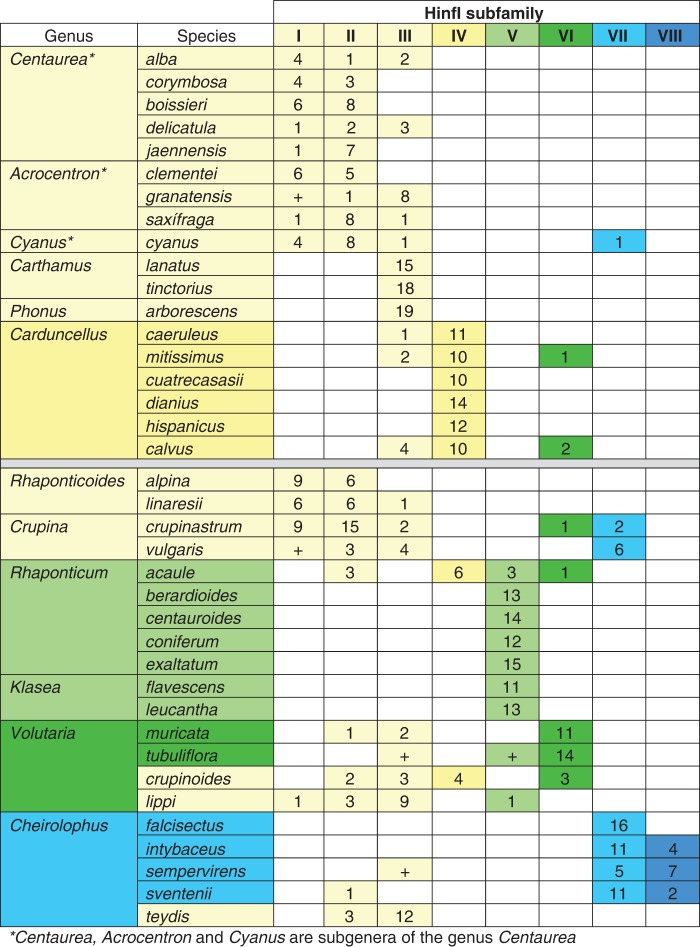
Figure 2 shows the distribution of HinfI subfamilies among species. Species of Centaurea and Rhaponticoides were characterized by the presence in their genomes of the HinfI sequences belonging to subfamilies I, II and III, some with sequences of two or the three subfamilies coexisting in the same species. In both Rhaponticoides spp. analysed, sequences belonged either to subfamily I or to subfamily II, with one sequence of R. linaresii belonging to subfamily III. In the case of Centaurea, subfamilies I and II were found in all species of subgenus Centaurea, with the presence of subfamily III in two species. In the species analysed of subgenus Cyanus (C. cyanus), we found eight out of 13 repeats belonging to subfamily II, but subfamilies I (four repeats) and III (one repeat) were also found. In addition, one of the sequences isolated from this species belonged to subfamily VII. In the case of subgenus Acrocentron, in C. clementei we did not find HinfI type III sequences, and subfamily I appeared to be absent from C. granatensis. The genomes of the two Crupina spp. analysed had sequences of subfamilies I, II and III and, additionally, we found up to six (out of 13) repeats of subfamily VII in C. vulgaris and three repeats (out of 29) of subfamilies VI (one) and VII (two) in C. crupinastrum (Table 1, Fig. 2). In the species of Carthamus and Phonus, we found only HinfI type III sequences. Subfamily IV is characteristic of Carduncellus. Nevertheless, some species of this genus also have low-copy repeats of subfamilies III and VI in their genomes. Subfamily V was found in Rhaponticum and Klasea. These species had only this type of HinfI repeat, except R. acaule, which also contained sequences of subfamilies II, IV and VI in its genome. We analysed four species of Volutaria (Table 1). The HinfI sequences of V. lippii belonged to subfamilies I, II, III or V, mainly to type III (nine sequences out of 14). In addition, most HinfI sequences of V. crupinoides belonged to subfamilies II, III and IV. However, most of the sequences of V. muricata and V. tubuliflora and three of V. crupinoides belonged to subfamily VI. Three species of Cheirolophus (C. intybaceus, C. sempervirens and C. sventenii) shared two types of HinfI monomers in their genomes, those of subfamilies VII and VIII. Cheirolophus sventenii also had a type II repeat. Cheirolophus falcisectus had only sequences of subfamily VII. In contrast, among the sequences isolated from C. teydis, 12 belonged to subfamily III and the other three to subfamily II (Fig. 2). We tested the reliability of the CenHinf1 and CenHinf2 primers to detect the eight subfamilies in these species. For this, we designed four additional primer pairs for the specific amplification of repeats of subfamilies I, III, V and VI, and probed them in a set of representative species. We obtained results similar to those obtained with the general primers. Nevertheless, some differences were found (Fig. 2; Supplementary Data Fig. S2). For example, subfamily I is present in Centaurea granatensis and Crupina vulgaris (the only species of Centaurea and Crupina for which we did not obtain repeats of subfamily I after sequencing of amplified product using the primers CenHinf1 and CenHinf2). The subfamily III is found in species of Volutaria and Cheirolophus other than those previously detected with the ‘general’ primers. However, the primers used were not able to amplify repeats of subfamily III in Carduncellus (even though we detected a few repeats of this type using the CenHinf1 and CenHinf2 primers). Subfamily V is found not only in Klasea and Rhaponticum but also in Volutaria, as we previously found by sequencing the CenHinf1/CenHinf2 amplified product. Subfamily VI was found only in Volutaria, as detected by CenHinf1 and CenHinf2 primers.
Fig. 2.
Survey of HinfI subfamilies in Centaureinae. Numbers indicate the number of sequences isolated of each subfamily from each species. The symbol ‘ + ’ indicates that the presence of the subfamily was detected in that species by PCR amplification using subfamily-specific primers. Colour codes indicate the subfamily type. A grey line separates the derived clade and the basal groups.
Phylogenetic reconstruction by ML and Bayesian inference resulted in similar trees. A Bayesian majority-rule consensus tree with Bayesian PPs is shown in Supplementary Data Fig. S3. A simplified tree layout without tip labels is shown in Fig. 3. The trees did not group sequences by specific or generic affinity. Although the tree grouped together the HinfI monomers of some particular genera or subtribal group, the most common view is that the repeats are grouped by HinfI subfamilies, which are conserved among different species, as classified in Figs 1 and 2. The tree represented in Fig. 3 shows indications of the correspondence between clades and HinfI subfamilies. We can distinguish two main clades. The first clade includes five subclades, each one corresponding to each of subfamilies I–V. The second clade includes three subclades, one for each of subfamilies VI–VIII.
Fig. 3.
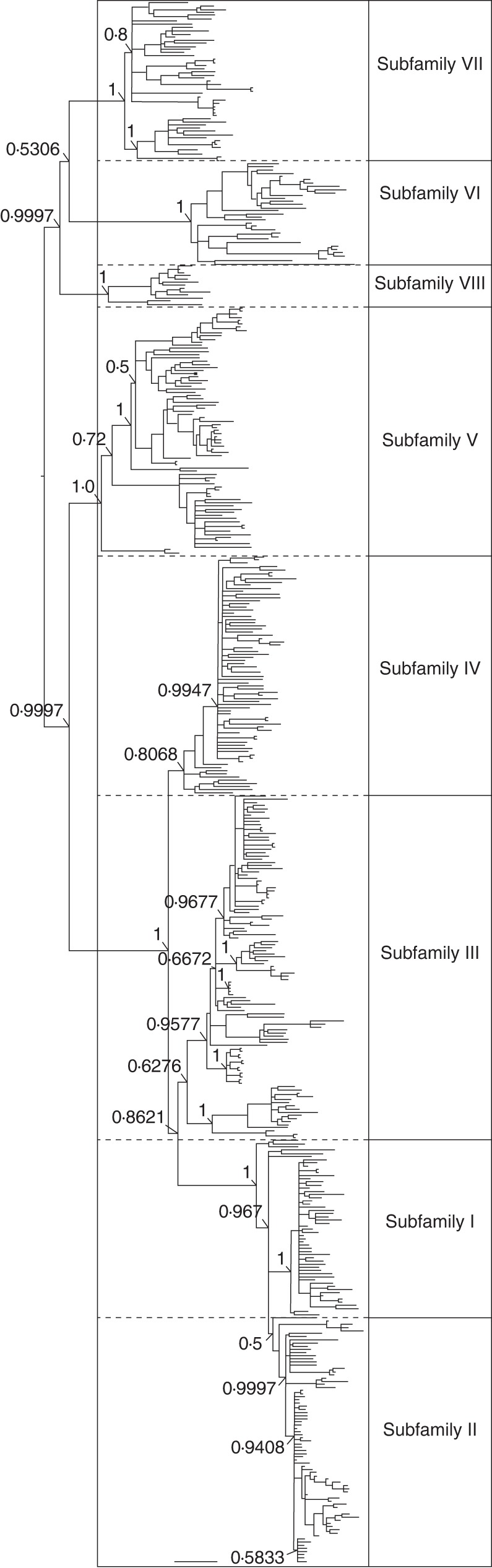
Majority-rule consensus tree based on Bayesian MCMC analysis of the HinfI repeat sequences. This figure represents a simplified tree layout without tip labels, indicating the correspondence between major clades and HinfI subfamilies. Bayesian posterior probability values for the main nodes are indicated. For a detailed version of this tree, refer to Supplementary Data Fig. S3.
In each clade, sequences of subfamilies I, II or III of the different species are intermingled in a species-independent manner (Supplementary Data Fig. S3). However, most sequences of subfamily III belonging to Phonus and Carthamus species tend to be grouped by taxonomic affinity, on one hand the sequences of Phonus arborescens and, on the other, the sequences of Carthamus tinctorius and those of Carthamus lanatus, although there are some intermixed sequences from each (Supplementary Data Fig. S3). In contrast, comparisons of subfamily III sequences of these species and low-copy counterparts of subfamily III in other species examined show high interspecific sequence conservation and the complete lack of any species-diagnostic mutations, and therefore they appear to be intermixed in the subfamily III clade (Supplementary Data Fig. S3). HinfI sequences of Carduncellus (subfamily IV) appear intermingled without separation by specific affinity (Supplementary Data Fig. S3). In the case of Rhaponticum and Klasea, sequences tend to be grouped by specific affinity (Supplementary Data Fig. S3). Sequences of subfamily VI of Volutaria are separated according to species of origin (Supplementary Data Fig. S3). However, the sequences of the two different subfamilies found in Cheirolophus (VII and VIII) are not grouped in phylogenetic trees by specific affinity and appear to be intermixed (Supplementary Data Fig. S3).
DISCUSSION
HinfI sequences have been found to be present in the genomes of all the species analysed of subtribe Centaureinae. These species are representative of the entire range of groups in this subtribe (Garcia-Jacas et al., 2001; Hellwig, 2004). The first phase of radiation of the subtribe might date to the late Oligocene and Miocene. Therefore, the HinfI satellite DNA would date to at least 28–23 million years ago (Garcia-Jacas et al., 2001; Hellwig, 2004). This is not common among satellite DNA families, specifically in plants, the most ancient found exceptionally in cycads (Cafasso et al., 2003).
We identified eight HinfI subfamilies. Our results suggest that the eight HinfI subfamilies were present in the common ancestor of Centaureinae, each one spreading differentially in different genera. The differential spreading accompanied the two main phases of radiation leading to two major groups in Centaureinae (Garcia-Jacas et al., 2001; Hellwig, 2004). Thus, subfamilies V–VIII are found to prevail in older genera (first phase of radiation in the subtribe, late Oligocene–Miocene), although a few repeats of subfamilies VI and VII were isolated from Carduncellus and Centaurea (derived clade). Subfamilies I–IV have expanded predominantly in the genomes of species belonging to the derived clade of Centaureinae (second phase of radiation, Pliocene to Pleistocene). Notably, there are several species of the early diverging groups having subfamilies I–III as the major representatives of HinfI sequences in their genomes. These data suggest that subfamilies I–IV have expanded recently, replacing other subfamilies in derived genera and in older genera.
The replacement of one sequence variant by another in different species is a common feature of satellite DNA that could be a consequence of the dynamics of satellite DNA evolution (Plohl et al., 2010, 2012). Molecular mechanisms of non-reciprocal exchange (unequal crossing-over, gene conversion, rolling-circle replication and re-insertion, and transposon-mediated exchange) would spread new sequence variants appearing in individual repeat units of a family of sequences and the changes are fixed in a population of randomly mating individuals by sexual reproduction according to a time-dependent two-step process called molecular drive, which leads to concerted evolution (Plohl et al., 2010, 2012; Pérez-Gutiérrez et al., 2012). Satellite DNA changes due to gradual accumulation of sequence divergence which results in divergence of satellite sequences in reproductively isolated groups of organisms (Plohl et al., 2012). However, given the scattering of subfamilies in each lineage of Centaureinae, an alternative but not mutually exclusive hypothesis could explain this differential distribution. According to the ‘library’ hypothesis, related taxa share a library of different conserved satellite DNA sequences (different satellite DNA families but also monomer variants or subfamilies of a satellite DNA family), which may be differentially amplified in each taxa. Variability can remain for long evolutionary periods by reduced action of molecular mechanisms of non-reciprocal exchange, and sequence variants persist as a library (Mravinac et al., 2002; Mestrovic et al., 2006) from which any of them may be differentially amplified in each taxon with the subsequent replacement of one sequence variant by another in different species. When this occurs, the study of unrelated species-specific dominant satellite DNA repeats reveals the presence of low-copy counterparts of each of them in other examined species, and comparisons of high-copy and low-copy monomer variants of these satellites show high interspecific sequence conservation and the complete lack of any species-diagnostic mutations, as found in Palorus (Meštrović et al., 1998). This hypothesis has been proved in insects (Meštrović et al., 1998; Mravinac et al., 2002; Cesari et al., 2003; Pons et al., 2004) and plants (Navajas-Pérez et al., 2009; Quesada del Bosque et al., 2011), and could explain the main observation made in Centaureinae concerning the scattering of HinfI types. Variation in satellite profiles is found in this case by changes in copy number (Plohl et al., 2012).
However, the overall variability profile of satellite DNA monomers in a genome is a complex feature that depends on several factors such as location, organization and repeat-copy number (Navajas-Pérez et al., 2005, 2009), time (Pérez-Gutiérrez et al., 2012), biological factors (Luchetti et al., 2003, 2006; Robles et al., 2004; Suárez-Santiago et al., 2007a) and functional constraints (Mravinac et al., 2005). Some patterns of HinfI repeat evolution in specific lineages of Centaureinae might result from the influence of some of these factors, discussed below.
Derived clade
In phylogenentic analyses of subtribe Centaureinae (Garcia-Jacas et al., 2001), in the derived clade, the Carthamus complex occupies the earliest diverging position, and subgenera Jacea and Cyanus of Centaurea, for which a sister relationship is firmly established, are the most derived (Susanna and Garcia-Jacas, 2009). Subgenus Acrocentron of Centaurea occupies an intermediate position, although connections between this subgenus and Jacea and Cyanus remain unclear (Garcia-Jacas et al., 2001; Susanna and Garcia-Jacas, 2009).
Two main observations can be emphasized with respect to the spread and evolution of HinfI sequences in this group. First, there are four HinfI subfamilies (I–IV) that have spread through the genomes of these species accompanying the major speciation processes. However, the four repeat subfamilies are not equally distributed in all the taxa analysed (Fig. 2). Subfamilies I and II spread secondarily as specific subfamilies of Centaurea, almost completely replacing subfamily III and completely replacing subfamily IV, whereas subfamily III expanded in Phonus and Carthamus and subfamily IV expanded in Carduncellus. Nevertheless, the four subfamilies are found in species belonging to the earliest diverging clade (first radiation). In addition to the presence of low-copy repeats of these four subfamilies in some species of the early diverging groups, in some of these species (Cheirolophus teydis, Volutaria lippii and Crupina spp.) these types are the main components of HinfI sequences in their genomes. These findings support the library hypothesis, as discussed above. In this sense, these subfamilies might be as old as the rest of the subfamilies studied in this paper, differentially expanding in different lineages. Regardless of phylogeny, in agreement with the library hypothesis, there is a convergence in the spread of some subfamily variants in different lineages. It should be remembered that the hypothesis does not predict whether any of the sequences of the library can be amplified into a major satellite family/subfamily or whether there is selective pressure favouring some sequences or the amplification mechanism involved (Fry and Salser, 1977).
Secondly, several comments on concerted evolution should be emphasized. Differential speciation pathways gave rise to differential patterns of sequence evolution in different lineages. Different subfamilies coexist in most of the taxa analysed in Centaurea. The presence of different subfamilies in their genomes was explained as a result of reticulate evolution in a part of this genus (Suárez-Santiago et al., 2007a, b). Hybridization should be a process that maintains different HinfI satellite DNA subfamilies in a particular genome. In addition, gene flow between taxa should reduce the amount of genetic differences between those taxa but should increase the amount of intraspecific variation. Thus, under this evolutionary scenario, contrary to the expectations on the concerted evolution model, we should find similar or even higher levels of intraspecific variation than interspecific divergence. Additionally, the library hypothesis might explain the existence of additional copies of some subfamilies in some species. Species of Carthamus and Phonus have type III sequences and they are almost differentiated in such a way that intraspecific variation is lower than interspecific divergence, a sign of concerted evolution. Phonus and Carthamus, formerly considered synonymous, are separate genera (Vilatersana et al., 2000). In fact, Phonus is closer to Carduncellus than to Carthamus and has differentiated biological traits and habitats and biogeographical and evolutionary stories (Vilatersana et al., 2000). Carthamus contains two rather different groups: Carthamus sensu stricto, which includes only section Carthamus (type species C. tinctorius); and section Atractylis (type species C. lanatus) (Vilatersana et al., 2005). Type III sequences of both species are differentiated in the tree in Supplementary Data Fig. S3. Concerted evolution is not found in Carduncellus, the species of which have subfamily IV. Intraspecific variability is greater than interspecific divergence in this genus, and HinfI sequences appear intermixed in the phylogenetic tree. Speciation in Carduncellus is a recent relative to other Centaureinae, and the HinfI divergence values such as those found in a different study for the ITS (internal transcribed spacer) divergence values were low (Vilatersana et al., 2000). Failure to resolve relationships in Carduncellus highlights the extremely low level of variation in many genera of recent origin (Nepokroeff and Sytsma, 1996; Susanna et al., 1999). In fact, in the absence of selective and biological constraints, the rate of concerted evolution of a family of satellite DNA sequences should depend basically on the divergence time between species (Pérez-Gutiérrez et al., 2012).
Early diverging groups
Major clades of early diverging Centaureinae have been established, but some genera could not be clearly classified in any group. Molecular phylogeny indicated that these genera constitute an old stock in the subtribe, suggesting that divergence between all these genera is old (Garcia-Jacas et al., 2001; Susanna and Garcia-Jacas, 2009). However, recent phylogenetic analyses (Susanna et al., 2011) suggested informal entities, as natural groups and phylogenetic relationships have been partially resolved. The Volutaria group is located close to the base of the phylogenetic trees and includes Volutaria and Amberboa, among others (Susanna et al., 2011). The crown node for the clade of basal Centaureinae received good support, but relationships among the main groups in that clade remain poorly resolved, forming a general trichotomy (Susanna et al., 2011). It includes a strongly supported clade of Cheirolophus, another equally supported clade which encompasses Rhaponticum (including former Acroptilon and Leuzea), Myopordon and Oligochaeta, and a third clade containing the remaining taxa. This latter clade comprises a clade which includes, among others, Klasea and Serratula, and, finally, an unsupported polytomy includes three clades (Susanna et al., 2011): the first contains Plectocephalus; the second contains Stizolophus; and the third is a moderately supported clade, containing Crupina and Rhaponticoides, among others.
Here, we also highlight two main observations with respect to HinfI sequence distribution and evolution in these species. First, there are four HinfI subfamilies (V–VIII) that have spread through the genomes of these species, accompanying the major speciation processes. In contrast to subfamilies I–IV, in this case each subfamily has spread almost exclusively within one or two specific genera: subfamily V is almost exclusive to Rhaponticum and Klasea (although we have also detected it in some species of Volutaria), subfamily VI spread in some species of Volutaria, and subfamilies VII and VIII spread in most species of Cheirolophus analysed. We can conclude in this case that these distributions are old and restricted to specific lineages. The presence of some copies of these types of HinfI repeats in a few additional species again supports the library hypothesis.
Secondly, referring to concerted evolution, we can conclude in this case also that differential speciation pathways gave rise to differential patterns of sequence evolution in different lineages. Sequences of subfamily V evolve in concert in the Klasea–Rhaponticum clade (Supplementary Data Fig. S3). Sequences are in general more similar within species than between species and they tend to be grouped according to species affinity, thus confirming that the spreading of this subfamily precedes the diversification of the genus and that this speciation is old in Centaureinae. Subfamily VI of HinfI sequences is found in Volutaria muricata and V. tubuliflora (and in low copy number in a few more species). The sequences of these species cluster separately, a sign of homogenization within species and differentiation between these two species (Supplementary Data Fig. S3), i.e. concerted evolution. However, sequences of subfamilies VII and VIII are distributed equally throughout all Cheirolophus spp. analysed in this study (except in C. teydis, as discussed above). In this genus, HinfI sequences of different species are more similar than when compared within species and they appear intermingled in the phylogenetic trees. As with Carduncellus, Susanna et al. (1999) found speciation in Cheirolophus to be recent relative to other Centaureinae, which might be a factor influencing the absence of concerted evolution (Pérez-Gutiérrez et al., 2012).
SUPPLEMENTARY DATA
LITERATURE CITED
- Cafasso D, Cozzolino S, De Luca P, Chinali G. An unusual satellite DNA from Zamia paucijuga (Cycadales) characterised by two different organisations of the repetitive unit in the plant genome. Gene. 2003;311:71–79. doi: 10.1016/s0378-1119(03)00555-9. [DOI] [PubMed] [Google Scholar]
- Cesari M, Luchetti A, Passamonti M, Scali V, Mantovani B. Polymerase chain reaction amplification of the Bag320 satellite family reveals the ancestral library and past gene conversion events in Bacillus rossius (Insecta Phasmatodea) Gene. 2003;312:289–295. doi: 10.1016/s0378-1119(03)00625-5. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fry K, Salser W. Nucleotide sequence of HS-α satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell. 1977;12:1069–1084. doi: 10.1016/0092-8674(77)90170-2. [DOI] [PubMed] [Google Scholar]
- Garcia-Jacas N, Susanna A, Garnatje T, Vilatersana R. Generic delimitation and phylogeny of the subtribe Centaureinae (Asteraceae): a combined nuclear and chloroplast DNA analysis. Annals of Botany. 2001;87:503–515. [Google Scholar]
- Hellwig FH. Centaureinae (Asteraceae) in the Mediterranean – history of ecogeographical radiation. Plant Systematics and Evolution. 2004;246:137–162. [Google Scholar]
- Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- López-Flores I, Garrido-Ramos MA. The repetitive DNA content of eukaryotic genomes. In: Garrido-Ramos MA, editor. Repetitive DNA. Genome dynamics. vol 7. Basel: Karger; 2012. pp. 1–28. [DOI] [PubMed] [Google Scholar]
- Luchetti A, Cesari M, Carrara G, et al. Unisexuality and molecular drive: Bag320 sequence diversity in Bacillus taxa (Insecta Phasmatodea) Journal of Molecular Evolution. 2003;56:587–596. doi: 10.1007/s00239-002-2427-9. [DOI] [PubMed] [Google Scholar]
- Luchetti A, Marini M, Mantovani B. Non-concerted evolution of RET76 satellite DNA family in Reticulitermes taxa (Insecta, Isoptera) Genetica. 2006;128:123–132. doi: 10.1007/s10709-005-5540-z. [DOI] [PubMed] [Google Scholar]
- Meštrović N, Plohl M, Mravinac B, Ugarkovic D. Evolution of satellite DNAs from the genus Palorus – experimental evidence for the library hypothesis. Molecular Biology and Evolution. 1998;15:1062–1068. doi: 10.1093/oxfordjournals.molbev.a026005. [DOI] [PubMed] [Google Scholar]
- Meštrović N, Castagnone-Sereno P, Plohl M. Interplay of selective pressure and stochastic events directs evolution of the MEL172 satellite DNA library in root-knot nematodes. Molecular Biology and Evolution. 2006;23:2316–2325. doi: 10.1093/molbev/msl119. [DOI] [PubMed] [Google Scholar]
- Meusel H, Jäger EJ. Jena: Fischer Verlag; 1992. Vergleichende Chorologieder zentraleuropäischen Flora 3. Band III (Text- und Kartenteil) [Google Scholar]
- Mravinac B, Plohl M, Meštrovi N, Ugarkovic D. Sequence of PRAT satellite DNA ‘frozen’ in some coleopteran species. Journal of Molecular Evolution. 2002;54:774–783. doi: 10.1007/s00239-001-0079-9. [DOI] [PubMed] [Google Scholar]
- Mravinac B, Plohl M, Ugarkovic D. Conserved patterns in the evolution of Tribolium satellite DNAs. Gene. 2004;332:169–177. doi: 10.1016/j.gene.2004.02.055. [DOI] [PubMed] [Google Scholar]
- Mravinac B, Plohl M, Ugarkovic D. Preservation and high sequence conservation of satellite DNAs indicate functional constraints. Journal of Molecular Evolution. 2005;61:542–550. doi: 10.1007/s00239-004-0342-y. [DOI] [PubMed] [Google Scholar]
- Navajas-Pérez R, de la Herrán R, Jamilena M, et al. Reduced rates of sequence evolution of Y-linked satellite DNA in Rumex (Polygonaceae) Journal of Molecular Evolution. 2005;60:391–399. doi: 10.1007/s00239-004-0199-0. [DOI] [PubMed] [Google Scholar]
- Navajas-Pérez R, Quesada del Bosque ME, Garrido-Ramos MA. Effect of location, organization and repeat-copy number in satellite-DNA evolution. Molecular Genetics and Genomics. 2009;282:395–406. doi: 10.1007/s00438-009-0472-4. [DOI] [PubMed] [Google Scholar]
- Nepokroeff M, Sytsma KJ. Systematics and patterns of speciation and colonization in Hawaiian Psychotria and relatives based on phylogenetic analysis of ITS sequence data. American Journal of Botany. 1996;83:181–182. [Google Scholar]
- Panero JL, Funk VA. The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed. Molecular Phylogenetics and Evolution. 2008;47:757–782. doi: 10.1016/j.ympev.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Pérez-Gutiérrez MA, Suárez-Santiago VN, López-Flores I, Romero AT, Garrido-Ramos MA. Concerted evolution of satellite DNA in Sarcocapnos: a matter of time. Plant Molecular Biology. 2012;78:19–29. doi: 10.1007/s11103-011-9848-z. [DOI] [PubMed] [Google Scholar]
- Plohl M, Petrović V, Luchetti A, et al. Long-term conservation vs high sequence divergence: the case of an extraordinarily old satellite DNA in bivalve mollusks. Heredity. 2010;104:543–551. doi: 10.1038/hdy.2009.141. [DOI] [PubMed] [Google Scholar]
- Plohl M, Meštrović N, Mravinac B. Satellite DNA evolution. In: Garrido-Ramos MA, editor. Repetitive DNA. Genome dynamics vol 7. Basel: Karger; 2012. pp. 126–152. [DOI] [PubMed] [Google Scholar]
- Pons J, Bruvo B, Petitpierre E, Plohl M, Ugarkovic D, Juan C. Complex structural features of satellite DNA sequences in the genus Pimelia (Coleoptera: Tenebrionidae): random differential amplification from a common ‘satellite DNA library. Heredity. 2004;92:418–427. doi: 10.1038/sj.hdy.6800436. [DOI] [PubMed] [Google Scholar]
- Quesada del Bosque ME, Navajas-Pérez R, Panero JL, Fernández-González A, Garrido-Ramos MA. A satellite DNA evolutionary analysis in the North American endemic dioecious plant Rumex hastatulus (Polygonaceae) Genome. 2011;54:253–260. doi: 10.1139/g10-115. [DOI] [PubMed] [Google Scholar]
- Robles F, de la Herrán R, Ludwig A, Ruiz-Rejón C, Ruiz-Rejón M, Garrido-Ramos MA. Evolution of ancient satellite DNAs in sturgeon genomes. Gene. 2004;338:133–142. doi: 10.1016/j.gene.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, et al. MrBayes 3·2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Santiago VN, Blanca G, Ruiz-Rejón M, Garrido-Ramos MA. Satellite-DNA evolutionary patterns under a complex evolutionay scenario: the case of Acrolophus subgroup (Centaurea L., Compositae) from the western Mediterranean. Gene. 2007a;404:80–92. doi: 10.1016/j.gene.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Suárez-Santiago VN, Salinas MJ, Garcia-Jacas N, Soltis PS, Douglas ES, Blanca G. Reticulate evolution in the Acrolophus subgroup (Centaurea L., Compositae) from the western Mediterranean: origin and diversification of section Willkommia Blanca. Molecular Phylogenetics and Evolution. 2007b;43:156–172. doi: 10.1016/j.ympev.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Susanna A, Garcia-Jacas N. Tribu Cardueae. In: Kadereit JW, Jeffrey C, editors. Flowering plants. Eudictos. Asterales (vol. 8, The families and genera of vascular plants. Berlin: Springer Verlag; 2007. pp. 123–146. [Google Scholar]
- Susanna A, Garcia-Jacas N. Cardueae (Carduoideae) In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, editors. Systematics, evolution, and biogeography of the Compositae. Vienna: IAPT; 2009. pp. 293–313. [Google Scholar]
- Susanna A, Garnatje T, Garcia-Jacas N. Molecular phylogeny of Cheirolophus (Asteraceae: Cardueae-Centaureinae) based on ITS sequences of nuclear ribosomal DNA. Plant Systematics and Evolution. 1999;214:147–160. [Google Scholar]
- Susanna A, Galbany-Casals M, Romaschenko K, Barres L, Martin J, Garcia-Jacas N. Lessons from Plectocephalus (Compositae, Cardueae-Centaureinae): ITS disorientation in annuals and Beringian dispersal as revealed by molecular analyses. Annals of Botany. 2011;108:263–277. doi: 10.1093/aob/mcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Masatoshi N, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilatersana R, Susanna A, Garcia-Jacas N, Garnatje T. Generic delimitation and phylogeny of the Carduncellus–Carthamus complex (Asteraceae) based on ITS sequences. Plant Systematics and Evolution. 2000;221:89–105. [Google Scholar]
- Vilatersana R, Garnatje T, Susanna A, Garcia-Jacas N. Taxonomic problems in Carthamus (Asteraceae): RAPD markers and sectional classification. Botanical Journal of the Linnaean Society. 2005;147:375–383. [Google Scholar]
- Wagenitz G. Centaurea in south-west Asia: patterns of distribution and diversity. Proceedings of the Royal Society of Edinburgh. 1986;89B:11–21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.