Abstract
Background and Aims
The Mob1 family includes a group of kinase regulators conserved throughout eukaryotes. In multicellular organisms, Mob1 is involved in cell proliferation and apoptosis, thus controlling appropriate cell number and organ size. These functions are also of great importance for plants, which employ co-ordinated growth processes to explore the surrounding environment and respond to changing external conditions. Therefore, this study set out to investigate the role of two Arabidopsis thaliana Mob1-like genes, namely Mob1A and Mob1B, in plant development.
Methods
A detailed spatio-temporal analysis of Mob1A and Mob1B gene expression was performed by means of bioinformatic tools, the generation of expression reporter lines and in situ hybridization of gene-specific probes. To explore the function of the two genes in plant development, knock-out and knock-down mutants were isolated and their phenotype quantitatively characterized.
Key Results
Transcripts of the two genes were detected in specific sets of cells in all plant organs. Mob1A was upregulated by several stress conditions as well as by abscisic acid and salicylic acid. A knock-out mutation in Mob1B did not cause any visible defect in plant development, whereas suppression of Mob1A expression affected organ growth and reproduction. In the primary root, reduced levels of Mob1A expression brought about severe defects in tissue patterning of the stem cell niche and columella and led to a decrease in meristem size. Moreover, loss of Mob1A function resulted in a higher sensitivity of root growth to abscisic acid.
Conclusions
Taken together, the results indicate that arabidopsis Mob1A is involved in the co-ordination of tissue patterning and organ growth, similarly to its orthologues in other multicellular eukaryotes. In addition, Mob1A serves a plant-specific function by contributing to growth adjustments in response to stress conditions.
Keywords: Mob1, Arabidopsis thaliana, plant development, root tip, stem cell niche, columella, tissue patterning, abscisic acid
INTRODUCTION
In plants, embryogenesis generates only a basic body organization with an apical–basal pattern rather than the complete organism, differently from what occurs in animals (reviewed by Jürgens, 2001; Willemsen and Scheres, 2004; Jenik et al., 2007). Most of the plant body is formed post-embryonically with the continuous generation of new tissues and structures throughout a plant's life. The process of organogenesis occurs in a reiterative form and depends on the activity of meristematic zones located at the tip of plant organs (reviewed by Bäurle and Laux, 2003; Jürgens, 2003). Meristems are constituted by cells that complete several rounds of cell division before undergoing expansion and differentiation. In turn, the replenishment of each meristem is ensured by a pool of stem cells that are maintained undifferentiated in a so-called ‘stem cell niche’. Thus, a tight regulation of cell cycle, mitosis and cell division (cytokinesis) and their co-ordination with cell differentiation are crucial to sustain organ outgrowth and, ultimately, to enable the plant to complete its developmental programme.
Eukaryotic cells evolved signalling components that co-ordinate exit from mitosis with cytokinesis and exit from the cell cycle with differentiation. Knowledge about these mechanisms mostly derives from extensive studies in the fission and budding yeasts, Schizosaccharomyces pombe and Saccharomyces cerevisiae, respectively. Schizosaccharomyces pombe cells divide by constriction of an actomyosin ring and concomitant assembly of a division septum, corresponding to a new cell wall (Gould and Simanis, 1997). Saccharomyces cerevisiae divides by forming a bud (Chant and Pringle, 1995). The onset of septation in S. pombe and budding in S. cerevisiae is signalled through the septation initiation network (SIN) and the mitotic exit network (MEN) signalling pathways, respectively (reviewed by Bardin and Amon, 2001). The SIN and MEN are similar signalling networks using orthologous proteins that control events at the end of mitosis. Both networks consist of a GTPase-activated kinase cascade. In the case of MEN, the activated form of the RAS-like GTPase Tem1 is thought to propagate a signal to the protein kinase Cdc15, which in turn activates the protein kinase Dbf2. It is known that Dbf2 kinase activity requires the Dbf2-associated factor Mob1 (Mah et al., 2001). The Mob1–Dbf2 interaction leads to release from the nucleolus and subsequent activation of Cdc14 phosphatase during anaphase (Stegmeier and Amon, 2004; Mohl et al., 2009). The release of Cdc14 from its inhibitor complex (Shou et al., 1999) promotes the inactivation of the mitotic Cdk1–cyclin B complex, finally driving exit from mitosis (Visintin et al., 1998). Besides its primary role as a promoter of mitotic exit, the MEN has been shown also to control cytokinesis (Lee et al., 2001; Lippincott et al., 2001; Luca et al., 2001). The SIN signalling cascade is organized similarly to the MEN, but its main role is to control cytokinesis by initiating contraction of the actin ring and synthesis of the septum (reviewed by Krapp and Simanis, 2008). In S. pombe, the orthologue of S. cerevisiae Dbf2 kinase is represented by Sid2, whose activity similarly requires the interaction with Mob1 (Hou et al., 2000). Yeast Mob1 proteins do not function solely as activators of Dbf2/Sid2, but are also required for Dbf2/Sid2 localization to activation sites (Frenz et al., 2000; Lee et al., 2001; Hou et al., 2004). Indeed, in agreement with their functions in mitosis exit and cytokinesis, Dbf2/Sid2–Mob1 complexes localize to the spindle pole body (SPB) in anaphase and move to the division site in late mitosis (Yoshida and Toh-e, 2001). Different conditional mutations of yeast Mob1 cause a late nuclear division arrest at the restrictive temperature and result in a quantal increase in ploidy at the permissive temperature (Luca and Winey, 1998). Several components of the MEN and SIN pathways are conserved among eukaryotes and are similarly involved in the regulation of cell division in multicellular organisms (Mailand et al., 2002; Bothos et al., 2005; Hergovich et al., 2006; Bedhomme et al., 2008). Studies in Drosophila have also related Mob proteins to a different signalling pathway that plays a crucial role in tissue growth and cell number control. The protein kinases Hippo (Hpo) and Warts (Wts)/large tumour suppressor (Lats), and the Hpo-scaffold proteins Salvador (Sav) and Mats (Mob as tumour suppressor, dMob1) are the key components of this pathway (Justice et al., 1995; Tapon et al., 2002; Harvey et al., 2003; Lai et al., 2005). Loss of any of these factors results in increased cell proliferation and decreased cell death, indicating that Sav, Hippo, Lats and Mats all function as tumour suppressors. The Dbf2-related Lats is phosphorylated by Hpo and needs to bind to its co-activator Mats to co-ordinate cell death and proliferation properly (reviewed by Pan, 2007). The components of the Hippo pathway are conserved from yeast to flies and humans, suggesting that this signalling cascade plays a fundamental role in cellular regulation.
Cell division is more complex in plants than in animals due to the presence of a rigid external cell wall. In contrast to yeast and animal cells, plant cells undergoing cell division display two unique cytoskeletal structures, namely the pre-prophase band (PPB) and the phragmoplast, which are necessary to ensure adequate positioning and assembly of a new cell wall between the separating sister nuclei (Verma, 2001). On the other hand, plants do not possess SPBs and centrosomes. Despite these differences, several components of the MEN/SIN pathways are conserved in plants (Bedhomme et al., 2008). In particular, several genes encoding putative proteins homologous to yeast Mob1 have been identified in different plant species (Vitulo et al., 2007). In Medicago sativa, Mob1-like genes were shown to be constitutively expressed with a maximum in proliferating tissues (Citterio et al., 2005). The Arabidopsis thaliana genome contains four different Mob1-like genes that can be divided into two sub-groups according to their similarity (Citterio et al., 2006; Vitulo et al., 2007). We have recently started the characterization of these genes investigating their role during gametophytic development (Galla et al., 2011). Here, we report a detailed expression pattern analysis of two arabidopsis Mob1-like genes (AtMob1A and AtMob1B) and demonstrate that AtMob1A function is required for proper plant development, the correct patterning of the root meristem and the control of root growth under stress conditions.
MATERIALS AND METHODS
Plasmid construction and plant transformation
The generation of the Mob1A (At5g45550) RNA interference (RNAi) construct has been described by Galla et al. (2011). In order to obtain the Mob1Apro::Mob1A-GUS construct, the genomic sequence including the Mob1A coding sequence with introns and a 1·9 kb region upstream of the start codon (forward primer 5′-CCTCCAAGGTGCAAGAGAAG-3′ and reverse primer 5′-ATAAGGTGAAATGATAGATT-3′) was cloned into the pENTR™/SD-TOPO® vector (Life Technologies, Carlsbad, CA, USA). Subsequently, it was transferred into the pMDC163 destination vector (Curtis and Grossniklaus, 2003) by Gateway recombination using the LR Clonase Enzyme Mix (Life Technologies). The resulting plasmid contained Mob1A in-frame with the β-glucuronidase (GUS) reporter gene driven by the Mob1A promoter and a kanamycin selection gene.
Similarly, a 1·8 kb promoter region of Mob1B (At4g19045), together with part of the first exon, was amplified from genomic DNA (forward primer 5′-ATCCGATGCAGAGAGCTTGT- 3′ and reverse primer 5′-TTCGCCTTCTTCAAACTCGT- 3′), cloned into the pDONR207 vector (Life Technologies) and transferred into the pMDC163 plasmid by recombination. The fidelity of all entry and destination clones was confirmed by both sequencing and restriction analyses.
Binary constructs were electroporated into Agrobacterium tumefaciens strain GV3101 pMP90. After clone verification, Agrobacterium-mediated transformation of arabidopsis plants was carried out using the floral dip method (Clough and Bent, 1998).
Plant material and growth conditions
Arabidopsis thaliana (L.) Heynh. (Col-0) was used as the wild type. The SALK_076775, SALK_062070 and GK719G04 lines were obtained from the Nottingham Arabidopsis Stock Centre (NASC) (Scholl et al., 2000). Homozygous plants for the T-DNA alleles were isolated by PCR using T-DNA left border primers and gene-specific primers listed in Supplementary Data Table S1. The Mob1A (At5g45550) RNAi lines used for root analyses were those described by Galla et al. (2011). The J2341 enhancer trap line belongs to the Hasselhoff collection and was provided by the NASC. The pWOX5::GFP (green fluorescent protein) line was described by Ditengou et al. (2008). Seeds were surface sterilized for 15 min with a solution of 5 % (w/v) calcium hypochlorite and 0·02 % Triton X-100. After three washes in sterile water, they were left to dry under sterile conditions. Seeds were sown on plates containing 1 % (w/v) sucrose, half-strength Murashige and Skoog (MS) salts (Duchefa, Biochemie BV, Haarlem, The Netherlands) and 12 g L−1 agar–agar (Carl Roth, Karlsruhe, Germany) (pH 5·8). After 2 d of stratification at 4 °C in darkness, plates were transferred to a growth chamber (16 h light/8 h darkness, 22 °C) for seed germination and were maintained in a vertical position.
RNA extraction, reverse transcription–PCR (RT–PCR) and quantitative PCR (q-PCR)
Total RNA was isolated from 1-week-old seedlings using the RNeasy plant kit (Qiagen, Venlo, The Netherlands) according to the manufacturer's protocol. Total RNA (1 µg) was first treated with DNase (Qiagen) and first-strand cDNA was subsequently synthesized using RevertAid™ M-MuLV Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) and oligo(dT) primer, according to the manufacturer's instructions. A 1·5 µL aliquot of first-strand cDNA was used as template for PCR amplification in a 25 µL reaction employing a home-made Taq DNA polymerase. Amplification reactions for semi-quantitative analysis were performed with an initial denaturation step at 95 °C for 30 s, followed by 72 °C annealing/extension for 1 min for 30 cycles, for Mob1A and Mob1B, and for 25 cycles, for ACTIN7. The ACTIN7 gene (At5g09810) was used as an internal control.
Quantitative PCR analysis was performed as described by Trevisan et al. (2011) on cDNAs synthesized as described by Manoli et al. (2012). Mob1A- and Mob1B-specific primers were chosen on the 3′-untranslated region (UTR) of the respective genes with the help of PRaTo (http://prato.daapv.unipd.it; Nonis et al., 2011). The primers for the reference gene ACTIN2 (At3g18780) have been previously described by Airoldi et al. (2010). All primer sequences used for q-PCR and RT–PCR analyses are reported in Supplementary Data Table S1.
Plant growth observations and root length measurements
After 1 week of growth in plates, seedlings were transferred to soil in pots. Images of rosettes, rosette leaves and siliques were taken with a digital camera. For the examination of seeds contained in siliques, the latter were dissected on a slide under a Zeiss Stemi SV11 Apo stereomicroscope (Carl Zeiss, Jena, Germany) and images were acquired with an AxioCam MRc camera (Carl Zeiss). Root observations and measurements were performed on seedlings 5 d after germination. To monitor the abscisic acid (ABA) effect on root growth, seedlings were transferred 3 d after germination from plates with control medium to new plates with control medium or medium supplemented with 250 nm ABA. Seedlings were then left to grow for an additional 4 d. For whole root length measurements, plates containing seedlings were scanned with a CanonScan 9950F scanner (Canon, Tokyo, Japan). For root tip observations and measurements, seedlings were incubated for 10 min in 10 µg mL−1 propidium iodide (PI) and mounted on slides in water. Images were subsequently acquired with a confocal scanning microscope, as described in the following paragraph.
Immunocytochemistry
For whole-mount immunolocalizations of PIN1, PIN2 and PIN4 in root cells, 4-day-old seedlings were fixed with 3 % (w/v) paraformaldehyde and 0·02 % Triton X-100 in MTSB (7·5 g L−1 PIPES, 0·95 g L−1 EGTA, 0·66 g L−1 MgSO4, 2·5 g L−1 KOH, pH 7·0) for 45 min and washed three times with dH2O. The subsequent steps were performed in an InsituPro VS robot (Intavis, Koeln, Germany). Briefly, tissue permeabilization was achieved by 30 min incubation in 0·15 % (w/v) driselase (Sigma-Aldrich, St Louis, MO, USA) and 0·15 % (w/v) macerozyme (Sigma-Aldrich) in 10 mm MES (pH 5·3) at 37 °C, followed by four washes in MTSB and two subsequent treatments of 20 min each with 10 % (v/v) dimethylsulfoxide (DMSO), 3 % (v/v) Nonidet-P40 (Sigma-Aldrich) in MTSB. After five washes in MTSB, blocking was performed with 3 % bovine serum albumin (BSA; Carl Roth) in MTSB for 1 h. Rabbit anti-PIN1 (Gälweiler et al., 1998) (1:400), guinea pig anti-PIN2 (Ditengou et al., 2008) (1:1000) and rabbit anti-PIN4 (Friml et al., 2002) (1:400) primary antibodies in 3 % BSA (in MTSB) were applied for 4 h at room temperature, followed by seven washes in MTSB. Goat anti-rabbit A555-conjugated and anti-guinea pig A488-conjugated secondary antibodies (1:600) (Life Techno-logies) were applied for 3 h at room temperature, followed by ten washes in MTSB. Samples were mounted in Prolong Gold antifade reagent containing 4′,6-diamidino-2-phenylindole (DAPI; Life Technologies).
Histochemical GUS staining
Histochemical localization of GUS activity was carried out on 3- to 4-day-old T1 and T2 generation seedlings. Samples from three and five independent lines of Mob1Apro::Mob1A-GUS and Mob1Bpro::GUS, respectively, were fixed in cold 90 % acetone for 30 min, rinsed in staining buffer (0·5 mm sodium phosphate buffer pH 7, 5 mm ferrocyanide, 5 mm ferricyanide) and then placed in a 0·5 mg mL−1 X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronide) staining solution. Tissues were vacuum-infiltrated for 10 min and incubated at 37 °C for 1·5 h. After incubation, plants were rinsed with water and cleared by an overnight incubation in chloral hydrate solution [chloral hydrate/glycerol/water 8:2:1 (w/v/v)]. Finally plants were mounted in 50 % glycerol on glass microscope slides and observed with an Olympus BX50 microscope (Olympus, Tokyo, Japan) equipped with differential interference contrast (DIC) optics. Imagines were captured with an Axiocam Zeiss MRc5 colour camera (Carl Zeiss), and processed with Adobe Photoshop CS4.
Plant fixation, embedding and in situ hybridization (ISH)
Tissue sections 7 µm thick were obtained from fixed and embedded 3- to 6-day-old arabidopsis seedlings as described by Begheldo et al. (2013). Antisense Mob1A riboprobes were labelled with digoxigenin-11-UTP using T3 polymerase following the protocol of the manufacturer (Roche, Basel, Switzerland). Probes were selected by PCR on leaf cDNA (forward 5′-CCCCAATAAATTAACGGTAAGAA-3′ and reverse 5′-TGCTTTTCACATGAACACACAT-3′ primers) and contained a 166 bp portion of the Mob1A 3′-UTR. De-waxed slides were obtained by immersion in Histo-Clear® (National Diagnostic, Atlanta, GA, USA) for 10 min. All ISH steps, with the exception of staining, were carried out using the Gene Paint suite accessories (Freedom EVO100, Tecan, Maennedorf, Switzerland) as described by Begheldo et al. (2013). The signal was developed with detection buffer containing NBT-BCiP (Roche) following the manufacturer's instructions.
Lugol staining
Five-day-old seedlings were dipped in Lugol's staining reagent (Carl Roth) for 10 min, rinsed twice in water and mounted on slides in chloral hydrate:glycerol:water (8:3:1, w/v/v). Images were acquired with an Axiovert 200M MAT Zeiss microscope equipped with DIC optics and an AxioCam ICc 1 camera (Carl Zeiss).
Simultaneous staining of cell borders and starch grains for confocal microscopy
A method allowing staining of cell borders and simultaneous sensitive detection of starch grains was employed by modifying the procedure described by Truernit et al. (2008) as follows. Seedling were fixed as described above for whole-mount immunolocalization, washed with dH2O, treated for 25 min in pure methanol and finally slowly rehydrated in dH2O. Subsequently, seedlings were incubated for 25 min in 1 % periodic acid, washed twice with dH2O and stained with 4 mg L−1 PI solution in 100 mm Na2SO3 (pH 1·4 adjusted with HCl, approx. 0·15 m final concentration). A chloral hydrate solution for microscopy analyses was used for mounting the samples on slides with a 100 µm spacer for preservation of the tissue structure.
Confocal microscopy
Images were acquired using a Zeiss LSM 510 NLO confocal scanning microscope. Excitation wavelengths were 488 nm (argon laser) for A488-conjugated antibodies and GFP and 543 nm (Helium-Neon-Laser) for A555-conjugated antibodies and PI staining. Emission was detected at 500–550 nm for A488-conjugated antibodies and GFP, and at >575 nm for A555-conjugated antibodies and PI staining. A two-photon module was used for imaging of DAPI with excitation at 730 nm and emission at 435–485 nm. All multilabelling signals were detected in multitracking mode to avoid fluorescence cross-talk. Images were analysed with the LSM image browser (Carl Zeiss) and Adobe Photoshop CS2.
Accession numbers
Sequence data of genes used in this article can be found in the EMBL/GenBank data libraries. The arabidopsis Mob1A and Mob1B genes refer to the At5g45550 and At4g19045 loci, respectively.
RESULTS
Bioinformatic analysis of Mob1-like gene expression in arabidopsis
Blastp analysis revealed that, among the four Mob1-like genes present in the arabidopsis genome (Vitulo et al., 2007), At5g45550 and At4g19045 encode predicted proteins with the highest similarity to S. cerevisiae Mob1 (E-values: 9e-51 and 1e-49, respectively). The two protein sequences both contain 215 amino acids and share 93 % identity (Supplementary Data Fig. S1). The two genes have been renamed Mob1A (At5g45550) and Mob1B (At4g19045), correcting the previous annotation of the Mob1B gene, which was formerly attributed to the At4g19050 locus and wrongly predicted to encode a protein of 1416 amino acids (Vitulo et al., 2007). Indeed, the At4g19045 locus has only recently been annotated as the predicted gene by The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/).
In a previous work, the expression of the Mob1A gene was documented by real-time PCR analysis in roots, leaves, flowers and siliques (Galla et al., 2011). Consistently, a search in Genevestigator V3 (Hruz et al., 2008) showed that Mob1A transcripts are detectable ubiquitously in arabidopsis tissues, with a maximum in seeds, especially in testa and suspensor (Supplementary Data Fig. S2]. In addition, Mob1A expression appeared to be upregulated by several abiotic (e.g. nitrogen starvation and hypoxia) and biotic stresses (e.g. inoculation with Pseudomonas syringae) as well as by two plant stress hormones, abscisic acid (ABA) and salicylic acid (Supplementary Data Fig. S3). Gene ontology (GO) analysis of the 54 genes showing the highest co-expression with Mob1A in response to 1662 perturbations (Genevestigator) revealed an over-representation of genes involved in protein localization, cellular lipid metabolic processes and oxidoreductase activity (Supplementary Data Fig. S4 and Table S3). Notably, GO analysis of the 55 genes with the highest co-expression with Mob1A in 74 anatomical parts supported the fact that the set of genes co-expressed with Mob1A was enriched with genes involved in protein localization (Supplementary Data Fig. S5 and Table S4). In addition, GO terms (biological processes) such as organelle localization, actin filament-based movement, vesicle docking, phosphorus metabolic processes, macromolecule modification and developmental cell growth were enriched in the set of genes co-regulated with Mob1A in different tissues (Supplementary Data Fig. S5).
Microarray data are not yet available for the newly annotated At4g19045 locus (Mob1B).
Tissue-specific expression analysis of Mob1A and Mob1B
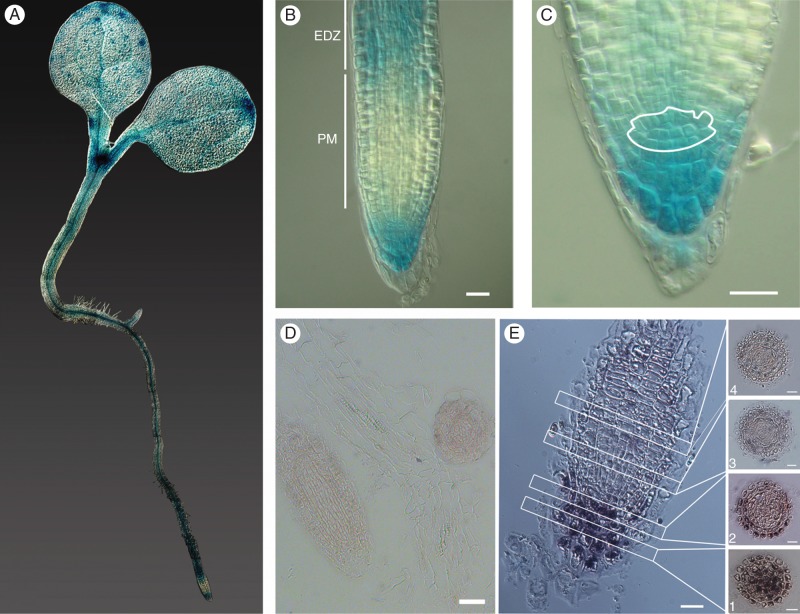
In order to obtain a more detailed spatial analysis of the Mob1A and Mob1B expression patterns in different tissues, we generated arabidopsis transgenic lines expressing Mob1A::GUS translational and Mob1B:GUS transcriptional fusions under the control of the Mob1A and Mob1B native promoters, respectively. The presence of the Mob1A–GUS fusion protein was confirmed in all organs, in agreement with a previous real-time PCR analysis (Galla et al., 2011). In 3- to 4-day-old seedlings, Mob1A expression was high in the shoot apical meristem and along the vasculature in cotyledons, hypocotyls and roots (Fig. 1A). At the root tip, Mob1A–GUS signal was detected in columella and lateral root cap cells as well as in the stem cell niche around the quiescent centre (QC) (Fig. 1B, C). The levels of Mob1A expression decreased progressively in the meristematic zone from the root tip towards the base of the root, becoming stronger again in the elongation zone. The specific gradient of Mob1A expression from the columella to the meristematic zone was further confirmed by ISH with digoxigenin-labelled probes, designed to target specifically a 166 bp non-conserved portion of the Mob1A 3′-UTR (Fig. 1D, E). In flowers, Mob1A transcription appeared localized in ovules and pollen, based on both GUS staining (Supplementary DataFig. S6A, D) and ISH data [Supplementary Data Fig. S6B, C, E, F]. This observation is consistent with the role of Mob1A in mega- and microgametogenesis evidenced by our previously reported results (Galla et al., 2011).
Fig. 1.
Spatial expression of Mob1A in arabidopsis seedlings. (A–C) GUS staining of 3- to 4-day-old pMob1A::GUS-Mob1A seedlings. In (B), the root proximal meristem (PM) and the beginning of the elongation–differentiation zone (EDZ) are labelled and indicated by white bars. In (C), the root stem cell niche is outlined in white. (D and E) In situ hybridization of specific sense (D) and antisense (E) Mob1A probes in longitudinal root sections. For (E) every second or every third 7 µm cross-section from the columella tissue up to the meristem is shown: (1) columella cells, (2) stem cell niche and (3 and 4) distal meristem cells. Scale bars are 10 µm in B and C, and 50 µm in D and E.
GUS expression driven by the promoter of the Mob1B gene was detected along the vasculature in cotyledons, hypocotyls and roots of 3- to 4-day-old seedlings (Supplementary Data Fig. S7). Along the root, the GUS signal of Mob1B was restricted to a more limited number of cell files when compared with that of Mob1A, being absent from columella cells and the root tip up to the transition zone (Supplementary Data Fig. S7G, H).
Isolation of T-DNA and RNAi lines for Mob1A and Mob1B
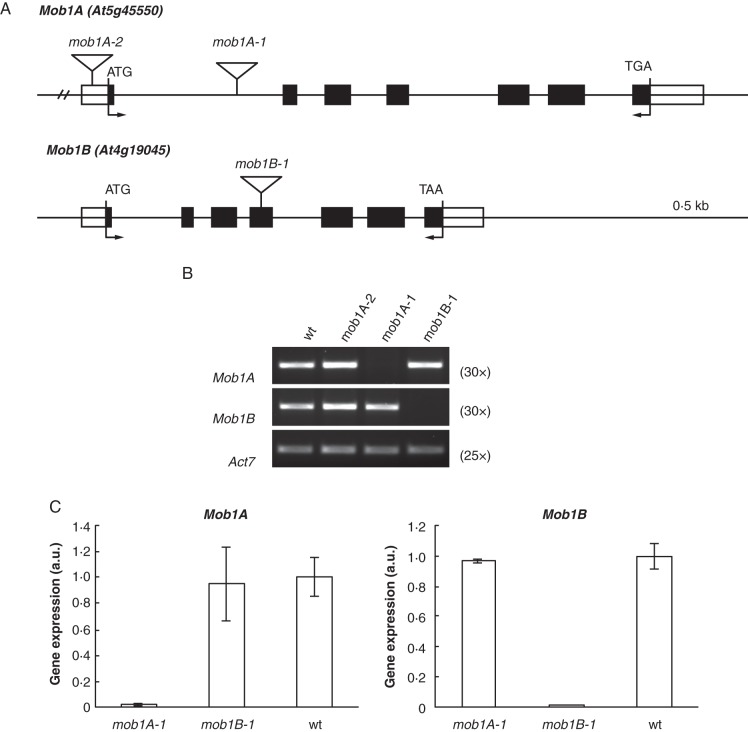
In order to assess the effects of a reduced or abolished expression of Mob1 genes in plants, we used both knock-down (Mob1A gene) and knock-out (Mob1A and Mob1B) approaches. To this end, putative T-DNA disruption mutants of the two genes were isolated. Concerning Mob1A, homozygous plants were selected from GABI-Kat (GK719G04; Rosso et al., 2003) and SALK (SALK_076775; Alonso et al., 2003) lines obtained from TAIR (Fig. 2A). The GK719G04 line carried a T-DNA insertion in the first intron of the gene and was renamed mob1A-1, while SALK_076775 had an insertion in the promoter region of the gene and was renamed mob1A-2. RT–PCR analysis performed on homozygous mob1A-2 plants with primers flanking the Mob1A coding sequence revealed a level of transcript similar to that of the wild type (Fig. 2B). In contrast, we could not detect Mob1A mRNA in homozygous mob1A-1 plants, suggesting that mob1A-1 represented a null mutant (Fig. 2B). For Mob1B, only one line (SALK_062070) was available and could be confirmed to contain a T-DNA insertion in the fourth exon (Fig. 2A). This T-DNA disruption allele was named mob1B-1. Analysis by RT–PCR on mob1B-1 homozygous plants with primers flanking the gene's open reading frame indicated the absence of the Mob1B transcript, suggesting mob1B-1 as a null allele (Fig. 2B). We analysed the expression of the two Mob1 genes in the reciprocal knock-out mutants by means of q-PCR. The abolished expression of Mob1A did not affect the transcript levels of Mob1B, and vice versa (Fig. 2C). As an additional approach, we studied Mob1A knock-down effects in three independent RNAi lines, in which the transcript level of the gene ranged between 50 and 70 % in comparison with that of the wild type, and whose generation has been recently described (Galla et al., 2011).
Fig. 2.
Expression analysis of Mob1A and Mob1B knock-out mutants. (A) Schematic representation of Mob1A and Mob1B intron–exon structure and T-DNA insertion sites of SALK and GABI Kat lines. Black and whitey boxes indicate exons and untranslated regions, respectively. Arrows indicate primers used for RT–PCR analysis of T-DNA mutants. (B) Semi-quantitative and (C) quantitative RT–PCR analysis of Mob1A and Mob1B expression in T-DNA mutants. In (B) the number of RT–PCR cycles is indicated in parentheses. Error bars in C represent the s.d.
Reduced Mob1A expression affects plant development and reproduction
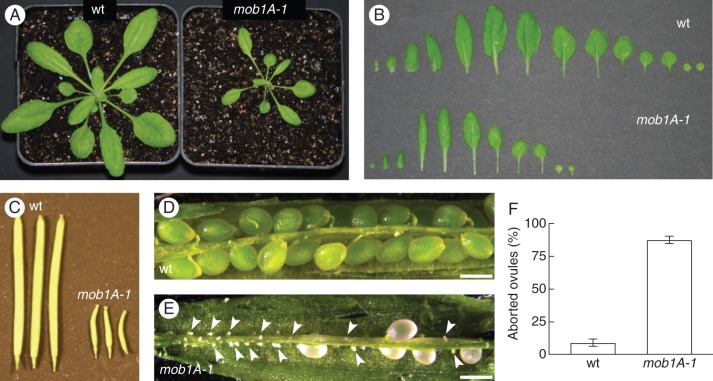
As an initial step to analyse the effects caused by the reduction of Mob1 gene expression, the phenotypes of mob1A-1 and mob1B-1 mutations were examined in plants grown in soil. mob1B-1 plants did not display any major defect in plant growth and development, and reproduced normally to the next generation (Supplementary Data Fig. S8A). As far as the mob1A-1 mutant is concerned, we first sought to confirm the developmental defects observed in Mob1A RNAi lines and described by Galla et al. (2011). Indeed, mob1A-1 exhibited severe defects in the growth of vegetative organs and in seed setting capacity, consistent with the reduced growth and defects in ovule development conferred by RNAi-mediated downregulation of Mob1A reported by Galla et al. (2011). Rosettes of mob1A-1 showed a reduction in size and number of leaves compared with wild-type plants of the same age (Fig. 3A, B). Although mob1A-1 carpels and stamens did not display any major morphological alteration (Supplementary Data Fig. S9), siliques collected 8–10 d after flowering presented a strongly reduced size (Fig. 3C) and a dramatically high proportion of aborted ovules (Fig. 3D–F).
Fig. 3.
mob1A-1 displays defects in plant growth and reproduction. (A) Rosette phenotype of 3-week-old wild-type Col-0 (wt) and mob1A-1 plants. Note the reduced size of the mob1A-1 mutant. (B) Arrangement of all leaves from 3-week-old wild-type and mob1A-1 plants reveals a reduced number of leaves for mob1A-1. (C) Siliques of wild-type and mob1A-1 plants 10 d after flower opening. mob1A-1 siliques display a clear reduction in size. (D and E) Morphology of seeds from wild-type and mob1A-1 plants. Shown are siliques at 8–10 d after flowering. mob1A-1 siliques contain several aborted (arrowheads) and pale seeds. (F) Proportion of aborted ovules from siliques of wild-type and mob1A-1 plants. Means ± s.d. are reported; n = 3 (ten siliques per plant; >370 total seeds were examined). Scale bars are 500 µm (D, E).
mob1A-1 roots display reduced size, shorter meristem and higher sensitivity to ABA
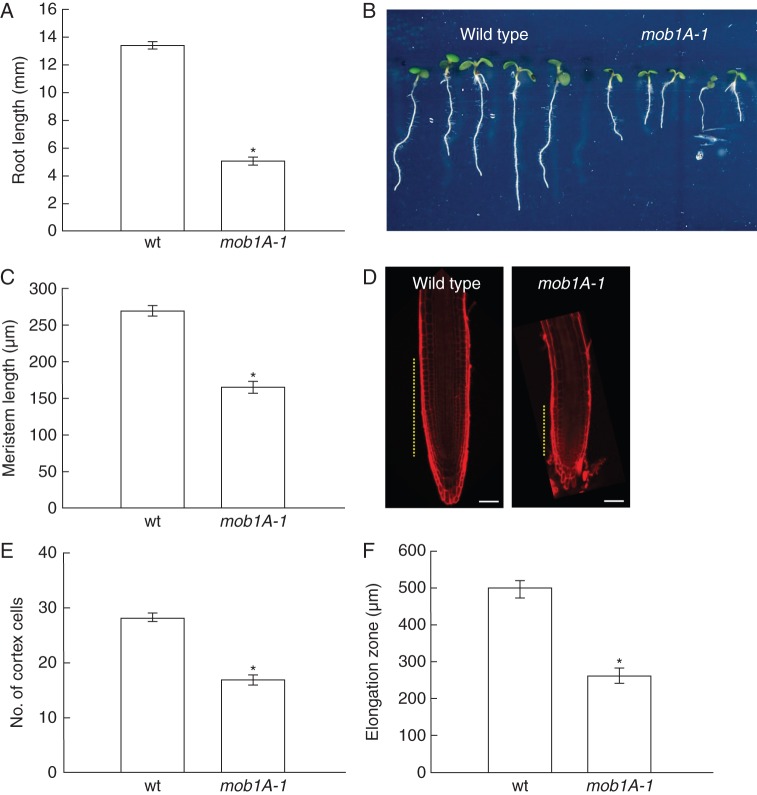
The expression pattern of Mob1A in the root tip is peculiar for the presence of high transcript levels in columella cells and the stem cell niche and for the low levels, nearly below detection, in the proliferating meristem. Therefore, we set out to investigate the specific effects of Mob1A knock-out and knock-down on root development. Five days after germination, the root meristem reaches its maximal size (Dello Ioio et al., 2007). At this age, mob1A-1 plants exhibited a shorter root length in comparison with wild-type seedlings (Fig. 4A, B). Microscopic measurements of mob1A-1 roots revealed a significant reduction in meristem size, number of cortical cells and size of the elongation zone (Fig. 4C–F). In contrast, mob1B-1 plants did not show any change in root length (Supplementary Data Fig. S8B). To test the growth response of seedlings to ABA, which up-regulates Mob1A expression, 3-day-old seedlings were transferred to a growth medium supplemented with 250 nm ABA. Interestingly, mob1A-1 roots displayed a reduced growth on ABA-containing medium, indicating a higher sensitivity to ABA in comparison with the wild type (Fig. 5).
Fig. 4.
Mob1A function is required for proper root development. (A and B) Whole root length analysis of wild-type (wt) and mob1A-1 plants (n = 42). (C and D) Length measurement (C) (n = 16) and size comparison (D) of wild-type and mob1A-1 root meristem. In D, meristem size is indicated by dashed yellow lines (scale bars are 50 µm). (E) Counting of cortex cell number (n = 16). (F) Length measurement of the root elongation zone (n = 16). All measurements and images were performed on seedlings 5 d after germination. In the graphs, the mean ± s.e. is reported (*P < 0·001, t-test).
Fig. 5.
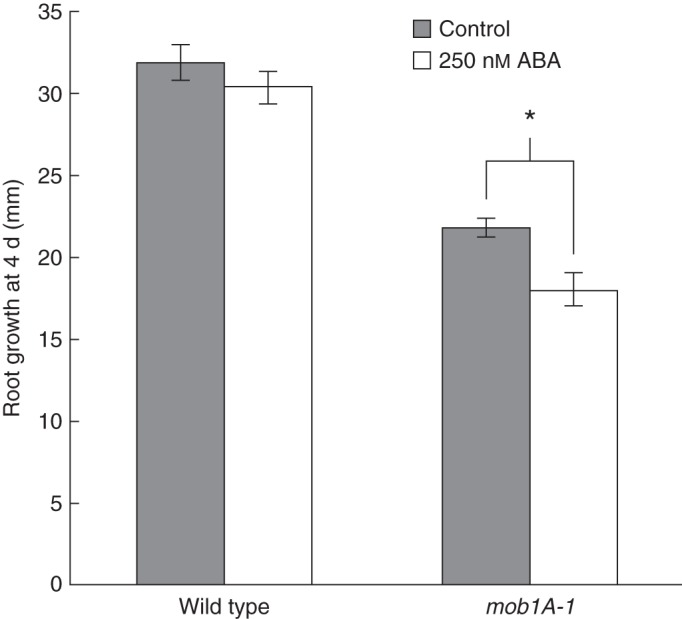
mob1A-1 roots are more sensitive to abscisic acid (AB). Four days root growth (n >10) of wild-type and mob1A-1 seedlings on control medium and medium containing 250 nM ABA. Seedlings were transfered in parallel to new plates 3 d after germination. The mean ± s.e. is reported (*P < 0·001, t-test).
Mob1A function is involved in tissue patterning of the root tip
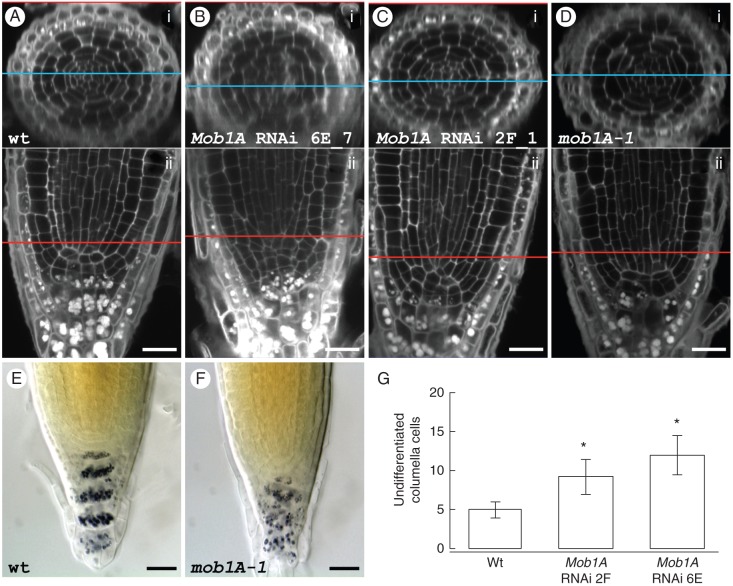
Root tip microscopic inspections of mob1A-1 seedlings and T2 plants from three independent Mob1A RNAi lines revealed severe defects in tissue patterning around the QC and the stem cell niche, consistent with Mob1A expression in this region. The root tip phenotype of the different lines was characterized by employing a simultaneous staining of cell borders and starch grains and through recording of z-stacks with a confocal microscope. The great majority (90 %) of wild-type root tips exhibited the typical ordered cellular organization, with the QC flanked by cortex and endodermis initials and, apically, by columella initials [columella stem cells (CSCs)] and columella cell layers (Fig. 6A). In contrast, the root tips of mob1A-1 and Mob1A RNAi lines showed a disordered cellular patterning with different degrees of misalignment of cell files and irregular cell shapes and division planes (Fig. 6B–D). The degree of penetrance of this phenotype ranged between 20 and 33·3 % in Mob1A RNAi lines, and was maximal (43·3 %) in the mob1A-1 null mutant (Table 1), consistent with the downregulation or knock-out of the gene, respectively. The starch granules in the columella region of these lines appeared completely misaligned, substantiating a lack of symmetry also in the cellular pattern of this tissue (Fig. 6A–F). The simultaneous staining of cell borders and starch grains enabled us to count the undifferentiated cells between the putative QC and the first columella cells containing starch granules throughout the whole z-stack of a root tip (Supplementary Data Video). In the roots of the RNAi lines that showed a distorted cellular pattern, the number of undifferentiated columella cells was significantly higher when compared with that in wild-type roots (Fig. 6G).
Fig. 6.
Reduced levels of Mob1A expression affect root meristem cellular pattern. (A–D) Staining of cell borders and starch grains in wild-type (A), Mob1A RNAi lines 6E_4 (B) and 2F_1 (C) and mob1A-1 (D) roots. The upper panel (i) of each figure shows a transversal section of the root, which was electronically reconstructed from a z-stack of longitudinal sections (ii). The blue and the red lines indicate the level in the root tip, to which the two sections correspond. (E and F) Lugol staining of wild-type (E) and mob1A-1 (F) root tips. Scale bars are 20 µm (A–F). (G) Counting of undifferentiated columella cells in wild-type (wt) roots and roots of Mob1A RNAi lines with a distorted cellular pattern (n = 4). Cells were counted throughout the planes of a confocal z-stack. The mean ± s.d. is reported (*P < 0·001, t-test). Scale bars are 20 µm (A–F).
Table 1.
Frequency of root tips with a distorted cellular pattern in the wild type, Mob1A RNAi lines and the mob1A-1 mutant line
| Wild type |
Mob1A RNAi lines |
mob1A-1 | |||
|---|---|---|---|---|---|
| 4G | 2F | 6E | |||
| Root tips with disordered cellular pattern | 3 | 6 | 8 | 10 | 13 |
| Observed root tips | 30 | 30 | 30 | 30 | 30 |
| Phenotype penetrance (%) | 10 | 20 | 26·7 | 33·3 | 43·3 |
To characterize further the cellular organization of the root tips, we employed markers specifically labelling the QC, the CSCs and their surrounding cells. A suitable marker for the plasma membrane of the cells around the QC is the auxin efflux carrier PIN4 (Friml et al., 2002). Indeed, immunolocalization analysis on Mob1A RNAi plants showed a misalignment of PIN4-labelled cell files around the QC and the stem cell niche (Fig. 7A, B). In addition, the typical PIN4 expression in wild-type columella initials was absent in Mob1A knocked-down lines. The localization pattern of PIN4 and of PIN1 and PIN2, two additional auxin efflux carriers (Gälweiler et al., 1998; Müller et al., 1998), in stele, endodermis, cortex and epidermis cells appeared normal (Fig. 7A, B) (Supplementary Data Fig. S10]. Mob1A RNAi lines were crossed with pWOX5::GFP (Ditengou et al., 2008) and the enhancer trap line J2341 (C24 background) (Sabatini et al., 2003), which show GFP expression in QC cells and columella initials, respectively. Analysis of an F2 segregating population revealed that the GFP expression domains were expanded or reflected the misalignment of cell files in root tips displaying a disordered cellular pattern (Fig. 7C–F).
Fig. 7.
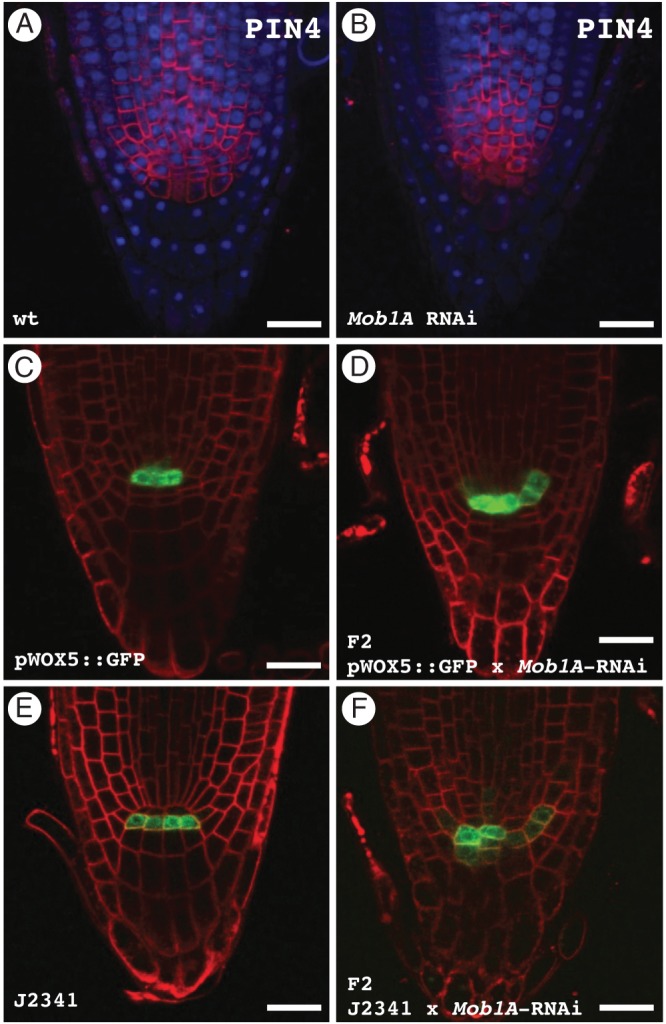
The expression pattern of different cellular markers is altered in Mob1A RNAi roots. (A and B) Immunolocalized PIN4 (red) and DAPI staining (blue) in wild-type (A) and Mob1A RNAi (B) root tips. (C and D) Root tip GFP distribution in the pWOX5::GFP reporter line (C) and in an F2 population from a pWOX5::GFP × Mob1A RNAi cross (D). Seedlings were incubated for 10 min in 4 µm FM4-64 prior to microscopy observation to visualize the cell plasma membrane. Note the expanded GFP expression in the sample from the F2 crossing population. (E and F) Root tip GFP distribution in the J2341 columella initials marker line (E) and in an F2 population from a J2341 × Mob1A RNAi cross (F). Plasma membranes were stained with FM4-64 as described above. Cells expressing GFP in the F2 crossing population failed to display the same alignment as in J2341 roots. Scale bars are 20 µm (A–F).
DISCUSSION
Developmental patterning and morphogenesis of multicellular organisms are determined by co-ordinated cell proliferation, cell differentiation and programmed cell death. Mob1 proteins are conserved among eukaryotes and are essential components of pathways that control fundamental cellular processes such as mitotic exit, cytokinesis and apoptosis (reviewed by Vitulo et al., 2007). Mob1A (At5g45550) and Mob1B (At4g19045) are the closest arabidopsis orthologues to S. cerevisiae Mob1, and their predicted protein sequences share a very high identity level (93 %). Yeast Mob1 is a key player in the MEN and SIN, two signalling pathways that co-ordinate exit from mitosis with cytokinesis in S. cerevisiae and S. pombe, respectively. Bedhomme et al. (2008) have shown that, for several MEN/SIN components, the arabidopsis genome contains two paralogues for each of the yeast orthologous genes and have suggested a certain level of functional redundancy. However, our q-PCR analysis of Mob1A and Mob1B knock-out mutants excluded a reciprocal regulation of the two Mob1 genes in arabidopsis seedlings. In addition, mob1A-1 plants exhibit a severe phenotype in contrast to the absence of visible growth and reproduction defects in mob1B-1 mutants. Thus, Mob1B cannot compensate for the loss of Mob1A function. This might be explained by the more restricted expression pattern of Mob1B in comparison with Mob1A or by the loss of Mob1B protein function.
The phenotype of mob1A-1 plants included defects in the number of rosette leaves, ovule development, root growth and root tip cellular organization. All these traits correlate with the documented expression of Mob1A in the respective tissues and can be potentially linked to the hypothesized function of arabidopsis Mob1A in the regulation of cell division and cell proliferation. The expression of Mob1A at the shoot apical meristem, where leaf primordia are formed (Byrne, 2012), supports a possible role for the gene in this process, as inferred by the reduction in leaf number upon Mob1A loss of function. The high proportion of aborted ovules observed for mob1A-1 has also been reported for Mob1A RNAi lines (Galla et al., 2011). In particular, post-transcriptional silencing of Mob1A affected the normal progression of both female meiosis and megagametogenesis, as well as pollen maturation. Indeed, we verified the expression of Mob1A in both ovules and pollen.
In this study, we focused particularly on the role of Mob1A during root development. The expression of the gene was documented along the root vasculature and displayed a specific pattern at the root tip. Mob1A transcript levels were high in columella and lateral root cap cells as well as in the stem cell niche, but appeared very low in the proximal meristem. The knock-out of Mob1A caused a significant reduction in the size of the root meristem. A decrease in meristem size can result from reduced stem cell activity, from loss of division potential of meristematic cells in the proximal meristem, or from a more rapid entry of meristematic cells into the elongation zone (Dello Ioio et al., 2007). Our data indicate that a reduction in stem cell activity affecting the replenishment of the meristem is likely to occur in mob1A-1 mutant seedlings. The distorted cellular pattern of the stem cell niche and the columella is a clear sign that the activity of the stem cells is altered. The disorganization in the cellular architecture of the root tip was common to both mob1A-1 mutant and Mob1A RNAi lines, although with a variable degree of phenotypic penetrance that was related to the level of downregulation of Mob1A expression. A probable alteration of stem cell activity in Mob1A knock-down roots is also suggested by the expansion and mispositioning of the expression patterns of proWOX5 and the promoter trap J2341, which identify the QC and CSCs, respectively.
The expression of Mob1A in the stem cell niche and columella cells together with the incorrect patterning of these tissues in Mob1A knock-out and knock-down lines indicate that the cellular function of Mob1A is critical for pattern formation in this region. The documented role of Mob1 orthologues in the control of cell division and cell proliferation in other eukaryotes can potentially also explain the root tip phenotype observed in arabidopsis mob1A-1 and Mob1A RNAi lines. Anomalous cell divisions in the stem cell niche might lead to the observed aberrations in the positioning of the QC and stem cells and, as a consequence, to the documented defects in root morphology. More detailed studies are required to verify which aspects of cell division are regulated by Mob1A in arabidopsis roots. The rate and the orientation of cell division as well as the completion of cytokinesis need to be taken into consideration.
The unco-ordinated growth of the columella tissue might also contribute to the phenotype of mob1A-1 and Mob1A RNAi root apexes that in some cases resembled ‘tumour-like’ structures. This phenotype is reminiscent of the effects caused by loss of Mats function in Drosophila, consisting of increased cell proliferation, defective apoptosis and induction of tissue overgrowth (Lai et al., 2005; Shimizu et al., 2008). Mats is an orthologue of yeast Mob1 and has been involved in the Hpo signalling pathway, which participates in the control of tissue growth (reviewed by Hariharan and Bilder, 2006; Harvey and Tapon, 2007). The morphology of the root tip is ensured by the programmed cell death of distal columella cell layers, which are progressively shed from the root cap. Similarly to the role of Mats in Drosophila, Mob1A might perform a critical function in the co-ordinated growth of the columella tissue. Support for this hypothesis derives from the observed increase in the number of undifferentiated columella cells in distorted Mob1A RNAi root tips. Hence, reduced protein levels could affect the correct balance between cell proliferation and programmed cell death.
Mob1A expression is upregulated by several biotic and abiotic stress conditions. This transcriptional response could be modulated by a mechanism involving plant stress hormones, as suggested by the induction of Mob1A expression also brought about by ABA and salicylic acid. Notably, mob1A-1 roots were more sensitive to ABA. These observations reveal a role for Mob1A in the response to ABA and suggest a specific function for a Mob1 gene in plants, namely the adjustment of plant growth in response to stress conditions. Such a function is fundamental to plants, which, in contrast to animals, cannot escape unfavourable life conditions.
In summary, our results provide new insights into the role of Mob1 proteins during plant development. It emerges that Mob1A function is required for proper organ development and is likely to be involved in the control of cell division and cell proliferation, similarly to other eukaryotes. In addition, Mob1A might potentially play a role in the adjustment of plant growth in response to stress conditions.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Dr A. Nonis (DAFNAE, University of Padova, Italy) for q-PCR analysis, Dr X. Li (Institute of Biology II, Faculty of Biology, Albert-Ludwigs-University of Freiburg, Germany) for the production of PIN1, PIN2 and PIN4 antibodies, Dr R. Nitschke and the Life Imaging Center (Center for Biological Systems Analysis, Albert-Ludwigs-University of Freiburg, Germany) for the use of confocal microscopes, and the Nottingham Arabidopsis Stock Centre for providing seed stocks. The authors are grateful to Katja Rapp and Bernd Gross (Institute of Biology II, Faculty of Biology, Albert-Ludwigs-University of Freiburg, Germany) for excellent technical assistance. This work was supported by SFB 746, the Excellence Initiative of the German Federal and State Governments (EXC 294), EU FP6 (‘AUTOSCREEN’, LSHG-CT-2007 – 037897), DLR and Bundesministerium für Bildung und Forschung (BMBF).
LITERATURE CITED
- Airoldi CA, Rovere FD, Falasca G, et al. The Arabidopsis BET bromodomain factor GTE4 is involved in maintenance of the mitotic cell cycle during plant development. Plant Physiology. 2010;152:1320–1334. doi: 10.1104/pp.109.150631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Amon A. Men and sin: what's the difference? Nature Reviews Molecular and Cell Biology. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Bäurle I, Laux T. Apical meristems: the plant's fountain of youth. Bioessays. 2003;25:961–970. doi: 10.1002/bies.10341. [DOI] [PubMed] [Google Scholar]
- Bedhomme M, Jouannic S, Champion A, Simanis V, Henry Y. Plants, MEN and SIN. Plant Physiology and Biochemistry. 2008;46:1–10. doi: 10.1016/j.plaphy.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Begheldo M, Ditengou FA, Cimoli G, et al. Whole-mount in situ detection of microRNAs on Arabidopsis tissues using Zip Nucleic Acid probes. Analytical Biochemistry. 2013;434:60–66. doi: 10.1016/j.ab.2012.10.039. [DOI] [PubMed] [Google Scholar]
- Bothos J, Tuttle RL, Ottey M, Luca FC, Halazonetis TD. Human LATS1 is a mitotic exit network kinase. Cancer Research. 2005;65:6568–6575. doi: 10.1158/0008-5472.CAN-05-0862. [DOI] [PubMed] [Google Scholar]
- Byrne ME. Making leaves. Current Opinion in Plant Biology. 2012;15:24–30. doi: 10.1016/j.pbi.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Chant J, Pringle JR. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. Journal of Cell Biology. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio S, Albertini E, Varotto S, et al. Alfalfa Mob 1-like genes are expressed in reproductive organs during meiosis and gametogenesis. Plant Molecular Biology. 2005;58:789–807. doi: 10.1007/s11103-005-8104-9. [DOI] [PubMed] [Google Scholar]
- Citterio S, Piatti S, Albertini E, Aina R, Varotto S, Barcaccia G. Alfalfa Mob1-like proteins are involved in cell proliferation and are localized in the cell division plane during cytokinesis. Experimental Cell Research. 2006;312:1050–1064. doi: 10.1016/j.yexcr.2005.12.032. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, et al. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Current Biology. 2007;17:678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, et al. Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2008;105:18818–18823. doi: 10.1073/pnas.0807814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenz LM, Lee SE, Fesquet D, Johnston LH. The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. Journal of Cell Science. 2000;113:3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell. 2002;108:661–673. doi: 10.1016/s0092-8674(02)00656-6. [DOI] [PubMed] [Google Scholar]
- Galla G, Zenoni S, Marconi G, et al. Sporophytic and gametophytic functions of the cell cycle-associated Mob1 gene in Arabidopsis thaliana L. Gene. 2011;484:1–12. doi: 10.1016/j.gene.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Gould KL, Simanis V. The control of septum formation in fission yeast. Genes and Development. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- Hariharan IK, Bilder D. Regulation of imaginal disc growth by tumor-suppressor genes in Drosophila. Annual Review of Genetics. 2006;40:335–361. doi: 10.1146/annurev.genet.39.073003.100738. [DOI] [PubMed] [Google Scholar]
- Harvey K, Tapon N. The Salvador–Warts–Hippo pathway – an emerging tumour-suppressor network. Nature Reviews Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nature Reviews Molecular and Cell Biology. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- Hou MC, Salek J, McCollum D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Current Biology. 2000;10:619–622. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- Hou M-C, Guertin DA, McCollum D. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p–Mob1p kinase complex. Molecular and Cellular Biology. 2004;24:3262–3276. doi: 10.1128/MCB.24.8.3262-3276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W. Embryonic patterning in Arabidopsis thaliana. Annual Review of Cell and Developmental Biology. 2007;23:207–236. doi: 10.1146/annurev.cellbio.22.011105.102609. [DOI] [PubMed] [Google Scholar]
- Jürgens G. Apical–basal pattern formation in Arabidopsis embryogenesis. EMBO Journal. 2001;20:3609–3616. doi: 10.1093/emboj/20.14.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. Growing up green: cellular basis of plant development. Mechanisms of Development. 2003;120:1395–1406. doi: 10.1016/j.mod.2003.03.001. [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes and Development. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Krapp A, Simanis V. An overview of the fission yeast septation initiation network (SIN) Biochemical Society Transactions. 2008;36:411–415. doi: 10.1042/BST0360411. [DOI] [PubMed] [Google Scholar]
- Lai Z-C, Wei X, Shimizu T, et al. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Lee SE, Frenz LM, Wells NJ, Johnson AL, Johnston LH. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Current Biology. 2001;11:784–788. doi: 10.1016/s0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- Lippincott J, Shannon KB, Shou W, Deshaies RJ, Li R. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. Jouyrnal of Cell Science. 2001;114:1379–1386. doi: 10.1242/jcs.114.7.1379. [DOI] [PubMed] [Google Scholar]
- Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Molecular and Cellular Biology. 2001;21:6972–6983. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca FC, Winey M. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Molecular Biology of the Cell. 1998;9:29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah AS, Jang J, Deshaies RJ. Protein kinase Cdc15 activates the Dbf2–Mob1 kinase complex. Proceedings of the National Academy of Sciences, USA. 2001;98:7325–7330. doi: 10.1073/pnas.141098998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Lukas C, Kaiser BK, Jackson PK, Bartek J, Lukas J. Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nature Cell Biology. 2002;4:317–322. doi: 10.1038/ncb777. [DOI] [PubMed] [Google Scholar]
- Manoli A, Sturaro A, Trevisan S, Quaggiotti S, Nonis A. Evaluation of candidate reference genes for qPCR in maize. Journal of Plant Physiology. 2012;169:807–815. doi: 10.1016/j.jplph.2012.01.019. [DOI] [PubMed] [Google Scholar]
- Mohl DA, Huddleston MJ, Collingwood TS, Annan RS, Deshaies RJ. Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. Journal of Cell Biology. 2009;184:527–539. doi: 10.1083/jcb.200812022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, et al. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO Journal. 1998;17:6903–6911. doi: 10.1093/emboj/17.23.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonis A, Scortegagna M, Nonis A, Ruperti B. PRaTo: a web-tool to select optimal primer pairs for qPCR. Biochemical and Biophysical Research Communications. 2011;415:707–708. doi: 10.1016/j.bbrc.2011.10.148. [DOI] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes and Development. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Molecular Biology. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes and Development. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH. Seed and molecular resources for Arabidopsis. Plant Physiology. 2000;124:1477–1480. doi: 10.1104/pp.124.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Ho L-L, Lai Z-C. The mob as tumor suppressor gene is essential for early development and regulates tissue growth in Drosophila. Genetics. 2008;178:957–965. doi: 10.1534/genetics.107.081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, et al. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annual Review of Genetics. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Trevisan S, Manoli A, Begheldo M, et al. Transcriptome analysis reveals coordinated spatiotemporal regulation of hemoglobin and nitrate reductase in response to nitrate in maize roots. New Phytologist. 2011;192:338–352. doi: 10.1111/j.1469-8137.2011.03822.x. [DOI] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, et al. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. The Plant Cell. 2008;20:1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma DPS. Cytokinesis and building of the cell plate in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:751–784. doi: 10.1146/annurev.arplant.52.1.751. [DOI] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Molecular Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Vitulo N, Vezzi A, Galla G, et al. Characterization and evolution of the cell cycle-associated mob domain-containing proteins in eukaryotes. Evolutionary Bioinformatics Online. 2007;3:121–158. [PMC free article] [PubMed] [Google Scholar]
- Willemsen V, Scheres B. Mechanisms of pattern formation in plant embryogenesis. Annual Review of Genetics. 2004;38:587–614. doi: 10.1146/annurev.genet.38.072902.092231. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Toh-e A. Regulation of the localization of Dbf2 and mob1 during cell division of Saccharomyces cerevisiae. Genes and Genetics Systems. 2001;76:141–147. doi: 10.1266/ggs.76.141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.