Abstract
Our aim was to evaluate the delivery of transposase-based vectors by Ultrasound Targeted Microbubble Destruction (UTMD) in mice. DNA vectors were attached to cationic lipid microbubbles (1-3 μm in diameter), injected intravenously, and delivered to the liver by destruction of the carrier bubbles with ultrasound in burst mode at a 1.0 MHz, 20 μs pulse duration, 10 Hz pulse repetition frequency, and acoustic peak negative pressure of ~1.3 MPa. We evaluated the expression and genomic integration of conventional (pcDNA3) and piggyBac transposase-based (pmGENIE) reporter vectors. In vivo, we observed UTMD-mediated liver-specific expression of pmGENIE for an average of 24 days, compared to 4 days with pcDNA3. Reporter expression was predominately located in proximity to blood vessels initially, while expression after three days was more evenly distributed through the parenchyma of the liver. We confirmed random genomic integration for pmGENIE in vitro, however, integration events for pmGENIE in vivo were targeted to specific areas of chromosome 14. Our results suggest that a combination of UTMD with nonviral DNA transposase vectors can mediate weeks of hepatic-specific gene transfer in vivo, and analyses performed by non-restrictive linear amplification-mediated (nrLAM) PCR, cloning, and sequencing identify an unexpected tropism for integration within a specific sequence on chromosome 14 in mice. UTMD delivery of transgenes may be useful for the treatment of hepatic gene deficiency disorders.
Keywords: Ultrasound, microbubbles, hepatic gene therapy, transposase, site-specific chromosomal integration
Introduction
The development of safe, minimally invasive methods for delivering therapeutic expression constructs to specific anatomical regions of interest would be valuable for improving gene therapy. Currently, many gene therapy trials rely on viral-based strategies using retroviruses, adenoviruses or adeno-associated viruses to mediate gene transfer (Nathwani et al. 2004; Tilemann et al. 2012). While viral-based therapies have high rates of transfection efficiency, there are many limitations associated with their safety and efficacy. These include: low tissue specificity, strong immunogenicity and inflammatory responses, hepatic toxicity, and size limitations for carrying transgenes and their regulatory elements (Edelstein et al. 2007; Huang et al. 2009; Kay 2011). As an alternative, nonviral strategies present many advantages over viruses, including: low toxicity, lower immune responses (which may permit multiple treatments), high tissue specificity, and larger construct carrying capacity. However, the potential clinical value of nonviral approaches is hindered by ineffective vector delivery, low transfection efficiency, and transient gene expression (Lindner 2004). In the present study, we demonstrate that a combined approach using Ultrasound Targeted Microbubble Destruction (UTMD) to deliver nonviral transposon-based vectors can address these limitations.
UTMD is minimally invasive and can mediate site-specific delivery of bioactive molecules, including therapeutic genes, to ultrasound-accessible target organs including the heart and liver. Briefly, in UTMD, a vector encoding a gene of interest is added to the shell of microbubbles; these are then injected intravenously and the construct is deposited at the target organ by acoustic cavitation at a resonant frequency of the bubbles (Walton et al. 2011). In previous studies, UTMD has been used to deliver luciferase, green fluorescent protein (GFP) and beta-galactosidase reporter genes and potentially therapeutic genes, such as vascular endothelial growth factor (VEGF), Hexokinase I, and Factor IX (Chen et al. 2003; Lindner 2004; Miao et al. 2005; Kay 2011). While these studies have demonstrated short-term gene expression and higher transfection efficiency than plasmid DNA alone, further enhancement of the technique is necessary to obtain therapeutic levels and duration of expression. An intrinsic advantage of UTMD is that as a platform technology, it can be combined with additional advances in vector technology and gene delivery, such as tissue-specific promoters and transposon-based vectors, to improve the efficacy of gene transfer (Chen et al. 2005).
Transposons are naturally occurring genetic elements that are capable of moving from one chromosomal location to another. DNA transposon systems facilitate genomic integration of transgenes into mammalian host genomes through a “cut- and-paste” mechanism and provide the potential to address the limitation of transient gene expression in gene transfer studies (Mates et al. 2009; Nakanishi et al. 2010). The DNA insertion sequences and transposition activities for the Sleeping Beauty, Tol2, Mos1, and piggyBac transposon systems have been evaluated and compared for mammalian gene delivery (Wu et al. 2006; Grabundzija et al. 2010). For gene transfer applications, these systems typically involve the delivery of separate donor and helper vectors that encode the expression cassettes for the transposon and transposase respectively. The transposase removes the transposon, encoding a transgene of interest, by recognizing and excising specific inverted repeat elements at the ends of the transposon. After the transposon is mobilized, the transposase further directs its integration into a host chromosomal region containing specific naturally occurring direct nucleotide repeat elements distinctive to the transposon. The Sleeping Beauty and piggyBac transposons have high transposition efficiencies in mammalian cells and have been adapted for gene therapy investigations (Wilson et al. 2007; Grabundzija et al. 2010). The Sleeping Beauty system of the Tc1/mariner superfamily of transposable elements is the most widely used model for preclinical studies (Saridey et al. 2009; Mates et al. 2009) and is currently the only transposon system being used in a small number of gene therapy clinical trials (Li et al. 2011). In addition, hyperactive Sleeping Beauty transposases have been generated and validated using various target cells through ex vivo and in vivo gene transfer experiments. The most active construct, SB100×, has displayed transposition activity up to ~100-fold higher than the original Sleeping Beauty transposase (Mates et al. 2009)
The piggyBac transposon system, originating from the genome of the cabbage looper moth, Trichoplusia ni, possesses many features that make it an attractive tool for mammalian gene transfer studies. PiggyBac has high transposition efficiency, can mobilize and integrate transposons as large as 100 kb, and codon optimized piggyBac transposase constructs demonstrate high levels of protein production and transposition (Li et al. 2011, Elick et al. 1996; Cadinanos and Bradley 2007; Doherty et al. 2012). In addition, the piggyBac system has minimal overproduction inhibition (that can result from high levels of transposase expression), which supports its use in a helper-independent single vector system (Wilson et al. 2007). These helper-independent piggyBac vectors encode the piggyBac transposase (pBt) and transposon elements in the same construct. This design eliminates the requirement for a conventional two-plasmid helper-donor system and ensures simultaneous delivery of both elements at a defined ratio. The pmGENIE2 and pmGENIE3-luc plasmids evaluated in this study utilize the helper-independent system and contain a transposase self-inactivation mechanism, which may enhance the overall safety profile of the vector (Urschitz et al. 2010).
In the present study, reporter gene expression from a conventional pcDNA3-luc plasmid, Sleeping Beauty, and piggyBac transposon vectors were compared in vitro. Based on these studies the piggyBac system was selected as the transposon model for in vivo evaluation with hepatic-targeted UTMD. This also allowed us to investigate the helper-independent piggyBac reporter in vivo. The vectors evaluated contain the cytomegalovirus (CMV) promoter which can drive high levels of transgene expression in many mammalian tissue types (Papadakis et al. 2004). Here, we tested the hypothesis that combining UTMD with the delivery of piggyBac transposon DNA vectors would enhance transfection efficiency and prolong transgene expression in the liver compared to the pcDNA3-luc reporter plasmid.
Materials & Methods
Plasmids
The pcDNA-luciferase (pcDNA3-luc) reporter construct drives constitutive expression with a cytomegalovirus (CMV) promoter (Addgene Inc, Cambridge, MA, USA). All pmGENIE plasmids have been created by author SM, and their constructions and transposition mechanisms have been described previously (Urschitz et al. 2010). Briefly, the pmGENIE2 and 3 piggyBac transposons contain a CMV-early-enhancer/chicken β-actin and β-globin intron promoter (CAG) driven mouse-codon optimized piggyBac transposase (pBt) and a CMV driven luciferase reporter cDNA that lies between the 5′ and 3′ terminal repeat elements (TREs). The 3′-TRE of the transposon is situated between the CAG promoter and the transposase cDNA to obtain self-inactivation of the vector after transposition. In addition, for pmGENIE3, the 3′-TRE is situated in an intron of the pBt gene and upon transposition, the pBt gene is left in a truncated form in the vector backbone resulting in further enzymatic inactivation. For our preliminary studies, a hyperactive Sleeping Beauty transposase plasmid, SB100× was kindly provided to us by Dr. Zsuzsanna Izsvák (Max Delbrück Center for Molecular Medicine, Berlin, Germany). The SB100× helper plasmid was delivered with the pT2/c-luciferase (pT2/c-luc) transposon reporter construct (Addgene Inc., Cambridge, MA) at 1:1 and 10:1 equimolar ratios of transposon to transposase plasmids.
Cell culture and transfection
Human Embryonic Kidney (HEK) 293 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, GIBCO, Carlsbad, CA, USA) supplemented with 10% Fetal Bovine Serum (HyClone Thermo Scientific, Logan, UT, USA) and 1% Penicillin/Streptomycin (Sigma-Aldrich Inc, St. Louis, MO, USA). Cells were stably transfected when 90% confluent using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) with equimolar amounts of DNA, according to the manufacturer’s protocol. The cells were incubated with the DNA vectors and transfection reagent for 48 hours and reseeded into new plates two times per week. Cell samples were intermittently assayed for protein concentration and transgene expression. Luciferase expression assays (Promega, Madison, WI, USA) were conducted on the cell lysates and measured in a Victor 2 plate reader (PerkinElmer, Waltham, MA, USA). The Bradford method was used to determine the protein concentrations of the samples to generate a final determination of relative luminescence per mg of protein.
Preparation of microbubbles
Lipid-stabilized microbubbles were prepared as previously described (Walton et al. 2011). Briefly, a stock solution of 200 mg of DPCC (DL-α-Phosphtidlycholine, Dipalmitolyl) (Sigma-Aldrich), 50 mg DPPE (DL-α-Phosphtidylethanolamine, Dipalmitolyl (Sigma-Aldrich), and 1 gram glucose mixed with Phosphate Buffered Saline (PBS) to a final volume of 10 ml was heated and mixed in a boiling water-bath for 30 minutes and then stored at 4°C. The plasmid DNA-loaded microbubbles were prepared by adding 250 ul of pre-warmed (40°C) cationic liposome stock solution to 50 ul glycerol and 1 mg plasmid DNA dissolved in a 500 ul volume of TE buffer. The microtube air space was replaced with perfluoropropane gas and then shaken vigorously for 20 seconds using a Vialmix dental amalgamator (Lantheus Medical Imaging, N. Billerica, MA, USA). The lipid-DNA microbubble solution was then washed three times with PBS. Between each wash, the solution was incubated on ice for 15 minutes before the subnatant (containing unbound plasmid DNA) was removed, leaving the DNA-bound microbubble supernatant. The final microbubble-DNA solution was diluted with PBS to a volume of 1 ml prior to in vivo infusion. Approximately 200 ug of plasmid DNA remains bound to bubbles using this protocol (as determined by UV spectroscopy of the subnatant containing the unbound plasmid DNA).
Animals
Eight-week old C57BL/6 male mice (n=49 total) were used to compare reporter vector performance. Fifteen and ten mice were used in the pmGENIE2-luc and pcDNA3-luc UTMD treatment groups, respectively (including controls). Eight mice were used for the histological evaluation of pmGENIE2-luc and pcDNA-luc expression (including controls). Sixteen mice, including controls, were used in the in vivo genomic integration analyses and were transfected with pmGENIE3-luc or pcDNA3-luc by UTMD. All animal research was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Hawaii.
Ultrasound system and calibration
The ultrasound signal was generated using an arbitrary function generator (Tektronix, Model AFG3021C) operating in burst mode. The sine wave signal was sent to a 55 dB gain radio frequency (RF) power amplifier (Electronics & Innovation, EI, Model A150, 150W) and then transmitted to a low frequency 1.0 MHz unfocused transducer (Olympus Panametrics-NDT model A303S 1.0 MHz, 0.5 inch nominal element size). Calibration of the ultrasound system was performed using a pre-calibrated 1 mm needle hydrophone with a 20 dB in-line attenuator (Precision Acoustics Ltd, United Kingdom) faced co-linear to the ultrasound probe in deionized and degassed water at a distance of 0.2 mm using a stereotactic apparatus. The hydrophone was connected to a DC coupler with a power supply and the final signal was sent into an oscilloscope (100 Mega samples per second digitizer, National Instruments Corporation, Austin, TX). The oscilloscope acquisition was triggered by function generation and acoustic signals were processed in Laboratory Virtual Instrumentation Engineering Workbench (LabVIEW, National Instruments Corporation). Acoustic pressure was determined using instructions from Precision Acoustics (Dorchester, UK) whereby the maximum voltage (Vmax) was taken from the time domain hydrophone signal and converted to acoustic peak negative pressure (PNP) in megapascals (MPa) for a hydrophone sensitivity at 1 Mhz (equation below) (AIUM 2000).
In vivo UTMD gene delivery
Prior to microbubble delivery and UTMD, mice were anesthetized IP with 100mg/kg ketamine and 5mg/kg xylazine. Under ultrasound guidance, 100ul of the DNA-loaded microbubble solution was injected into the left ventricle of the heart as previously described (Walton et al. 2011). The microbubble bolus was visualized using a VisualSonics high frequency 30 MHz ultrasound transducer (VisualSonics 2100, transducer model RMV707B, Ontario, Canada) clamped above the thorax of the mouse in a long-axis view. Immediately following the injection, microbubble destruction was carried out for 5 minutes using a second, low frequency 1.0 MHz unfocused transducer (Olympus Panametrics-NDT model A303S 1.0 MHz/0.5 inch) that was manually moved several times over the skin above the entire surface of the liver, traversing approximately 2.3 cm, coupled with diagnostic ultrasound gel, targeting destruction of bubbles across this region. The ultrasound was administered to the liver in burst mode with an acoustic working frequency of 1.0 MHz, 20 μs pulse duration, and 10 Hz pulse repetition rate. Using these settings, the acoustic peak negative pressure was 1.3 MPa at 0.2 mm and 1.79 MPa at 3.4 mm from the transducer face as determined by the calibration methods and displayed in Supplementary Figure 1. Control mice were given intraventricular injections of equivalent doses of plasmid diluted in PBS without microbubbles and otherwise treated identically.
In vivo bioluminescence evaluation
Bioluminescence resulting from luciferase expression was monitored in mice transfected with pcDNA3-luc and pmGENIE2-luc plasmids using the Xenogen In Vivo Imaging System (IVIS), (Caliper Life Sciences, Hopkinton, MA, USA). Images were taken of all mice the first day after UTMD-mediated transfection and subsequently every three to four days until expression was no longer detectable. Livers were harvested from mice upon loss of expression. An additional set of mice (n=12) were transfected with the pmGENIE3-luc reporter plasmid for the integration site analysis and were evaluated for bioluminescence day 1 and 3 after UTMD. To measure bioluminescence in vivo, mice first received an IP injection (150 mg/kg dose) of the luciferase reporter probe D-luciferin diluted in sterile PBS. Mice were anesthetized 3 minutes later as above. Biodistribution of the D-luciferin substrate was allowed to proceed for 10 minutes before full body IVIS image scans were obtained using a five minute acquisition time. The IVIS system software was used to measure and quantify the visible luminescent region of interest as the maximum photons per second per centimeter squared per steradian for each mouse at each time point. These data were then normalized to a background measurement for each image.
Histological evaluation of transgene expression
Livers and hearts were obtained from UTMD and control mice after in situ perfusion with PBS followed by 4% paraformaldehyde. These tissues were collected from pcDNA3-luc and pmGENIE2-luc transfected mice three days after UTMD. Control groups included tissues from mice transfected with naked plasmid DNA and PBS for both pcDNA3-luc and pmGENIE2-luc. Explanted livers were embedded in Optimal Cutting Temperature (OCT) compound and 10 um sections were evaluated by immunofluorescence using a mouse monoclonal primary antibody to luciferase (Invitrogen). The 4′-6-Diamidino-2-phenylindole (DAPI) fluorescent stain (Vector Laboratories Inc., Burlingame, CA, USA) and the Alexa Fluor 568 fluorescent dye conjugate (Invitrogen) were used as probes for nuclei and luciferase respectively. Immunofluorescent images were obtained using a Zeiss Axioscop 2 Plus microscope with AxioVision Release 4.7 software (Carl Zeiss MicroImaging, LLC, Thornwood, NY, USA). Image J software was used to quantify the number of immuno-positive cells. Transgene expression was quantified for the number of positive cells inside and outside of a five cell radius from major hepatic vessels. Five images surrounding different vessels were evaluated for each plasmid at each time point (day 3 and > day 14 after UTMD-mediated transgene delivery).
Integration Site Analysis
To analyze genomic integration of the piggyBac transposon in vitro, mouse 3T3 cells were transfected with a pmGENIE3-eGFP (enhanced Green Fluorescent Protein) reporter plasmid or control plasmid. Forty-eight hours after transfection, eGFP positive cells (from pmGENIE3-eGFP transfections) were collected using Fluorescence Activated Cell Sorting (FACS) and seeded into 96 well plates by serial dilution to obtain single eGFP-expressing cells. Isolated clones were then subcultured in 24 well plates. Genomic DNA was purified from samples using the Qiagen AllPrep DNA Mini Kit (Valencia, CA, USA) according to the manufacturer’s protocol. Integration site analyses were performed with genomic DNA samples using pmGENIE3-eGFP 5′ and 3′-TRE-based nonrestrictive linear amplification-mediated (nrLAM) PCR as previously described (Paruzynski et al. 2010). The PCR reactions were carried out using 5′ or 3′-biotinylated primers, 5′-TRE-CGACTACGCACTAGCCAACA and 3′-TRE-GGTGCACGAGGTAAGAGAGAGG, for the linear PCR reactions, and 5′-TRE-CTTACCGCATTGACAAGCAC and 3′-TRE-GGGAGTCCCTCTCACCAAA for the first exponential PCR reactions. The following non-biotinylated primers were used for the second exponential PCR reactions, 5′-TRE-ACGGATTCGCGCTATTTAGA and 3′-TRE-CCGATAAAACACATGCGTCA.
For in vivo integration site analyses, genomic DNA from livers transfected with, pmGENIE3-luc and pcDNA3-luc by UTMD was isolated as above and nrLAM PCR was conducted using the same primers as the in vitro studies. The pmGENIE3-luc and pcDNA3-luc liver samples were all collected 3 days after UTMD (n=13 for pmGENIE3-luc and n=3 for pcDNA3-luc). The nrLAM PCR amplicons obtained from the in vitro and in vivo samples were cloned into the pCR4-TOPO TA vector (Invitrogen), and sequenced by dideoxy chain termination using an Applied Biosystems 3730×l DNA Analyzer. Sequencing data was evaluated to determine integration sites using the ApE v2.0.39 plasmid editor program (University of Utah) and the NCBI Basic Local Alignment Search Tool (BLAST) genome browser. Genomic DNA sequences from piggyBac integration sites were identified by locating the known nrLAM PCR priming site in the piggyBac 3′ TRE and the priming site of the DNA linker sequence ligated to the genomic DNA during the nrLAM PCR protocol. Sequences that mapped to the mouse genome (GRCm38) with >95% similarity were accepted as authentic in vivo integration sites and the surrounding sequence was further analyzed in 2 kilobase windows for AT and TTAA content.
Statistical Analysis
Data were analyzed using the GraphPad Prism program (Version 5.0b). Mean values were calculated for experimental groups and error bars indicate ± SEM. The Student’s t-test was used to analyze differences in luciferase expression between groups and ANOVA was used for multiple group comparisons. P-value of <0.05 was considered to be statistically significant.
Results
Preliminary evaluation of piggyBac and hyperactive Sleeping Beauty reporter expression in mammalian cells
In preliminary studies, (Supplementary Figure 2) we compared the transfection efficiency and transgene expression levels between the hyperactive Sleeping Beauty and helper-independent piggyBac transposase systems in HEK293 cells. We compared the reporter activity from cells transfected with the conventional pcDNA3-luc, piggyBac helper-independent pmGENIE2-luc and pmGENIE3-luc, and Sleeping Beauty transposase SB100×, (delivered in trans with a pT2/c-luciferase transposon plasmid). All plasmids were transfected at equimolar ratios and the Sleeping Beauty co-transfections with SB100× and pT2/c-luciferase were evaluated at transposon to transposase ratios ranging from 1:1 to 10:1. On average, we observed between 10- and 100-fold stronger reporter expression from the pmGENIE2-luc and pmGENIE3-luc plasmids after 21 days compared to SB100× delivered with pT2/c-luc. This difference was statistically significant for pmGENIE2-luc compared to the SB100× and pT2/c-luc co-transfections for both the 1:1 and 1:10 ratios (p=0.02). We therefore proceeded with helper-independent pmGENIE piggyBac reporter constructs for long-term in vitro and in vivo experiments.
Long-term evaluation of expression from pmGENIE and pcDNA3-luc plasmids in a mammalian cell line
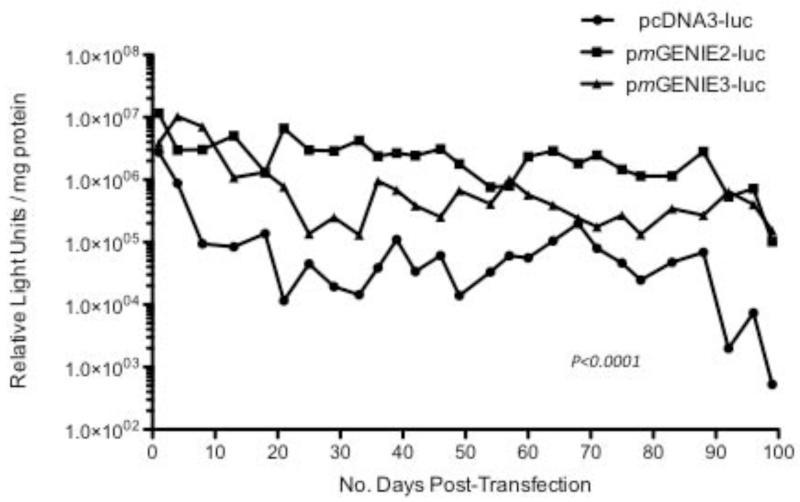
We evaluated the expression patterns of pcDNA3-luc, and pmGENIE2 and pmGENIE3 luciferase plasmids, all with CMV promoters, in HEK293 cells. Expression for pmGENIE2-luc and pmGENIE3-luc ranged from 10 to 100-fold stronger than pcDNA3-luc expression over 100 days of evaluation (Figure 1). The expression patterns for both pmGENIE transposase reporter plasmids are consistent and display similar activities through 100 days. These data demonstrate that pmGENIE helper-independent transposon plasmids mediate stronger and more stable transgene expression in vitro than a conventional CMV-driven luciferase plasmid.
Figure 1.
Luciferase expression measured in HEK 293 cells transfected with the pcDNA3-luc plasmid (CMV promoter) and two piggyBac transposon luciferase plasmids (pmGENIE2-luc, pmGENIE3-luc, with the CMV promoter). HEK: human embryonic kidney; luc: luciferase; CMV: cytomegalovirus. P<0.0001, pmGENIE2 and 3-luc expression compared to pcDNA3-luc.
Comparison of the duration and intensity of transgene expression in mice
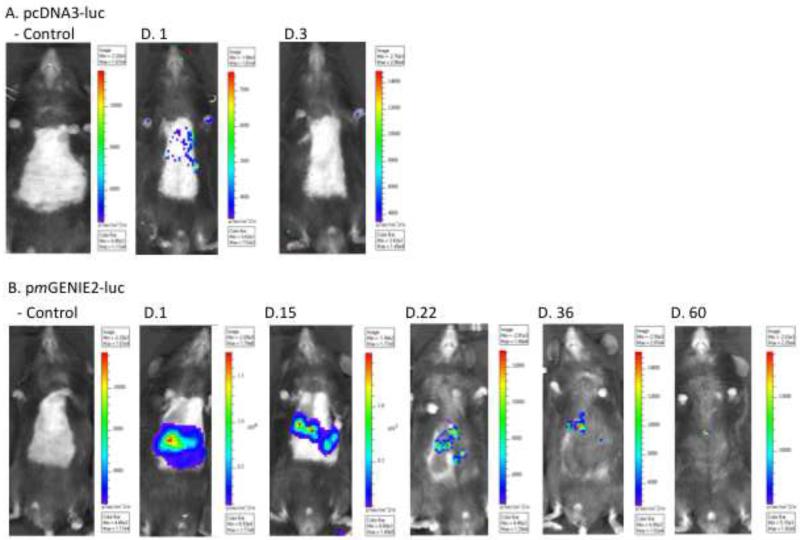
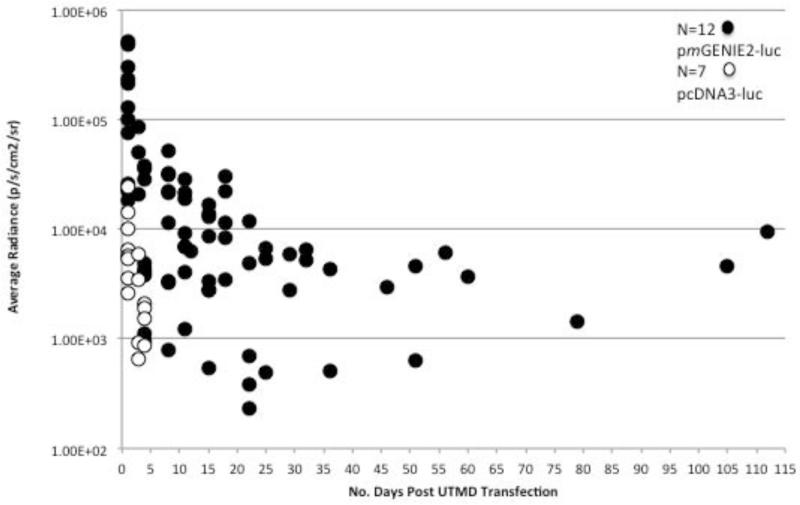
Mice were transfected by UTMD with microbubbles containing pcDNA3-luc (n=7 mice) and pmGENIE2-luc (n=12). Bioluminescent images demonstrate hepatic transgene expression at day 1 for all mice transfected with either plasmid. However, between day 3 and day 5, hepatic expression became undetectable in mice transfected with pcDNA3-luc (Figure 2a,b). We observed substantial variability in transgene expression patterns between mice transfected with pmGENIE2-luc. Specifically, one mouse from the pmGENIE2-luc transfection group in C57Bl/6 mice demonstrated loss of transgene expression by day 4, whereas another mouse displayed continuous luciferase expression for 105 days post-transfection. Quantitative measurements of luciferase activity were obtained at each time point for each mouse (Figure 3). Overall, the measured radiance of luciferase activity is higher in mice transfected with pmGENIE2-luc than pcDNA3-luc even during the first five days after UTMD transfection.
Figure 2.
In vivo luciferase expression over time in C57BL/6 mice livers. IVIS Xenogen bioluminescence imaging of UTMD treated mice. (a) pcDNA3-luc control mouse: plasmid with PBS followed by liver directed UTMD, pcDNA3-luc treatment mouse (Day 1 and 3 post UTMD): plasmid with microbubbles followed by liver directed UTMD. (b) pmGENIE2-luc control mouse: plasmid with PBS followed by liver directed UTMD, pmGENIE2-luc treatment mouse (Day 1, 15, 22, 36, 60 post UTMD): plasmid with microbubbles followed by liver directed UTMD. IVIS: In Vivo Imaging System; UTMD: Ultrasound Targeted Microbubble Destruction; luc: luciferase.
Figure 3.
In vivo comparison of luciferase expression over time in mice transfected with pcDNA3-luc (n=7) or pmGENIE2-luc (n=12). Luciferase expression (luc) is quantified as the average radiance in units of photons/second/cm2/steradian. IVIS Xenogen bioluminescence optical imaging was used to acquire each image (Figure 2). Living Image 3.0 software was used to measure and quantify the visible luminescent region of interest. The values for the average radiance of luciferase expression on a specific day for each mouse are shown. A background region of interest measurement was taken for each image and the average radiance data shown was normalized to this value. IVIS: In Vivo Imaging System.
Ex vivo histological validation of in vivo transgene expression
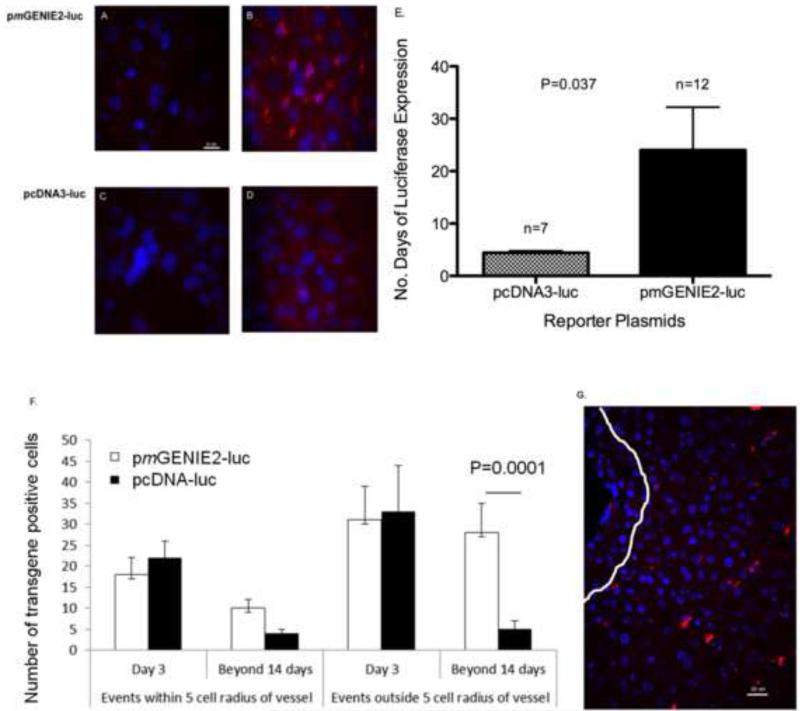
By immunofluorescence, liver samples collected three days after UTMD displayed more robust luciferase expression in mice transfected with pmGENIE2-luc compared to pcDNA3-luc (Figure 4a-d). The average number of days of transgene expression for the pcDNA3-luc and pmGENIE2-luc plasmid groups was 4.4 ± 0.37 days, and 24.4 ± 8.2 days, respectively (Figure 4e, p=0.037). Specifically, we observed increased luciferase expression in the peri-nuclear region of hepatocytes, with increased abundance around the hepatic blood vessels. Images of sections from livers of mice three days after UTMD, as well as sections from livers expressing longer than 14 days, were also evaluated for the anatomical distribution of luciferase expression (Figure 4f,g). Long-term reporter expression is associated with a more diffuse distribution of signal throughout the liver and is less localized to the cells nearest to hepatic blood vessels, as observed at Day 3. These data suggest stable transgene expression is associated with transfection of hepatocytes deeper in the parenchyma. TUNEL staining assays were conducted on liver tissue sections prepared from samples collected three days after UTMD to evaluate DNA damage and showed no increase in apoptosis from UTMD using either the pcDNA3-luc or pmGENIE2-luc plasmids compared to a non-sonicated control (data not shown). In addition, we evaluated potential inflammation in the liver by staining slides for macrophage infiltration, and performed assays for immune responses including cytokine levels using LUMINEX Inflammatory and Th1/Th2 cytokine assays. The assays were performed on samples from mice representing days 3, 4, 36, and 56 after UTMD treatment with pmGENIE-luc and days 3 and 5 (longest time point for expression) after UTMD treatment with pcDNA3-luc. There was no significant increase in either the number of monocytes or tissue cytokine levels after UTMD with the parameters we used (data not shown).
Figure 4.
Immunofluorescence evaluation of luciferase expression in the livers of mice transfected with CMV-luciferase (pcDNA3-luc) or piggyBac transposon luciferase (pmGENIE2-luc) plasmids. Primary mouse monoclonal antibody to luciferase with a secondary AlexaFluor 568 mouse antibody (red), and DAPI stain for nuclei (blue) is displayed in panels A-D. (a) Negative Control: pmGENIE2-luc with PBS only, Day 3 post UTMD, scale bar = 10 um. (b) pmGENIE2-luc with microbubbles, Day 3 post UTMD, scale bar as in (a). (c) Negative Control: pcDNA3-luc with PBS only, Day 3 post UTMD, scale bar as in (a). (d) pcDNA3-luc with microbubbles, Day 3 post UTMD, scale bar as in (a). Panels (a) and (c) show no expression after transfection without microbubbles, panel (b) shows substantial luciferase immunoreactivity with the pmGENIE2-luc plasmid compared to modest, diffuse signal with pcDNA3-luc (d). (e) The average number of days of luciferase expression in the liver after UTMD in mice transfected with the CMV-luciferase plasmid (pcDNA3-luc) or piggyBac transposon luciferase plasmid, pmGENIE2-luc. Luciferase expression was evaluated using Xenogen IVIS bioluminescence optical imaging. P<0.05, number of days of pmGENIE2-luc expression compared to pcDNA3-luc. Error bars indicate, SEM. (f), (g) Quantification of luciferase expression distribution from IHC data. (f) Five random images were evaluated for each time point and the number of positive events were averaged. P<0.05, number of pmGENIE2-luc positive events outside radius after 14 days compared to pcDNA-luc. Error bars indicate, SEM. (g) Example of analysis. IHC image of pmGENIE2-luc 475 post-UTMD. White line delineates 5 cell diameters away from vessel, scale bar = 20 um. luc: luciferase; CMV: cytomegalovirus; DAPI: 4:6-diamidino-2-phenylindole; IVIS: In Vivo Imaging System.
Genomic integration analysis
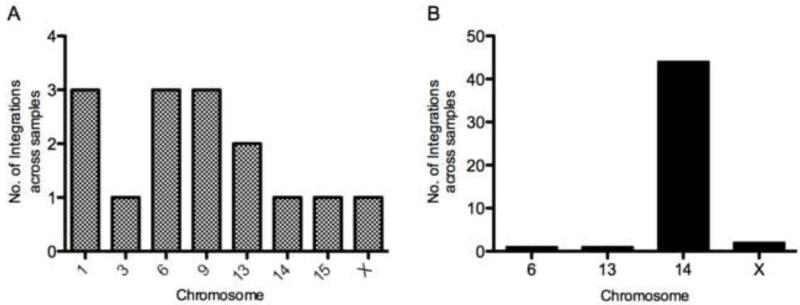
To evaluate genomic integration of the pmGENIE plasmids in vitro, we conducted nrLAM PCR with genomic DNA isolated from mouse 3T3 cells expressing pmGENIE3-eGFP (Supplementary Table 1). Mouse 3T3 cells were used for the in vitro genomic integration evaluation to maintain consistency in species for our genomic analyses in mice. We also evaluated sequences obtained from nrLAM PCR performed on genomic DNA extracted from livers collected 3 days after C57Bl/6 mice were transfected with pmGENIE3-luc (Supplementary Table 2). Twelve samples from the in vitro study were evaluated by nrLAM PCR, and sequence data characterizing genomic integration aligned to the mouse genome on chromosomes 1, 3, 6, 9, 13, 14, 15, and X with > 95% similarity (Supplementary Table 1). Sequences ranged in size from 111 bp (Chr. 13) to 963 bp (Chr. 6) with an average nrLAM PCR read length of 464 bp. Sixteen samples (n=13 mice transfected with pmGENIE3-luc, n=3 controls, transfected with pcDNA3-luc) were evaluated for the in vivo integration analysis, and sites aligning to the mouse genome were observed for chromosomes 6, 13, 14, and X for the C57Bl/6 mice (Supplementary Table 2) transfected with pmGENIE3-luc plasmid. No chromosomal integration or sequence alignment to the mouse genome was detected for the liver samples transfected with pcDNA3-luc. For the in vivo integration analysis, sequences with >95% similarity ranged in size from 111 bp to 773 bp with an average length of 386 bp. While sequences aligning with >95% similarity were included (Galvan et al. 2009), on average, we observed >98% similarity in our alignments. The fifteen piggyBac transposase-mediated in vitro genomic integration events evaluated appeared to be randomly distributed among several chromosomes (Figure 5a). However, the genomic integration events observed in vivo aligned to specific loci from a small number of chromosomes (Figure 5b, n=48 genomic integration events in the 13 mice tested). Interestingly, we observed a high frequency of in vivo integration events on chromosome 14 in which sequences generated from nrLAM PCR aligned to a 120 bp sequence. This particular sequence is repeated on chromosome 14 forty-eight times (with >95% similarity) between the 14B-14C2 nucleotide region of the chromosome. Previous studies have reported piggyBac transposition tendencies, including preferential integration for AT and TTAA nucleotide rich regions of the genome (Wilson et al. 2007; Galvan et al. 2009). In our analyses of the AT and TTAA content within the 2 kb sequence windows surrounding the integration events, we observed an average of ~59% AT content and 4.3 TTAA sites for the in vivo integration sites. In comparison, the AT content of the whole mouse genome is 58% and the frequency of TTAA sequences is 1.06 (Waterston et al. 2002; Galvan et al. 2009). The AT and TTAA content surrounding the observed integration on chromosome X was particularly rich in TTAA sites in which 26 TTAA sequences were observed in 2 kb of sequence (Supplementary Figure 3).
Figure 5.
Chromosomal distribution of piggyBac integration events in vitro and in vivo. (a) pmGENIE3-eGFP integration events in MEF 3T3 cells. A total of 15 pmGENIE3-eGFP chromosome integration events were observed in nrLAM PCR generated amplicons from MEF 3T3 cells. pmGENIE3-eGFP positive single cell events were observed for ~65 samples (Data shown is for 12 of these samples that were analyzed for integration). (b) pmGENIE3-luc integration events in livers of C57Bl/6 mice (n=13). A total of 48 samples from 13 livers transfected with pmGENIE3-luc (C57Bl/6 mice) were analyzed and demonstrated integration into the mouse genome. At least 4 transformed colony samples per liver were evaluated and the data shown describes 48 of these events. The frequency distribution represents the number of times an integration in a particular chromosome occurred within each sample and the values shown have been pooled for all livers. eGFP: enhanced green fluorescent protein; MEF: mouse embryonic fibroblast; nrLAM PCR: non restrictive linear amplification mediated polymerase chain reaction; luc: luciferase.
Discussion
The use of nonviral DNA vectors in clinical gene therapy trials is increasing (Edelstein et al. 2007). Nonviral vectors have better safety profiles and lower production costs than viruses; however, low transfection efficiency limits their applicability for the treatment of genetic diseases that require substantial and sustained gene expression. In this study we applied UTMD to improve anatomic specificity and transfection efficacy of nonviral vectors in the liver. We demonstrated that UTMD can target the delivery of piggyBac helper-independent vectors to the liver and provide increased intensity and duration of hepatic luciferase transgene expression compared to a conventional CMV-driven construct. This enhanced gene expression reflects the combination of the UTMD-mediated mechanism for hepatic transfection with the genomic integration capacity of the piggyBac transposon vector. We have shown the anatomical specificity of this approach using bioluminescence imaging, and conducted a histological evaluation of the heart, kidney and spleen from liver targeted mice, as negative expression control tissues (Supplementary Figure 4), which revealed high hepatic specificity for the UTMD targeting (Figure 4). Minimal luciferase expression was detected in the kidneys of mice transfected with pmGENIE-luc, which were included in the field of insonation. We anticipate that a liver-specific promoter would further enhance the anatomical site-specificity in future studies. We have also presented histological data that suggests stable transgene expression is associated with transfection of hepatocytes located deeper in the parenchyma. Immediately after UTMD, we expect that cells surrounding the hepatic artery and portal vein will be transfected more easily because they come in contact with plasmids first. However, these cells may also be subjected to greater immune surveillance, with an increased likelihood of immunological-based silencing and/or loss of DNA due to cellular damage resulting from inflammatory responses (Chen et al. 2008). The anatomic architecture of the liver may be particularly amenable to deep transfection of the tissue. If microbubbles can traverse the fenestrated endothelium then the structure of this microvasculature may additionally facilitate stable transfection to hepatocytes further from vascular structures (Shen et al. 2008).
Recently, the potential of piggyBac transposon technology for gene transfer has been recognized (Doherty et al. 2012). We have shown that piggyBac vectors can mediate stronger and more persistent reporter expression in the liver compared to a conventional reporter plasmid. The nrLAM PCR-based integration analyses we conducted confirm genomic integration of these vectors in transfected cells and the livers of treated mice. Surprisingly, the approach used in this study showed a strong degree of integration into a specific genomic region. Our in vitro integration site evaluation demonstrates an apparently random pattern of chromosomal target events in mouse fibroblast 3T3 cells, while a less random pattern was observed in vivo, specifically with particular tropism for a 120 bp repeated sequence on chromosome 14. Previous studies have shown a broad range of integration of piggyBac in vitro (Wilson et al. 2007; Galvan et al. 2009; Meir et al. 2011) albeit with a few suggested “hotspots”. Preferential integration regions have been reported for the piggyBac transposon in primary human T cells (Galvan et al. 2009) and HEK 293 cells (Meir et al. 2011). Specifically, piggyBac integration in primary human T cells demonstrated preference for TTAA-rich regions in the genome, as does our data from mouse 3T3 cells (Supplementary Figure 2). Data from in vitro studies have favored integration loci for chromosomes 3, 14, 16, 17, and 20 (Meir et al. 2011). We also observed piggyBac integration in chromosomes 3 and 14, at different loci in vitro (with additional sites occurring on chromosomes 1, 6, 9, 13, 15, and X).
Presently, few studies have presented data on in vivo integration analyses (Saridey et al. 2009; Doherty et al. 2012). By using a relatively non-toxic, rapid, and minimally invasive method for gene delivery, we were able to evaluate a large number of in vivo integration events compared to previous studies. We found a high tropism for a specific repeated sequence in many independent liver samples. In addition to the sites in chromosome 14, we also found single integration events in chromosomes 6 and 13, and double integrations in X. The 120 bp repeated sequence captured from our nrLAM PCR experiments was sequenced and analyzed and was mapped to forty-eight specific loci between the 14B and 14C2 region of the BLAST-generated chromosome 14 ideogram, with at least 95% similarity, and is conserved between mouse strains. In an effort to further characterize this finding, we analyzed the 2 kb window of chromosomal sequence surrounding these repeated sites and found them to be part of larger segments of repetitive DNA (displaying >95% similarity among regions) located throughout this particular region within chromosome 14. We found no syntenic similarity between this sequence in the mouse and the human genome. We observed the greatest significance for the abundance of TTAA sites, previously associated with piggyBac integration, in our analyses of chromosome X (Supplementary Figure 2). Further analysis of mouse chromosome 14 showed known repeat elements within and around this repeated sequence. These elements included two of the four types of repeat classes known for mice, the long interspersed nucleotide element (LINE)-like element, L1, and the endogenous retrovirus-like (ERV) element with long terminal repeats (LTRs, of the Class III, ORR1 MaLR type) (Waterston et al. 2002). We also observed the TAGTG simple repeat sequence in these analyses (Waterston et al. 2002). L1 repeats comprise the largest proportion of interspersed repeat sequences in the mouse genome where they have continued to remain highly active sites of transposition over evolutionary time; however their activity in humans has declined In addition, the ERV elements have also been reported to be almost extinct in humans while remaining active in mice (Waterston et al. 2002). These characteristics suggest that the repeated integration site in chromosome 14 will be limited to the mouse genome. In general, LINE repeats have been found to be more common in A+T-rich regions of the genome, which is also consistent with our observation of piggyBac integration in regions of higher AT and TTAA content and LINE repeats in the genome.
We hypothesize that the variable integration observed from the CMV-driven transgene is the result of silencing of transgene expression in the liver. While the CMV promoter and enhancer can drive high levels of transgene expression in mammalian tissues, they can can also stimulate humoral and cell-mediated immune responses to transgene elements (Papadakis et al. 2004). Inflammatory responses to transferred DNA can lead to loss of transgene expression and may be associated with unmethylated cytosine-guanine dinucleotide (CpG) sequences in the CMV promoter (Papadakis et al. 2004). It has been shown that the type of promoter used to drive transgene expression also plays an important role in the epigenetic CpG methylation status of transposon transgene elements (Chen et al. 2008). The incorporation of a mammalian liver-specific promoter, such as the apolipoprotein E enhancer/alpha1-antitrypsin (hATT) promoter into nonviral transposon-based vectors may be advantageous for avoiding immune responses to delivered transgenes while further enhancing the specificity of a UTMD targeted approach (Miao et al. 2001). In future research, we are interested in evaluating the vectors used in this study with liver-specific (hATT) mammalian promoters to further enhance UTMD-mediated transgene expression in the mouse liver.
Overall, our results suggest that UTMD is a useful, minimally invasive, method for hepatic gene delivery. Although we used intracardiac injection for convenience, there is compelling evidence in previous studies that any intravenous route will suffice. We have shown substantial and persistent transgene expression when combined with transposon-based vectors. It is likely that one could augment the site-specificity of this combined approach further by introducing tissue-specific promoters into vectors, as well as optimizing sonographic parameters of UTMD for each target organ. In a previous study, it was shown that UTMD-mediated gene transfer efficiency in the liver is dependent on the peak negative acoustic pressure (Shen et al. 2008). Interrogation of acoustic parameters may identify a better balance of transfection efficiency with minimization of tissue damage. However, the protocol evaluated in this study provides easily detectable, persistent expression with negligible tissue damage.
Here we describe reporter gene expression to quantitate the efficacy of various gene expression vectors; our ultimate goal will be to use UTMD with transposon-based vectors to deliver therapeutic transgenes, such as those for the coagulation factors deficient in hemophilia, to the liver. A recent clinical trial highlights the possible therapeutic value of gene therapy for this disease (Nathwani et al. 2011). Our study demonstrates that the UTMD technique presently permits the rapid evaluation of novel vectors for gene delivery in vivo, and we expect this approach to accelerate progress toward therapeutic applications.
Supplementary Material
Acknowledgements
The authors would like to thank Steffen Oeser from the JABSOM Genomics Core Facility for sequencing support, Miyoko Bellinger from the JABSOM Histology Core for histology processing support, the Imaging Core for microscopy resources, and Aaron Tuia from the JABSOM Mouse Phenotyping Core for animal husbandry. These studies were supported by NIH grants HL080532, HL073449, RR016453, and their supplements (to RVS), GM08315804 and RR018727 (to SM), RR003061 and MD007601 for Histology and Imaging Core resources, as well as AHA WSA grant 12PRE12040462 (to CDA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Potential Conflict of Interest
Stefan Moisyadi, one of the authors of this manuscript, is the owner of Manoa BioSciences, which holds two patents for the pmGENIE constructs.
References
- AIUM Mechanical Bioeffects from Diagnostic Ultrasound: AIUM Consensus Statements, Section 7--discussion of the mechanical index and other exposure parameters. J Ultrasound Med. 2000;19:143–8. 54–68. doi: 10.7863/jum.2000.19.2.143. American Institute of Ultrasound in Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyros O, Wong SP, Niceta M, Waddington SN, Howe SJ, Coutelle C, Miller AD, Harbottle RP. Persistent episomal transgene expression in liver following delivery of a scaffold/matrix attachment region containing non-viral vector. Gene Ther. 2008;15:1593–605. doi: 10.1038/gt.2008.113. [DOI] [PubMed] [Google Scholar]
- Bekeredjian R, Chen S, Frenkel PA, Grayburn PA, Shohet RV. Ultrasound-targeted microbubble destruction can repeatedly direct highly specific plasmid expression to the heart. Circulation. 2003;108:1022–6. doi: 10.1161/01.CIR.0000084535.35435.AE. [DOI] [PubMed] [Google Scholar]
- Bekeredjian R, Grayburn PA, Shohet RV. Use of ultrasound contrast agents for gene or drug delivery in cardiovascular medicine. J Am Coll Cardiol. 2005;45:329–35. doi: 10.1016/j.jacc.2004.08.067. [DOI] [PubMed] [Google Scholar]
- Brooks AR, Harkins RN, Wang P, Qian HS, Liu P, Rubanyi GM. Transcriptional silencing is associated with extensive methylation of the CMV promoter following adenoviral gene delivery to muscle. J Gene Med. 2004;6:395–404. doi: 10.1002/jgm.516. [DOI] [PubMed] [Google Scholar]
- Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, He CY, Ehrhardt A, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther. 2003;8:495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- Chen ZY, He CY, Kay MA. Improved production and purification of minicircle DNA vector free of plasmid bacterial sequences and capable of persistent transgene expression in vivo. Hum Gene Ther. 2005;16:126–31. doi: 10.1089/hum.2005.16.126. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Riu E, He CY, Xu H, Kay MA. Silencing of episomal transgene expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol Ther. 2008;16:548–56. doi: 10.1038/sj.mt.6300399. [DOI] [PubMed] [Google Scholar]
- Doherty JE, Huye LE, Yusa K, Zhou L, Craig NL, Wilson MH. Hyperactive piggyBac gene transfer in human cells and in vivo. Hum Gene Ther. 2012;23:311–20. doi: 10.1089/hum.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2007--an update. J Gene Med. 2007;9:833–42. doi: 10.1002/jgm.1100. [DOI] [PubMed] [Google Scholar]
- Elick TA, Bauser CA, Fraser MJ. Excision of the piggyBac transposable element in vitro is a precise event that is enhanced by the expression of its encoded transposase. Genetica. 1996;98:33–41. doi: 10.1007/BF00120216. [DOI] [PubMed] [Google Scholar]
- Galvan DL, Nakazawa Y, Kaja A, Kettlun C, Cooper LJ, Rooney CM, Wilson MH. Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. J Immunother. 2009;32:837–44. doi: 10.1097/CJI.0b013e3181b2914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabundzija I, Irgang M, Mates L, Belay E, Matrai J, Gogol-Doring A, Kawakami K, Chen W, Ruiz P, Chuah MK, VandenDriessche T, Izsvak Z, Ivics Z. Comparative analysis of transposable element vector systems in human cells. Mol Ther. 2010;18:1200–9. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Chen Z, Hu S, Jia F, Li Z, Hoyt G, Robbins RC, Kay MA, Wu JC. Novel minicircle vector for gene therapy in murine myocardial infarction. Circulation. 2009;120:S230–7. doi: 10.1161/CIRCULATIONAHA.108.841155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay MA. State-of-the-art gene-based therapies: the road ahead. Nat Rev Genet. 2011;12:316–28. doi: 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- Li MA, Turner DJ, Ning Z, Yusa K, Liang Q, Eckert S, Rad L, Fitzgerald TW, Craig NL, Bradley A. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011;39:e148. doi: 10.1093/nar/gkr764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner JR. Microbubbles in medical imaging: current applications and future directions. Nat Rev Drug Discov. 2004;3:527–32. doi: 10.1038/nrd1417. [DOI] [PubMed] [Google Scholar]
- Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, Ma L, Samara-Kuko E, Gysemans C, Pryputniewicz D, Miskey C, Fletcher B, VandenDriessche T, Ivics Z, Izsvak Z. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–61. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- Meir YJ, Weirauch MT, Yang HS, Chung PC, Yu RK, Wu SC. Genome-wide target profiling of piggyBac and Tol2 in HEK 293: pros and cons for gene discovery and gene therapy. BMC Biotechnol. 2011;11:28. doi: 10.1186/1472-6750-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao CH, Brayman AA, Loeb KR, Ye P, Zhou L, Mourad P, Crum LA. Ultrasound enhances gene delivery of human factor IX plasmid. Hum Gene Ther. 2005;16:893–905. doi: 10.1089/hum.2005.16.893. [DOI] [PubMed] [Google Scholar]
- Miao CH, Thompson AR, Loeb K, Ye X. Long-term and therapeutic-level hepatic gene expression of human factor IX after naked plasmid transfer in vivo. Mol Ther. 2001;3:947–57. doi: 10.1006/mthe.2001.0333. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Higuchi Y, Kawakami S, Yamashita F, Hashida M. piggyBac transposon-mediated long-term gene expression in mice. Mol Ther. 2010;18:707–14. doi: 10.1038/mt.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani AC, Davidoff AM, Tuddenham EG. Prospects for gene therapy of haemophilia. Haemophilia. 2004;10:309–18. doi: 10.1111/j.1365-2516.2004.00926.x. [DOI] [PubMed] [Google Scholar]
- Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, O’Beirne J, Smith K, Pasi J, Glader B, Rustagi P, Ng CY, Kay MA, Zhou J, Spence Y, Morton CL, Allay J, Coleman J, Sleep S, Cunningham JM, Srivastava D, Basner-Tschakarjan E, Mingozzi F, High KA, Gray JT, Reiss UM, Nienhuis AW, Davidoff AM. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365:2357–65. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakis ED, Nicklin SA, Baker AH, White SJ. Promoters and control elements: designing expression cassettes for gene therapy. Curr Gene Ther. 2004;4:89–113. doi: 10.2174/1566523044578077. [DOI] [PubMed] [Google Scholar]
- Paruzynski A, Arens A, Gabriel R, Bartholomae CC, Scholz S, Wang W, Wolf S, Glimm H, Schmidt M, von Kalle C. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat Protoc. 2010;5:1379–95. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- Saridey SK, Liu L, Doherty JE, Kaja A, Galvan DL, Fletcher BS, Wilson MH. PiggyBac transposon-based inducible gene expression in vivo after somatic cell gene transfer. Mol Ther. 2009;17:2115–20. doi: 10.1038/mt.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen ZP, Brayman AA, Chen L, Miao CH. Ultrasound with microbubbles enhances gene expression of plasmid DNA in the liver via intraportal delivery. Gene Ther. 2008;15:1147–55. doi: 10.1038/gt.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilemann L, Ishikawa K, Weber T, Hajjar RJ. Gene therapy for heart failure. Circ Res. 2012;110:777–93. doi: 10.1161/CIRCRESAHA.111.252981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urschitz J, Kawasumi M, Owens J, Morozumi K, Yamashiro H, Stoytchev I, Marh J, Dee JA, Kawamoto K, Coates CJ, Kaminski JM, Pelczar P, Yanagimachi R, Moisyadi S. Helper-independent piggyBac plasmids for gene delivery approaches: strategies for avoiding potential genotoxic effects. Proc Natl Acad Sci U S A. 2010;107:8117–22. doi: 10.1073/pnas.1003674107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton CB, Anderson CD, Boulay R, Shohet RV. Introduction to the ultrasound targeted microbubble destruction technique. J Vis Exp. 2011 doi: 10.3791/2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton CB, Shohet RV. Tiny bubbles and endocytosis? Circ Res. 2009;104:563–5. doi: 10.1161/CIRCRESAHA.109.195651. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wilson MH, Coates CJ, George AL., Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–45. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, Kaminski JM. PiggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci U S A. 2006;103:15008–13. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.