Abstract
One the most important discoveries of the post-genomic era is that a large fraction of the genome transcribes a heterogeneous population of noncoding RNAs (ncRNA). ncRNAs shorter than 200 nucleotides are usually identified as short/small ncRNAs—examples include PIWI-interacting RNAs, small interfering RNAs, and microRNAs (miRNAs)—whereas those longer than 200 nucleotides are classified as long ncRNAs (lncRNAs). These molecules are emerging as important regulators of cellular process, such as development, differentiation, and metabolism. Not surprisingly, ncRNAs are involved also in human diseases, such as cancer and metabolic and neuronal disorders. Although the role of miRNAs is being largely investigated in cardiovascular biology, little is known about other classes of ncRNA in this field. However, recent reports have started to reveal the importance of lncRNA in heart development and suggest also an involvement in heart failure. Here, we will discuss these reports and the therapeutic potential of lncRNA for heart failure.
Keywords: Long noncoding RNA, Heart, Hypertrophy, Heart failure, Cardiomyocytes
Introduction
Heart failure (HF) is the ultimate outcome of many cardiovascular pathologies and a leading cause of morbidity and mortality [1]. It is underlain by gene expression reprogramming, wherein certain fetal genes are upregulated while adult genes become downregulated HF [2, 3]. This reprogramming causes cardiac hypertrophy, a pathological event that is ultimately responsible for a deterioration in heart function. Mechanisms similar to those that govern gene expression in heart development are at the basis of those that occur in HF [4]. The study of the mechanisms that control cardiac gene expression in HF and heart development provides information conducive to the better understanding of HF pathogenesis, and could lead to new diagnostic and therapeutic tools [5].
Over the last few years, one of the greatest surprises of high-throughput analysis of the transcriptome has been the discovery of noncoding RNAs (ncRNAs): these are a variety of RNAs that are not translated into a protein, but rather influence gene expression through various mechanisms—such as RNA interference—at the level of transcription or translation. Based on size, ncRNAs can be subdivided into two groups: (1) small ncRNAs (<200 nt), which include microRNA (miRNA), PIWI-interacting RNA (piRNA), and endogenous small interfering RNA (siRNA), and (2) long ncRNAs (lncRNAs), which have a length between 0.2 and 2 kb and, thus, a coding potential of less than 100 amino acids [6–9].
So far, a large range of functions have been attributed to ncRNA, such as cell proliferation, apoptosis, and cell invasion and imprinting, indicating that these molecules may represent a major regulatory component of the eukaryotic genome. Not surprisingly, ncRNAs are emerging as important players in several human pathologies, including cardiovascular diseases [6–9]. Indeed, miRNAs play a key role in driving the gene expression changes of HF, atherosclerosis, and cardiac ischemic/reperfusion injury [10]. However, we are just beginning to understand the biology of piRNA, siRNA, and lncRNA in the cardiovascular system. Here, we will give an overview of recent discoveries that support an important role of lncRNA in heart development and heart failure.
Biological Roles of lncRNAs
lncRNAs are currently defined as transcripts greater than 200 nt without known protein-coding function [11]. Though thousands of lncRNAs have been identified across eukaryotes, many are species specific and appear less conserved than protein-coding genes [12, 13].
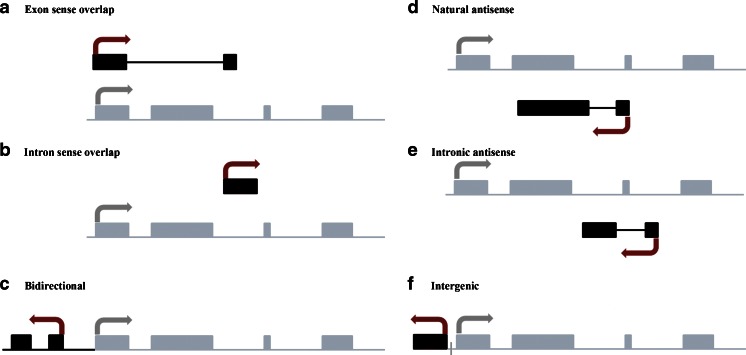
Based on genomic distribution, lncRNAs can be classified into one or more of the following five categories: (1) sense, when overlapping one or more exons of another transcript on the same strand; (2) antisense, when overlapping one or more exons of another transcript on the opposite strand; (3) bidirectional, when expression of the lncRNA and a neighboring coding transcript on the opposite strand is initiated in close genomic proximity, (4) intronic, when derived from an intron of a second transcript; and/or (5) intergenic, when it lies as an independent unit within the genomic interval between two genes (Fig. 1).
Fig. 1.
Biogenesis of long noncoding RNAs. Schematic diagram depicting the network of long noncoding transcript (black box) associated with a representative gene (gray box). a Sense lncRNA: the lncRNA sequence overlaps with the sense strand of a protein-coding gene; b intronic lncRNA: the lncRNA sequence derives exclusively from within an intron of another transcript. This may be either a true independent transcript or a product of pre-mRNA processing; c bidirectional lncRNA: the lncRNA sequence is located on the opposite strand to that of a protein-coding gene whose transcription is initiated <1,000 base pairs away; d, e antisense lncRNA: the lncRNA sequence overlaps with the antisense strand of a protein-coding gene; f intergenic lncRNA: the lncRNA sequence is not located near any other protein-coding loci
Although only a very small percentage of identified lncRNAs have been explored experimentally, they are known to be implicated in many biological processes, such as X chromosome inactivation and genomic imprinting, nuclear compartmentalization and architecture, cell fate specification, RNA splicing, translational control, and chromatin modification [14]. Below we summarize the main recent findings on these functions:
X chromosome Inactivation and Genomic Imprinting
One of the best-studied lncRNAs to date is Xist. Discovered in 1991, it is widely accepted to be required for the silencing of hundreds of genes on the inactive X chromosome in female somatic cells by favoring the formation of a chromatin structure with an epigenetic profile linked to transcription repression, a process known as X chromosome inactivation (XCI) [15]. However, despite having been studied extensively, the exact mechanism of Xist-mediated XCI is yet to be fully elucidated [14].
lncRNAs are also implicated in genomic imprinting, a process that inactivates either the maternally or the paternally inherited allele. Imprinted genes play crucial roles in mammalian development, and so their expression must be highly regulated. Intriguingly, many imprinted gene loci express, in addition to mRNAs, a significant number of lncRNAs (e.g., Air, H19, and Kcnq1ot1) that appear to play major roles in regulating the expression of neighboring imprinted protein-coding genes in cis [16].
Nuclear Compartmentalization and Architecture
The contribution of lncRNAs in the building of a cell’s architecture is well known. A clear example comes from paraspeckles—ribonucleoprotein bodies found in the interchromatin space of mammalian cell nuclei—which play a central role in regulating gene expression during cell differentiation. These structures control many aspects of transcription and RNA processing, such as transcription initiation, coactivation and corepression, RNA splicing, and transcription termination, through retention of RNA in the nucleus [17]. Given the large numbers of long noncoding transcripts currently being discovered through whole-transcriptome analysis, paraspeckles may be a paradigm for a class of sub-nuclear bodies formed around lncRNA.
Cell Fate Specification
It is well accepted that many lncRNAs provide an additional layer of regulation in the specification of cellular identities through the regulation of gene expression at transcriptional and translational levels [18]. Recently, the muscle-specific lncRNA linc-MD1 was identified as a regulator of muscle cell differentiation, by acting as a competing endogenous RNA in mouse and human myoblasts. Downregulation and over-expression of linc-MD1 correlate with retardation and anticipation of the muscle differentiation program, respectively [19].
RNA Splicing and Translational Control
Alternative splicing of pre-mRNAs increases the diversity of the proteome by generating different protein products having non-overlapping functions from a single mRNA. MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) was originally identified as a gene that was specifically upregulated in metastatic non-small cell lung cancer cells, but has subsequently been re-characterized as an lncRNA that accumulates in the nucleus and is now referred to as NEAT2 (nuclear-enriched noncoding transcript 2) [20]. This lncRNA regulates alternative splicing through its interaction with serine-/arginine-rich (SR) nuclear phosphoproteins involved in the splicing machinery [21].
Chromatin Modification
lncRNAs have been proposed to regulate transcription by recruiting chromatin-remodeling complexes, which in turn mediate epigenetic changes. For example, Kcnq1ot1, Airn, Xist, and HOTAIR are four lncRNAs that act to promote the formation of repressive chromatin structure across large genomic regions and even entire chromosomes. This is achieved through recruitment of epigenetic enzymes, such as polycomb repressive complex 1 (PRC1) and polycomb repressive complex 2 (PRC2), which mediate mono-ubiquitinylation of lysine 119 on histone 2A and trimethylation of lysine 27 on histone H3, respectively [22, 23].
Another class of ncRNA that has received much emphasis is found expressed at enhancer regions: this type of RNA has been named enhancer RNA (eRNA). The size of eRNA ranges from 0.1 to 9 kb, comprising an average of 800 nt. Most promote transcription of neighboring genes. An example is ncRNA-a7, which acts in cis to induce transcriptional activation of the nearby gene Snai1 [24].
There are probably many other functions of lncRNAs awaiting discovery. For example, the lncRNA NRON has been shown to regulate nuclear trafficking of the transcription factor nuclear factor of activated T cells, and the observation that many lncRNAs are located in the cytoplasm suggests that they might have undiscovered roles in cell biology [25].
Molecular Mechanisms of lncRNAs
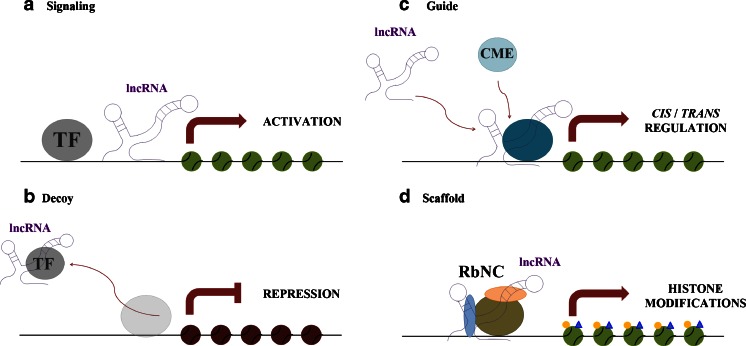
The precise mechanism of how lncRNAs function still remains largely unknown and is the subject of intense investigation. Wang and Chang have proposed to classify the molecular mechanisms of lncRNAs into four main categories (Fig. 2) [8]:
Fig. 2.
Molecular mechanism of long noncoding RNAs. Schematic diagram illustrating the four molecular mechanisms of lncRNAs. a Signaling lncRNA: the combination of lncRNAs and transcription factors may affect gene regulation in space and time; b decoy lncRNA: lncRNAs can displace transcription factors and other complexes away from the chromatin, leading to the silencing of a nearby gene; c guide lncRNA: lncRNAs may guide chromatin-modifying enzymes to their site of action, either in cis or in trans; d scaffold lncRNA: lncRNAs can induce the assembly of multiple proteins, forming ribonucleoprotein complexes affecting histone modifications. TF transcription factor, CME chromatin-modifying enzymes, RbNC ribonucleoprotein complexes
Signal lncRNAs
These lncRNAs can serve as molecular signals because transcription of individual lncRNAs occurs at very specific times and places in order to interpret cellular context or to respond to diverse stimuli. For instance, Air and Kcnq1ot1 mediate the transcriptional silencing of multiple genes by interacting with chromatin and recruiting the chromatin-modifying machinery [8, 26].
Decoy lncRNAs
The molecular decoy type of activity takes place when specific lncRNAs are transcribed and then bind to and titrate away protein factors. Decoy lncRNAs can “sponge” protein factors such as transcription factors and chromatin modifiers. This leads to broad changes in the cell’s transcriptome. One of the most abundant nuclear lncRNAs in mammalian cells is NEAT2, which is localized in nuclear speckles. NEAT2 binds to and sequesters several SR splicing factors to nuclear speckles. Depletion of NEAT2 alters splicing-factor localization and activity, leading to an altered pattern of alternative splicing for a set of pre-mRNAs [27].
Guide lncRNAs
These can act as molecular chaperons by localizing particular ribonucleoprotein complexes to specific chromatin targets. This activity can cause changes in the gene expression of neighboring (cis) or distantly located (trans) genes that cannot be easily predicted by just the lncRNA sequence itself. Examples are HOTAIR and HOTTIP, both transcribed within the human HOX clusters.
HOTTIP may organize chromatin domains to coordinate long-range gene activation by serving as key intermediates that transmit information from higher order chromosomal looping into chromatin modifications. In contrast, lncRNAs such as HOTAIR are able to alter and regulate epigenetic states in cells through their targeting of chromatin-modifying complex occupancy/localization/enzymatic activity in trans [28].
Scaffold lncRNAs
Some lncRNAs possess multiple domains that bind distinct proteins. The complexes formed perform functions such as transcriptional activation or repression. Thus, the lncRNA serves as an adaptor to form the functional protein complexes. The telomerase RNA TERC (TERRA), which is part of telomeric heterochromatin in addition to being present in the nucleoplasm, is a classic example of an RNA scaffold and is essential for telomerase function [29].
Are lncRNAs Important Players of Heart Development?
Over recent years, next-generation sequencing technologies have enabled the study of transcriptomes in various cell types at different developmental stages. These studies have revealed that the expression of many lncRNAs is regulated during development and that their role ranges from the control of pluripotency to lineage specification [29–31]. For instance, XCI is tightly correlated with both early embryonic development and pluripotency in embryonic stem cells and induced pluripotent stem cells [32]. Knockdown of dozens of lncRNAs causes either exit from the pluripotent state or upregulation of lineage commitment programs, demonstrating that lncRNAs play key roles in the circuitry controlling the embryonic stem cell state [33]. The role of lncRNA in heart development is only starting to emerge. Indeed, two recent reports demonstrated that two lncRNAs, Fendrr and Braveheart (Bvht), are involved in defining the gene transcription program underlying the development of lateral mesoderm in heart and the differentiation of cardiomyocytes, respectively [34, 35].
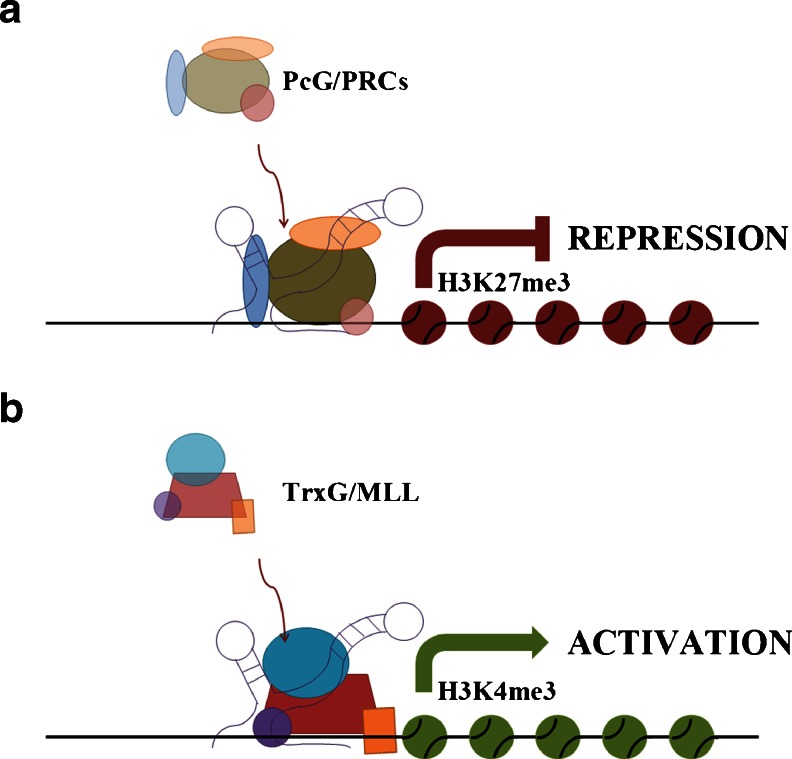
Fendrr is specifically expressed in nascent lateral plate mesoderm and is required for the correct development of this tissue in heart and the body wall. Indeed, Fendrr-deficient mice die around E13.75 due to abnormal functioning of the heart. The heart of these mice presents with a thin ventricular wall, caused by cardiac hypoplasia linked to an altered proliferation of cardiac myocytes at later stages of heart development. Fendrr regulates the expression of important transcription factors, such GATA-6, NKX2-5, FOXF1, TBX3, IRX3, and PITX2, by controlling the epigenetic profile of the promoters of these genes. In fact, Fendrr can bind either to PRC2 or to the Trithorax group/MLL protein complex (TrxG/MLL), which induces trimethylation of lysine 27—an H3 histone marker associated with transcription activation—and trimethylation of lysine 4 on histone H3—a marker associated with transcription activation—respectively [34] (Fig. 3). This lncRNA–protein interaction targets the complexes to the promoters of specific genes (Table 1).
Fig. 3.
Schematic representation of the mechanism of action of Fendrr in heart development. This lncRNA can either induce gene transcription repression or promote gene transcription through the recruitment of PRC2 (a) or TrxG/MLL (b), respectively, on promoters of gene targets during heart development
Table 1.
Long noncoding RNAs with a possible association with heart diseases
| lncRNA | Cardiac function | Heart disease |
|---|---|---|
| Fendrr | Required for the development of lateral plate mesoderm regulating the expression of GATA-6, NKX2-5, FOXF1, TBX3, IRX3, and PITX2 genes | None described [34] |
| Bvht | Required for cardiomyocyte differentiation | None described [35] |
| MIAT | Unknown | Myocardial infarction [36] |
| Kcnq1ot1 | Regulates the expression of the potassium channel KCNQ1 | None described [38] |
| ANRIL | Represses the expression of oncosuppressor genes INK4b, ARF, and INK4a | Coronary heart disease [39] |
The lncRNA Braveheart (Bvht) is required for cardiomyocyte differentiation in vitro and for maintaining the cardiac phenotype in neonatal cardiomyocytes. Bvht is required for expression of core gene regulatory networks involved in defining cardiovascular cell fate and acts upstream of mesoderm posterior (MesP1), a master gene of cardiovascular lineage commitment. In a similar way to Fendrr, Bvht regulates cardiomyocyte differentiation by modulating the epigenetic profile of cells through interaction with SUZ12, a component of PRC2 [35].
lncRNA: a Possible Player in Heart Failure
If we are only at the beginning of understanding the role of lncRNA in heart development, we are even further away from comprehending the function of these molecules in cardiovascular disease. However, recent studies have started to investigate their role in this area. A genome-wide association study (GWAS) found six single-nucleotide polymorphisms (SNP) in the lncRNA MIAT (myocardial infarction-associated transcript) associated with myocardial infarction; one SNP (A11741G) caused a 1.3-fold increase in MIAT transcription in vitro [36].
Another study revealed 15 lncRNAs modulated in the heart of mice subjected to aortic constriction—a procedure that, through the creation of pressure overload, induces first compensated hypertrophy and then HF [37].
Moreover, transcription in the heart of Kcnq1—a gene encoding a potassium channel—depends on the expression of the lncRNA Kcnq1ot1. This is an unspliced, ∼60 kb lncRNA whose transcription starts at intron 10 of Kcnq1 and in the opposite direction to that of the host gene. Mice that express a truncated Kcnq1ot1 have an increased expression of Kcnq1. Because correct potassium channel activity is required for normal cardiac functioning, any alteration of the Kcnq1ot1-mediated control of Kcnq1 could be responsible for abnormal heart function [38].
The involvement of lncRNA in HF is also suggested by the role of ANRIL (antisense noncoding RNA in the INK4 locus) in coronary heart disease [39]. This lncRNA is an antisense noncoding RNA transcribed with the INK4b–ARF–INK4a gene cluster, which encodes oncosuppressor proteins p15INK4b, p14 ARF, and p16INK4a. GWAS revealed that many SNPs mapped to this genomic locus and, in particular, to the gene encoding ANRIL and were associated with increased susceptibility to coronary artery disease and several other diseases, including cancer. ANRIL regulates the expression of the INK4b–ARF–INK4a gene cluster through recruitment of both PRC1 and PRC2, promoting the formation of a repressive chromatin structure [40]. In the light of these findings, it was proposed that loss-of-function mutations in this lncRNA lead to altered expression of p15INK4b, p14 ARF, and p16INK4a, influencing thus the susceptibility to coronary heart disease and cancer.
lncRNA: a Possible Therapeutic Target for Heart Failure?
A therapeutic strategy for HF is to cure the sick heart cells by interfering with the gene expression program that underlies HF. To this end, novel therapeutic opportunities might arise from RNA-based strategies that consist in the use of synthetic RNAs capable of programming biological function. The potential of this strategy for HF is supported by the discovery of the role of miRNAs in triggering the gene expression program of HF and by the possibility of manipulating ncRNAs in vivo. Indeed, anti-miRNAs and antisense oligonucleotides are being employed to inhibit the expression of specific miRNAs in vitro and in vivo for investigative and clinical purposes [41, 42]. The use of the same strategies for lncRNA could open new therapeutic avenues for HF, overcoming problems linked to the use of miRNA-based molecules, such as low specificity and poor pharmacokinetics. Indeed, future studies on the role of lncRNA in HF and heart development will improve our understanding of the ncRNA network involved in regulating gene expression changes underlying HF and, thus, allow the development of specific therapeutic strategies based on the interference not only of miRNAs but also of lncRNA important for HF. These studies will greatly benefit from the combination of next-generation sequencing technologies applied to RNA (RNA-seq) with bioinformatics tools developed to identify lncRNAs that are differentially expressed in different biological conditions and for the prediction of their mechanism of action. Table 2 lists some bioinformatics tools used today to study lncRNAs.
Table 2.
A list of bioinformatics tools for lncRNA discovery and investigation
| Tool | Description | Link |
|---|---|---|
| MMSEQ | Haplotype- and isoform-specific expression estimation using multi-mapping RNA-seq reads | bgx.org.uk/software/mmseq.html [43] |
| Cufflinks/Cuffdiff/Cuffcompare | Cufflinks assembles transcripts, estimates their abundances, and tests for differential expression and regulation in RNA-Seq samples | cufflinks.cbcb.umd.edu/ [44] |
| RNAsnp | Predicting SNP effects on local RNA secondary structure | rth.dk/resources/rnasnp/download.html [45] |
| RIsearch | RIsearch is a program for fast RNA–RNA interaction search | rth.dk/resources/research/ [46] |
| RNAz | RNAz is a program for predicting structurally conserved and thermodynamically stable RNA secondary structures | tbi.univie.ac.at/∼wash/RNAz/ [47] |
| RNAclust | RNAclust is a perl script summarizing all the single steps required for clustering of structured RNA motifs | bioinf.uni-leipzig.de/∼kristin/Software/RNAclust/ |
| ORF Finder | The ORF Finder is a tool which finds all open reading frames of a selectable minimum size in a user's sequence | bioinformatics.org/sms2/orf_find.html [48] |
| Pfam | The Pfam database is a large collection of protein families, each represented by multiple sequence alignments and hidden Markov models | pfam.sanger.ac.uk/ [49] |
| Rfam | The Rfam database is a collection of RNA families, each represented by multiple sequence alignments, consensus secondary structures, and covariance models | rfam.sanger.ac.uk/ [50] |
Conclusion
Considering the growing interest in synthetic ncRNA for therapeutic application and the diagnostic potential of miRNAs as biomarkers, the identification and characterization of lncRNAs for HF could lead to realistic therapeutic opportunities. However, before these opportunities can become real, it is necessary that future studies define the function of lncRNAs in driving the gene expression changes underlying HF and their capacity to predict this pathology.
Acknowledgments
This work was possible thanks to an “Advanced” grant (CardioEpigen) from the European Research Council and grants from Fondation LeDucq and Fondazione CARIPLO (12-4-5157157-31) to GC and grant EPIGEN (progetto bandiera epigenomica) to RP.
Conflict of Interest
None.
Glossary
Abbreviations and acronyms
- HF
Heart failure
- ncRNA
Noncoding RNA
- piRNA
PIWI-interacting RNA
- miRNA
MicroRNA
- siRNA
Small interfering RNA
- eRNA
Enhancer RNA
- MALAT1
Metastasis-associated lung adenocarcinoma transcript 1
- NEAT2
Nuclear-enriched noncoding transcript 2
- PRC1
Polycomb repressive complex 1
- PRC2
Polycomb repressive complex 2
- Bvht
Braveheart
- MIAT
Myocardial infarction-associated transcript
Contributor Information
Roberto Papait, Email: roberto.papait@humanitasresearch.it.
Gianluigi Condorelli, Email: gianluigi.condorelli@humanitasresearch.it.
References
- 1.McMurray, J. J. (2010). Systolic heart failure. Clinical practice. The New England Journal of Medicine, 362(3), 228–238. [DOI] [PubMed]
- 2.Lompre AM, Schwartz K, D'albis A, Lacombe G, Van Thiem N, Swynghedauw B. Myosin isoenzyme redistribution in chronic heart overload. Nature. 1979;282:105–107. doi: 10.1038/282105a0. [DOI] [PubMed] [Google Scholar]
- 3.Izumo S, Nadal-Ginard B, Mahdavi V. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:339–343. doi: 10.1073/pnas.85.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter JJ, Chien KR. Signaling pathways for cardiac hypertrophy and failure. The New England Journal of Medicine. 1999;341:1276–1283. doi: 10.1056/NEJM199910213411706. [DOI] [PubMed] [Google Scholar]
- 5.Han P, Hang CT, Yang J, Chang CP. Chromatin remodeling in cardiovascular development and physiology. Circulation Research. 2011;108:378–396. doi: 10.1161/CIRCRESAHA.110.224287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nature Reviews Neuroscience. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitra SA, Mitra AP, Triche TJ. A central role for long non-coding RNA in cancer. Frontiers in Genetics. 2012;3:17. doi: 10.3389/fgene.2012.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saxena A, Carninci P. Long non-coding RNA modifies chromatin: epigenetic silencing by long non-coding RNAs. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology. 2011;33:830–839. doi: 10.1002/bies.201100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nature Reviews Cardiology. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 11.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wutz A, Gribnau J. X inactivation Xplained. Current Opinion in Genetics & Development. 2007;17:387–393. doi: 10.1016/j.gde.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/S0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 16.Reik W, Murrell A. Genomic imprinting. Silence across the border. Nature. 2000;405:408–409. doi: 10.1038/35013178. [DOI] [PubMed] [Google Scholar]
- 17.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. The Journal of Cell Biology. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Reports. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, et al. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Letters. 2011;585:671–676. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terranova R, Yokobayashi S, Stadler MB, Otte AP, Van Lohuizen M, Orkin SH, et al. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Developmental Cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 23.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Molecular Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 24.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 26.Mohammad F, Mondal T, Kanduri C. Epigenetics of imprinted long noncoding RNAs. Epigenetics: Official Journal of the DNA Methylation Society. 2009;4:277–286. doi: 10.4161/epi.4.5.9242. [DOI] [PubMed] [Google Scholar]
- 27.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins K. Physiological assembly and activity of human telomerase complexes. Mechanisms of Ageing and Development. 2008;129:91–98. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PloS One. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deuve JL, Avner P. The coupling of X-chromosome inactivation to pluripotency. Annual Review of Cell and Developmental Biology. 2011;27:611–629. doi: 10.1146/annurev-cellbio-092910-154020. [DOI] [PubMed] [Google Scholar]
- 33.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Developmental Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. Journal of Human Genetics. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 37.Lee JH, Gao C, Peng G, Greer C, Ren S, Wang Y, et al. Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circulation Research. 2011;109:1332–1341. doi: 10.1161/CIRCRESAHA.111.249433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korostowski L, Sedlak N, Engel N. The Kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart. PLoS Genetics. 2012;8:e1002956. doi: 10.1371/journal.pgen.1002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mcpherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Molecular Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nature Medicine. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 42.Van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circulation Research. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turro E, Su SY, Goncalves A, Coin LJ, Richardson S, Lewin A. Haplotype and isoform specific expression estimation using multi-mapping RNA-seq reads. Genome Biology. 2011;12:R13. doi: 10.1186/gb-2011-12-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabarinathan, R., Tafer, H., Seemann, S. E., Hofacker, I. L., Stadler, P. F., & Gorodkin, J. (2013). The RNAsnp web server: predicting SNP effects on local RNA secondary structure. Nucleic Acids Research. [DOI] [PMC free article] [PubMed]
- 46.Wenzel A, Akbasli E, Gorodkin J. RIsearch: fast RNA–RNA interaction search using a simplified nearest-neighbor energy model. Bioinformatics. 2012;28:2738–2746. doi: 10.1093/bioinformatics/bts519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gruber AR, Neubock R, Hofacker IL, Washietl S. The RNAz web server: prediction of thermodynamically stable and evolutionarily conserved RNA structures. Nucleic Acids Research. 2007;35:W335–W338. doi: 10.1093/nar/gkm222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28(1102):1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 49.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Research. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Research. 2013;41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]