Highlights
-
•
Analysis of brittle star regenerating arms using differentiation markers.
-
•
Identification of the early segregation of skeletal and muscle progenitor cells.
-
•
Expression of skeletal and non-skeletal genes at different stages of regeneration.
-
•
Combinatorial role of TF genes in early specification of skeletal cells.
-
•
Same TF genes identify different skeletal structures later in regeneration.
Keywords: Regeneration, Brittle star, Transcription factors, Skeleton, ets1/2, tbr, gataC, foxB, alx1
Abstract
The brittle star Amphiura filiformis, which regenerates its arms post autotomy, is emerging as a useful model for studying the molecular underpinnings of regeneration, aided by the recent availability of some molecular resources. During regeneration a blastema initially is formed distally to the amputation site, and then a rapid rebuild is obtained by adding metameric units, which will eventually differentiate and become fully functional. In this work we first characterize the developmental process of the regenerating arms using two differentiation markers for muscle and skeletal structures – Afi-trop-1 and Afi-αcoll. Both genes are not expressed in the blastema and newly added undifferentiated metameric units. Their expression at different regenerating stages shows an early segregation of muscle and skeletal cells during the regenerating process, long before the metameric units become functional. We then studied the expression of a set of genes orthologous of the sea urchin transcription factors involved in the development of skeletal and non-skeletal mesoderm: Afi-ets1/2, Afi-alx1, Afi-tbr, Afi-foxB and Afi-gataC. We found that Afi-ets1/2, Afi-alx1, Afi-foxB and Afi-gataC are all expressed at the blastemal stage. As regeneration progresses those genes are expressed in a similar small undifferentiated domain beneath the distal growth cap, while in more advanced metameric units they become restricted to different skeletal domains. Afi-foxB becomes expressed in non-skeletal structures. This suggests that they might play a combinatorial role only in the early cell specification process and that subsequently they function independently in the differentiation of different structures. Afi-tbr is not present in the adult arm tissue at any stage of regeneration. In situ hybridization results have been confirmed with a new strategy for quantitative PCR (QPCR), using a subdivision of the three stages of regeneration into proximal (differentiated) and distal (undifferentiated) arm segments.
Echinoderms have been well known to possess extensive regenerative capabilities, ranging from growing and re-patterning of lost tissues (Carnevali, 2006; Edmondson, 1935; Torrele, 1909) all the way to forming a new organism, resembling a mode of asexual reproduction as observed in sea stars (Rubilar et al., 2005). Most echinoderms can regenerate almost any adult tissue and it is these qualities that make them a compelling system for studying this biological process. Furthermore, the regenerative potential of echinoderms is not limited to adult tissues, having also been reported in larvae (Vickery et al., 2001). Echinoderms are deuterostomes (Adoutte et al., 2000; Cameron et al., 2000; Philippe et al., 2005; Turbeville et al., 1994; Winchell et al., 2002) and they share many genes with chordates as shown by the analysis of the sea urchin genome sequence (Sodergren et al., 2006). This phylogenetic position and close evolutionary relationship with vertebrates makes them an attractive system for studying regeneration with the possibility to understand why they regenerate completely whilst so few vertebrates possess the potential for such extensive regeneration. Most studies of echinoderm regeneration have been carried out on visceral regeneration in holothurians (Dolmatov and Ginanova, 2001; García-Arrarás and Greenberg, 2001), spine regeneration in sea urchins (Dubois and Ameye, 2001; Gorzelak et al., 2011) and arm regeneration in crinoids and ophiuroids (Carnevali and Bonasoro, 2001). The focus of these previous works was mainly on the description of morphogenetic and cellular processes associated with events of regeneration of different adult tissues. It seems that the most significant issue in preventing echinoderms in being widely recognized as models for regeneration studies so far is the lack of appropriate molecular tools and data.
One class of echinoderms in which the study of molecular mechanisms can be carried out are brittle stars (Class: Ophiuroidea) that can completely regenerate their arms post autotomy in just few weeks. However, not all species of brittle stars are equally easy to manipulate, maintain and use for experimental procedures. It has been shown that rate of regeneration depends on different environmental conditions, feeding habits and species of brittle star (Bowmer and Keegan, 1983; Nilsson and Sköld, 1996). Amphiura filiformis is a burrowing brittle star that regenerates its arms within only five weeks and can be easily maintained in a laboratory environment. Research into the regeneration mechanism of this particular species has so far focused largely on histogenesis and morphogenesis of the early phases of regeneration (Biressi et al., 2010) and its rate of growth and differentiation (Dupont and Thorndyke, 2006). Recently, the compilation of A. filiformis (Afi) cDNA clone microarray allowed scientists to study molecular mechanisms during the regeneration of the adult arm (Burns et al., 2012, 2011). These studies used microarray and QPCR to measure the differential expression of genes during the three pre-established stages of regeneration – blastema, 50% differentiation and 95% differentiation stage (Dupont and Thorndyke, 2006). Amongst all adult regenerating echinoderms spatial expression data has been so far obtained for three genes implicated in embryogenesis and cancer progression in sea cucumbers (Mashanov et al., 2012), the bmp2/4 gene in crinoids (Patruno et al., 2003), and two signalling molecules from the BMP family in brittle stars (Bannister et al., 2005, 2008).
To determine the suitability of this organism to study the molecular basis of regeneration, it is important to be able to draw comparisons to well-known and thoroughly studied systems. One such well-characterized molecular pathway is the sea urchin gene regulatory network (GRN) for the specification of the skeletogenic lineage in the developing embryo (Oliveri et al., 2008). Throughout the echinoderm phylum only echinoids and ophiuroids contain a larval skeleton (Strathmann, 1993). It has also been shown that many of the embryonic genes are also expressed during skeleton development in the juvenile, potentially as a result of an evolutionary adaptation where the embryonic skeleton was co-opted from the adult (Gao and Davidson, 2008). A few genes not present during the formation of the Strongylocentrotus purpuratus (Sp) adult skeleton (including transcription factors Sp-foxB, Sp-foxO and Sp-tbr) could be strictly related to the autonomously specified micromere lineage specific to sea urchin embryos (Gao and Davidson, 2008). During arm regeneration in brittle star new skeletal elements are formed, thus, the sea urchin skeletogenic GRN can provide a good source of transcription factors, signalling molecules and downstream genes with a potential role in skeletogenesis during the brittle star regeneration process.
In this study we describe the expression pattern of a cohort of gene orthologs of sea urchin skeletogenic genes at different stages of A. filiformis arm regeneration. We show that some of the key transcription factors involved in the specification of the sea urchin embryonic skeletogenic cell lineage are likely to be also co-expressed in early phases of regeneration in the brittle star adult. These genes then identify different skeletal structures during final differentiation of the arm. Finally, we show that Afi-t-brain (Afi-tbr) and Afi-foxB are not implicated in adult brittle star skeletogenesis similarly to sea urchin juvenile spine formation. Our results show that A. filiformis can be employed to study the molecular mechanism underlying the vastly interesting and complex biological process that is regeneration.
1. Results and discussion
1.1. Adult brittle star arms: morphology of fully developed and regenerating tissues
The fully developed arm of the brittle star is formed by metameric units, each containing muscles and endoskeleton components, elements of the nervous and water vascular systems all covered by epidermis (Brusca and Brusca, 1990). There are five calcerous structures of the endoskeleton and two muscular structures in each arm segment (Fig. 1a–c and S1a–d). These tissues are divided into externally projecting spines and a pair of orally located podia, the superficial skeletal arm shields (oral, aboral and lateral), the internally localized vertebrae and a set of two intervertebral muscles. The localization of muscle tissue has been confirmed by phalloidin staining in both regenerating and non-regenerating arms (Fig. 1d), which shows strong staining of the intervertebral muscles and podia. The order in which these structures are established during the regenerative process is difficult to assess, however it seems clear from microscopic observations that the externally protruding spines and podia formed from initial buds just below the distal growth zone. Formation of the podia is specifically linked to the elongation of the radial water canal establishing a connection to the remaining water vascular system, which is the main body cavity of the animal (Brusca and Brusca, 1990). The initial phases of the regenerative process, from wound healing to blastema formation, in the brittle star A. filiformis have been previously described in some detail and shown to contain features of both epimorphic and morphallactic processes as identified by microscopic observations of histological sections. In fact, both undifferentiated pluripotent cells and the dedifferentiated myocytes contribute to the formation of the highly proliferative blastema structure, which will generate a new arm (Biressi et al., 2010). So far, little attention has been brought to either the morphogenetic and developmental processes after blastema formation or the gross anatomy of the regenerating arm. Furthermore, the well-understood anatomy of skeletogenic structures in the non-regenerating arm is not sufficient to correctly interpret any potential spatial expression patterns in the regenerating structure. In order to understand the results of a molecular study of gene expression in the process of regeneration, we first observed the initial early stages of regeneration to define the discreet organization of the blastema as well as the development of muscle and skeletal structures. Fig. S2 shows the three commonly used regenerative phases in the brittle star (Dupont and Thorndyke, 2006), which presents its growth and acquiring functional identity along the proximal–distal axis in time. In standard laboratory conditions approximately one week post amputation the blastemal bud is visible distally to the autotomy site. When the 50% differentiation stage is reached, a number of metameric units are added at the proximal end and differentiate in a gradient-like manner, while the distal tip remains highly proliferative and undifferentiated. Therefore, at this stage the miniature arm is constituted by several metameric units each of them at a slightly different developmental stage along the proximal–distal axis. The 95% stage is defined by the appearance of pronounced muscle and skeletal structures along the full length of the new arm with the exception of a few distal-most segments. The final regenerated structure contains all of the constituents of the adult non-regenerating arm. Consistent with previous findings (Biressi et al., 2010), throughout the whole process the distal-most growth zone remains proliferative and undifferentiated until the completion of regeneration. These observations lead us to subdivide the 50% and 95% stage arms into proximal and distal regions containing several segments (as shown by the blue dashed lines in Fig. S2) allowing us to better distinguish between the specification and differentiation events.
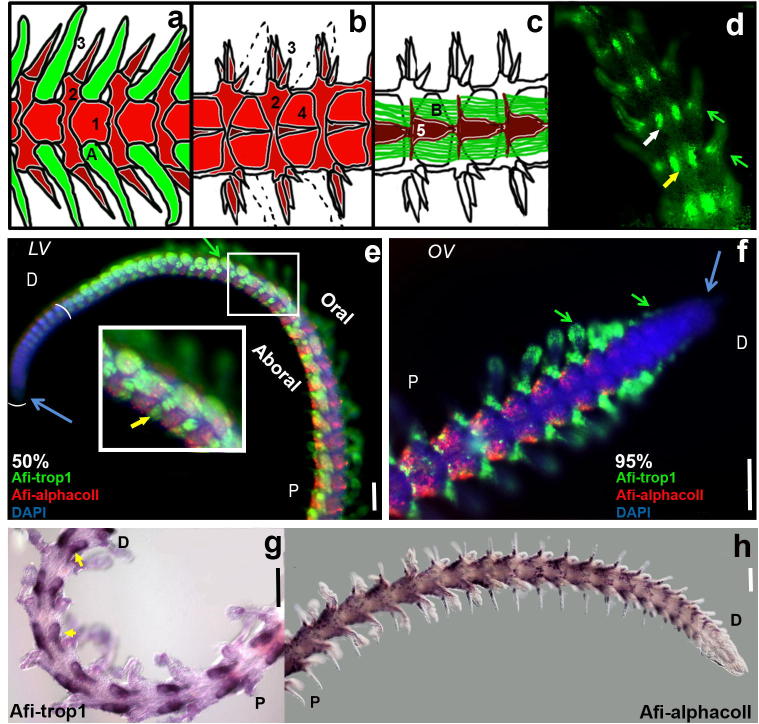
Fig. 1.
(a–d) Schematic diagram of muscle and skeletal structures within the brittle star arm. (a) Oral view showing the spines, podia, and lateral and oral arm shields. (b) Aboral view showing spines, and lateral and aboral arm shields. (c) View of internal structures including the vertebrae and the intervertebral muscles. (d) Phalloidin staining revealing muscle structures in the proximal part of the 95% regenerating arm (aboral view). (e, f) Double fluorescent in situ hybridization showing Afi-trop1 localized to the podia (green arrows) at the oral side and intervertebral muscles (yellow arrow) at the aboral side of the 50% (e) and 95% (f) regenerating arm. Afi-αcoll is restricted to lateral arm shields and the base of the spines. The two differentiation markers are not co-expressed and neither is found at the distal tip (blue arrow) of the regenerating arm which confirms their use as markers for specified structures. (g,h) Chromogenic single in situ hybridization clearly showing expression in 95% regenerating arms of Afi-trop1 in the intervertebral muscles (yellow arrows) as well as the podia and Afi-αcoll in the lateral shields and base of the spines. Scale bars – 100 μm. Green – muscle structures, red – skeletal structures, 1 – oral arm shield, 2 – lateral arm shield, 3 – spines, 4 – aboral arm shields, 5 – vertebrae, A – podia, B – intervertebral muscles. Green arrows – podia, yellow arrows – intervertebral muscles, blue arrows – distal tip. OV – oral view, LV – lateral view. P – proximal, D – distal.
1.2. Afi-tropomyosin-1 and Afi-alpha-collagen mark muscle and skeletal structures in the regenerating arm of A. filiformis
Determining the expression pattern of genes during development and regeneration processes has been instrumental for understanding the dynamics of the events and their genetic programme. So far, there has been limited spatial expression data available not only for this species of brittle star but for adult regeneration in echinoderms overall, while fluorescent in situ hybridization has not, to our knowledge, ever been employed for any adult echinoderms. The whole-mount in situ hybridization (WMISH) techniques used in this study have been compiled and optimized specifically for this species (see Methods).
In order to understand how muscle and skeletal structures arise during the regeneration process in brittle star we first studied the expression of two conserved differentiation markers: tropomyosin-1 (Afi-trop1) and alpha-collagen (Afi-αcoll). The two genes were selected from the Amphiura publicly available sequences (NCBI) on highest sequence similarity to genes with homologous roles in other animals. Both tropomyosin-1 and alpha-collagen are widely conserved genes primarily expressed in muscle and skeletal structures respectively (Lee and Im, 2012; Steinmetz et al., 2012; Tu et al., 2012). They were thus used in this study to initially confirm the applicability of the experimental procedures (Fig. 1e–h). Vertebrate TROPOMYOSIN-1 (TPM1) is an actin-binding protein found in striated muscle (Perry, 2001). We identified the homologue of vertebrate TPM1 in the brittle star by BLAST analysis considering the highest similarity score. In proximal segments of both 50% and 95% regenerating arms, Afi-trop1 shows expression in the muscle tissue only as also identified by phalloidin staining (compare Fig. 1e, f and g with Fig. 1d), namely the podia and intervertebral muscles. Notably, the intervertebral muscles are clearly visible in the fully differentiated segments of the 95% regenerating arm (Fig. 1g). In addition, the expression of Afi-trop1 in more distal segments is visible in primordial buds of podia. These metameric units are likely to be already specified, yet showing no fully differentiated structures (see first 6–7 segments expressing Afi-trop1 in Fig. 1e). The other marker is the sea urchin Sp-fcoIII/II/III/f gene, homologous to the vertebrate col2A1 (type II collagen alpha 1) gene that has been shown to be restricted to the skeletogenic cells at late developmental stages in the embryo (Livingston et al., 2006). We identified the homologue of the sea urchin Sp-fcoIII/II/III/f in A. filiformis (Afi-αcoll). Afi-αcoll expression is localized to spines and lateral arm shields of fully differentiated arm segment as shown by both single chromogenic (Fig. 1h and S3f) and double fluorescent WMISH (Fig. 1e, f). The chromogenic in situ (Fig. 1h, Fig. S3f) shows a pattern of single cells, which is consistent with the mesh-like structure of the echinoderm skeletal elements (i.e. stereomes and spines) in which the cells are embedded (Gorzelak et al., 2011). Double fluorescent in situ of Afi-αcoll and Afi-trop1 reveals no co-localization of these two genes throughout the whole length of the regenerating arm (Fig. 1e, f) at any analysed stage, suggesting an early segregation of skeletogenic and muscle precursor cells during the regeneration process. In the distal half of the 50% regenerating arm, DAPI staining additionally reveals the presence of cells that do not express either Afi-αcoll or Afi-trop1. This is consistent with the presence of other cell types and an early specification and segregation of all various cell types within the metameric unit. In accordance with the differentiation role of these genes, neither Afi-αcoll nor Afi-trop1 is expressed in the distal-most unspecified growth zone and the first few distal segments of the 50% regenerating arm (Fig. 1e, brackets). In the 95% regenerating arms only the tip does not express either gene, while Afi-trop1 is already visible in the first distinguishable primordial of podia of the distal segment (Fig. 1f). These data support the fact that these markers are appropriate for identifying muscle and skeletal structures during the regeneration process.
1.3. Dynamic expression of transcription factors in discreet domains
We then proceeded to study a set of transcription factor orthologs of genes involved in the specification and differentiation of skeletogenic cells in both larval and adult sea urchins (Oliveri et al., 2008; Gao and Davidson, 2008). This analysis will determine whether the role of any of these regulatory genes in skeletogenesis have been conserved not only in evolution but also between embryonic development and adult regeneration.
The genes used in these experiments are: Afi-alx1 (Ettensohn et al., 2003), Afi-foxB (Tu et al., 2006), Afi-tbr, Afi-ets1/2 (Koga et al., 2010; Oliveri et al., 2008) and Afi-gataC (Davidson et al., 2002; Solek et al., 2013). In the sea urchin GRN Sp-alx1, Sp-ets1/2 and Sp-tbr are early zygotic transcription factors that first define the skeletogenic micromere regulatory state downstream of the double-negative gate formed by Sp-hesC and Sp-pmar1 (Oliveri et al., 2002). Sp-foxB on the other hand, begins to be expressed at later stages when the skeletogenic mesenchyme ingresses and is directly upstream of the differentiation gene cassette (Minokawa et al., 2004). It has been shown that there is no expression of Sp-tbr or Sp-foxB in sea urchin juvenile skeletogenic centres (Gao and Davidson, 2008). Sp-gataC is primarily a non-skeletogenic mesodermal gene that shows some skeletogenic specificity in the aboral subpopulation of skeletogenic cells at late stages in sea urchin development (Materna and Davidson, 2012; Solek et al., 2013).
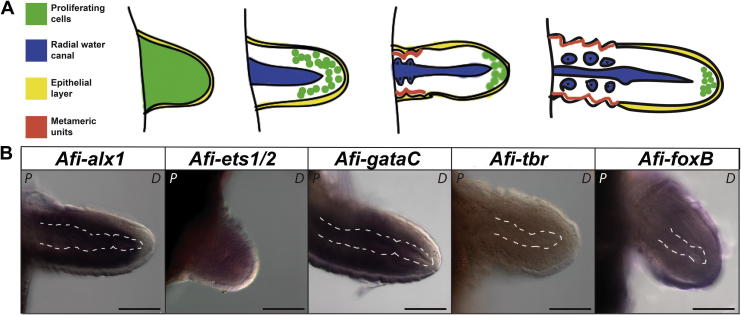
The expression of these genes has first been analysed at the early regenerative phase (Fig. 2, Fig. S3a–c). After wound healing, in brittle stars regeneration is marked by the formation of the blastema. In its earliest stage, the blastema consists of two layers – the epithelial layer and the inner layer composed of a heterogeneous population of proliferative cells (Fig. 2a; Biressi et al., 2010). According to previously published data (Biressi et al., 2010) and personal observations, as regeneration continues, the radial water canal becomes visible within the blastema and the zone of proliferation shifts to the distal end. When metameric units begin to appear distally to the amputation site, the undifferentiated growth zone is restricted to the distal tip of the blastema and remains there until the end of regeneration. To control for variability each gene expression pattern has been analysed in multiple arm samples per each stage and in at least two independent batches. Only the results that have been repeated are shown here. Interestingly, three of the four skeletogenic genes are expressed in the blastema, with the exception of Afi-tbr (Fig. 2). Afi-foxB is expressed in the outer epithelium of the blastema (Fig. 2, Fig. S3b). Afi-alx1 and Afi-gataC are both localized to the inner layers of the blastema and are absent from the outer epithelium (Fig 2, Fig. S3a, c). Afi-ets1/2 shows expression in the inner layer of the blastema (Fig. 2). Sampling variability should be accounted for, as collected blastemas often vary in length and amount of distinguishable metameric units (Fig. 2).
Fig. 2.
Structure of the blastema and expression of transcription factors. (A) Formation of metameric units and the specification of the blastema. At the very early regenerative phase the blastema is composed of two layers – an epithelium and the mass of proliferative and unspecified cells, of a total 50–100 cells. As the blastema elongates the proliferative area is becoming restricted to the distal-most tip of the newly forming arm and the blastema is organized into a three-layered structure, formed by the radial water canal, the midlayer and the epithelium. When metameric units defined by the appearance of pronounced podia and spines are being formed at the proximal end of the regenerate, the unspecified proliferative cells are only localized to the distal-most tip forming a growth zone that remains undifferentiated until the end of regeneration. (B) WMISH revealing differential expression patterns of transcription factors Afi-alx1, Afi-ets1/2, Afi-gataC, Afi-tbr and Afi-foxB in the blastema. All genes except for Afi-tbr are expressed in the blastema.Afi-foxB is localized to the epithelium, Afi-ets1/2 is ubiquitous and Afi-alx1 and Afi-gataC are both expressed within the thick midlayer of the blastema but not the distal-most tip. Scale bar: 100 μm, P – proximal, D – distal. White dashed lines – radial water canal.
Further whole-mount analysis of gene expression during later stages of regeneration (Fig. 3, Fig. S3d) reveals that Afi-tbr is in fact not expressed at any stage in any tissue of the regenerating arm, consistently with QPCR results (see below). On the contrary, Afi-alx1, Afi-ets1/2, Afi-foxB and Afi-gataC are all highly abundant in the distal arm segments directly in proximity to the undifferentiated growth zone in the intermediate thick layer of both 50% and 95% regenerating arms (Fig. 3 – Distal panels), with Afi-ets1/2 extending more broadly and distally compared to the other genes. This expression pattern implies that these genes are likely to be co-expressed in at least a subset of cells. This result is consistent with the hypothesis that they might have an early combined role in the initial specification of mesodermally derived cell populations and more specifically in the subpopulation that will give rise to the skeleton. On the contrary, in segmental units of the more advanced regeneration stage (e.g. proximal segments) Afi-alx1, Afi-ets1/2, Afi-foxB and Afi-gataC acquire differential expression patterns. Afi-alx1 in 50% proximal segments is restricted to four discrete patches that correspond to the emerging podia and spines; while in the 95% arms, Afi-alx1 is restricted to the base of the spines (effectively the lateral shield) and the whole length of the podia with higher staining at the base and tip (Fig. 3 – Proximal panels). In differentiating metameric units Afi-gataC has a strong expression in the lateral shields and becomes restricted to small groups of cells at the base of spines only at the 95% stage. Afi-ets1/2 contributes early in vertebra development of the 50% regenerating arms, where it has a strongly repetitive, patterned expression (Fig. 3, S3d). Later on, it becomes downregulated and retains a faint expression at the site of vertebrae of the 95% arms. Expression of Afi-foxB only around the bases of podia both in the 50% and 95% arms suggests that it does not play a role in regenerating any of the skeletal structures in the brittle star. The absence of expression of Afi-tbr in regenerative arms and the exclusion of Afi-foxB from skeletal structures is consistent with previous studies in other echinoderms where it has been shown that tbr and foxB are not implicated in building the juvenile skeleton (Gao and Davidson, 2008).
Fig. 3.
Spatial expression of transcription factors Afi-alx1, Afi-ets1/2, Afi-foxB, Afi-tbr and Afi-gataC in the regenerating arm. Afi-tbr is not expressed at all in the regenerating arm. All four remaining genes are strongly upregulated directly beneath the undifferentiated distal cap. Proximal expression of Afi-alx1 is localized to the bases of the spines and podia and Afi-gataC only to the base of the spines, both at the 50% and 95% differentiation stages. Afi-ets1/2 shows a repetitive pattern of expression restricted to the site of developing vertebrae in the proximal regions of the regenerating arm. Expression of Afi-foxB is localized to the non-skeletogenic structures (around the base of the podia). Scale bar: 100 μm. P – proximal, D – distal. Arrowheads – site of developing spines, arrows – site of developing podia, asterisks – intervertebral muscles.
We show here that in the proximal segments of adult brittle star regenerating arm, those transcription factors acquire specific expression domains at the sites of the various calcerous structures. This could implicate that after initial specification of mesodermal progenitors, when all the genes are expressed together; they become involved in the differentiation of distinct skeletal tissues.
1.4. Quantitative analysis of expression
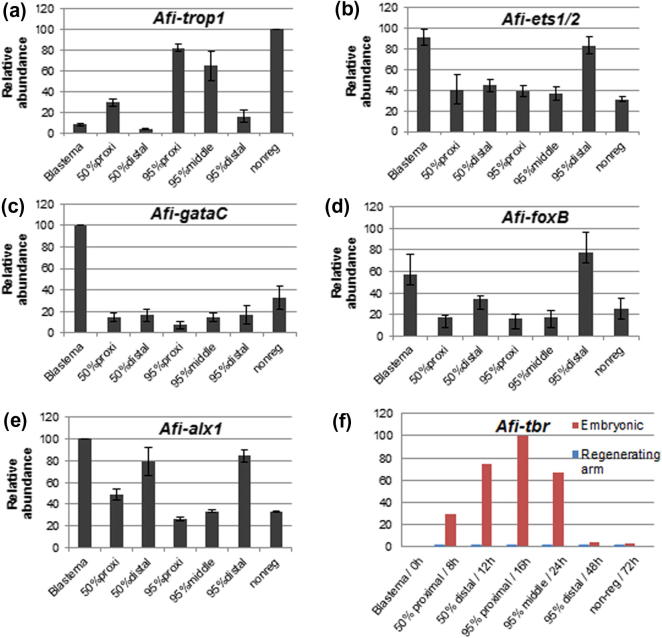
Quantification of mRNA transcripts has been previously carried out on complete blastemas, 50% and 95% regenerating arms of A. filiformis that have been pooled together (Burns et al., 2011). This study aimed to find genes associated with specific stages of regeneration, although those results could not be used to identify the possible roles of mRNAs in the specification or differentiation of distinct structures. We thus subdivided the arms at the 50% and 95% regeneration stage into more refined domains with the intention to separate the proximal differentiated segments from the distal undifferentiated or less differentiated segments (see Fig. S2). This partition was used on samples collected for QPCR and resulted in a more sensitive experimental procedure to identify dynamic changes in expression patterns throughout the arm regeneration process and to validate the observed expression pattern. It needs to be noted that the proximal and middle samples of the regenerates have a larger heterogeneity than the blastemas due to the intrinsic individual variability of the regeneration process. Fig. 4 shows the quantification plots for the downstream marker Afi-trop1, along with the upstream transcription factors Afi-alx1, Afi-ets1/2, Afi-gataC, Afi-foxB and Afi-tbr. The data are expressed in relative abundance of transcripts for at least two independent sample batches. Afi-trop1 temporal expression complements the observable spatial pattern showing upregulation (from 10% in the blastema to 80% in proximal segments of 95% arms and 100% in non-regenerating arms) in the tissues containing pronounced muscle structures (non-regenerating, proximal and middle sections of 95% arms) and correspondingly little or no expression in the blastema and distally developing segments of 50% and 95% regenerating arms (Fig. 4a) as expected from our in situ data (Fig. 1e–g). Afi-alx1, Afi-ets1/2, Afi-foxB and Afi-gataC transcripts on the other hand, have opposite dynamics of expression compared to Afi-trop1, supporting their important role in the early specification events (Fig. 4b–e). All three mRNAs are most abundant in the blastema where broad expression is observed (Fig. 2) and become gradually downregulated during the elongation and differentiation of the regenerating arm when their expression becomes localized in small number of cells (Fig. 3). Afi-alx1, Afi-foxB and Afi-ets1/2 are additionally highly expressed in the distal segments of 50% and 95% arms in agreement with the in situ results (compare Fig. 4b, d and e and Fig. 3). Our QPCR data shows that Afi-gataC is downregulated (5–10 folds reduction) after the blastema stage (Fig. 4c). This is partially consistent with the WMISH data and is likely caused by the expression being restricted to a population of cells at the base of the spines, which represent a very small percentage of the cells of the regenerative arms. Thus, the number of transcripts within the total RNA of the samples is highly diluted, even if single cells express the Afi-gataC gene at high levels detectable by in situs. Undeviating from WMISH studies, Afi-tbr expression is not detected by this method further confirming its complete lack of involvement in adult skeletogenesis. To confirm that Afi-tbr is truly not expressed and that this result is not an artefact of improper primer design or a technical problem, we conducted QPCR of Afi-tbr on cDNA from embryonic stages (8, 12, 16, 24, 48 and 72 h), which showed clear expression of this gene (Fig. 4f). Our results indicate that a finer subdivision of regenerates for QPCR can be successfully employed for assessing the dynamics of expression during the regeneration process and can also serve as a precise validation tool for in situ hybridization experiments. More importantly, we show that many changes in levels and localization of gene expression occur during the whole process of regeneration from the initial stage in which cells are proliferative and undifferentiated to full differentiation of arm structures.
Fig. 4.
Quantification of gene expression during brittle star arm regeneration. (a) The differentiation marker Afi-trop1 shows an upregulation in proximal, differentiated and non-regenerating tissue of the arm. (b,d,e) Afi-ets1/2, Afi-foxB and Afi-alx1 are also highly expressed in distal segments of the 50% and 95% differentiated arms. (c) Afi-gataC is downregulated after the blastema stage. (f) Afi-tbr is not expressed in the adult brittle star; a control using embryonic cDNA stages shows that the primers worked. All other transcription factors are most abundantly expressed in the blastema. Error bars represent standard deviation between two biological replicas (n = 15 animals per cDNA batch). At least four experimental replicas were used for each combination of cDNA and primers in each batch. h - hours post fertilisation.
2. Experimental procedures
2.1. A. filiformis animals and samples
Animals and regenerating arm samples were obtained from the Sven Lovén Centre for Marine Biology in Kristineberg, Sweden. Maintenance and collection was performed as previously described (Bannister et al., 2005). The animals were left to regenerate post-acclimatisation for approximately 1 week (for blastemas), 3 weeks (50% differentiation index) and 5 weeks (95% differentiation index) to obtain standardized stages (Dupont and Thorndyke, 2006), after which the arm samples were collected for RNA extraction or in situ hybridization according to the scheme in Fig. S2. Thirty individual arm samples (2arms/animal) were pooled together and stored in 1 ml RLT buffer (Qiagen) at −80 °C for later RNA extraction. Samples for in situ hybridization were fixed in 4% paraformaldehyde (PFA) in 1× phosphate-buffered saline (PBS) overnight at 4 °C, then washed three times with phosphate-buffered Tween-20 (PBT), and finally transferred to 100% methanol for long-term storage at −20 °C.
2.2. RNA extraction, cDNA synthesis and antisense RNA probe synthesizing
Total RNA was extracted from blastemas, 50% proximal and distal, 95% proximal, middle and distal segments (n = 30) and 10 whole non-regenerating arms using the RNeasy® micro kit (Qiagen) in accordance with manufacturer′s protocol. First strand cDNA was synthesised using the equivalent of 10 arms total RNA with the iScript™ cDNA synthesis kit (BIO-RAD), in 20 μl reaction according to manufacturer′s guidelines. Specific PCR fragments were amplified using the High fidelity PCR system (Roche) or by rapid amplification of cDNA ends (RACE) using KAPA HiFi DNA polymerase (Kapa Biosystems). The RACE library was constructed using the First Choice RLM-RACE kit (Invitrogen) from 5, 27 and 72 h cDNA stages. PCR products were then ligated respectively into pGEM®-T Easy (Promega) or pCR®-Blunt II-TOPO® vectors (Invitrogen). Identity and orientation of cloned fragments has been verified by sequencing. All primers were designed for A. filiformis sequences using Primer3 software (http://frodo.wi.mit.edu/), all sequences can be found in NCBI Genbank under accession numbers listed in Supplementary material Fig. s4.
Stable cDNA was transcribed in the presence of DIG or fluorescein-conjugated nucleotides to produce RNA antisense probes for fluorescent and chromogenic in situ hybridization (probe sizes ranged from 300 bp to 1.5k bp).
2.3. Whole mount in situ hybridization (WMISH)
The WMISH protocol was optimized from previously described protocols used in different species (Harkey et al., 1988; Tautz and Pfeifle, 1989; Zhu et al., 2001). Three to five regenerating arms were re-hydrated at room temperature (RT) with descending concentrations of ethanol/H2O (70%, 50% and 30%) and then washed 5 times in PBT. The samples were pre-hybridized for 1 h at 45 °C in hybridization buffer (HB) (50% Deionized formamide, 10% PEG, 0.6 M NaCl, 0.02 M Tris pH 7.5, 0.5 mg/ml yeast tRNA, 0.01% Tween-20, 0.05 M EDTA, 1× Denhardst, DEPC-treated H2O) then transferred to hybridization buffer containing 0.02 ng/μl of labelled probe and hybridized overnight at 45 °C. Excess of probe was washed 1× with 75% HB in PBT, 50% HB in PBT and 25% HB in PBT at 45 °C, 2× more in PBT and then three times in 0.1× SSC pH 7.0 (0.015 M NaCl, 0.0015 M sodium citrate, DEPC-treated H2O) at room temperature. The hybridized probe was detected with an appropriate antibody diluted 1:1000 for 1 h at RT. For chromogenic detection the anti-DIG-AP or anti-Fluorescein-AP FAB fragments (Roche) were used as previously described (Minokawa et al., 2004). The progression of staining was frequently checked under the stereomicroscope. For fluorescent staining the Tyramide Signal Amplification (TSA) system (Perkin Elmer; POD-TSA detection system) was used as described in (Croce and McClay, 2006). In both cases, following staining the regenerating arms were washed with 1× PBT containing 0.05 M EDTA and 4× in PBT and stored at 4 °C.
Phalloidin staining was conducted using fixed arms but before storage in methanol. Samples were washed 2× in PBT and 3× in PBS then incubated in 1:500 dilution of phalloidin-Atto 488 (Sigma) solution at 4 °C overnight. The next day the samples were washed 4× in PBS. All images were obtained using the AxioImager A1 fluorescent microscope (Zeiss).
2.4. Qpcr
Quantitative PCR was carried out on cDNA synthesized from regenerating arm samples in accordance to the method employed for sea urchin embryos (Rast et al., 2002). At least four experimental replicas were used for each combination of cDNA and primers and data were considered only if the replicas have a standard deviation less than 1.00. 18S rRNA was used as the internal standard and negative controls were included in each run. Power SYBR® Green QPCR master mix (Applied Biosystems) was employed along with primers (2.5 pmol/μl) designed as described before (Rast et al., 2002). The QPCR was run on a 384-well plate using the 7900HT Fast Real-Time PCR system (Applied Biosystems).
3. Conclusion
In this study we characterize the dynamics of the developmental processes during A. filiformis arm regeneration and the spatio-temporal expression pattern of several genes. The data described here shows that brittle stars can be employed for studying molecular regulation of the regenerative process.
Firstly, in situ hybridization analysis reveals that upstream transcription factors orthologous to sea urchin embryonic skeletogenesis genes (Afi-alx1 and Afi-ets1/2) are also expressed during the formation of skeletal structures (e.g. vertebrae, spines and shields) in the brittle star regenerating arm. The only exception is Afi-tbr, which is not expressed in A. filiformis arm regeneration. This is consistent with the idea that the co-option of tbr in the skeleton forming cells only is specific for sea urchin embryos and its ancestral regulatory role is not in specification or differentiation of the skeletogenic cells. In fact, Sp-tbr is generally expressed in endomesodermal cell types in other echinoderms including sand dollars (Minemura et al., 2009) and it is not expressed in juvenile skeletogenesis (Gao and Davidson, 2008), which represents the plesiomorphic character of the Echinodermata. Regulatory network analysis in S. purpuratus (Oliveri et al., 2008; Rafiq et al., 2012) shows that Sp-tbr gene only inputs in a few “skeletogenic” genes (e.g. p16rel1 and 2) in agreement with a relatively recent role during echinoid skeletogenesis. The remaining transcription factor genes (Afi-ets1/2, Afi-alx1, Afi-gataC and Afi-foxB) are highly upregulated in the blastema and the distal, differentiating segments of the 50% and 95% arms. Although Afi-foxB is present in adult regenerating tissue, it is not localized in skeletogenic tissue/cells during differentiation. This result complements previous studies, which show the lack of expression of this gene during the building of the adult skeleton (Gao and Davidson, 2008). The expression of Afi-alx1, Afi-ets1/2 and Afi-gataC beneath the growth cap suggests that these genes could have a role in early specification of mesodermal cell types and more specifically in skeletogenic lineage progenitors. On the contrary, in proximal segments characterized by a more advanced developmental stage and the appearance of muscle, skeletal and other more differentiated structures, the three genes acquire specific expression patterns and mark distinct calcerous tissue suggesting a non-combinatorial role of these genes. Afl-alx1 is specifically expressed in the base of spines and podia, while Afi-ets1/2 is highly transcribed in cells that coincide with the shape and position of the vertebras. Afi-gataC is primarily localized to the lateral shields and just the base of spines in the late regenerative phase. On the contrary, the combinatorial regulatory role of Afi-alx1 and Afi-ets1/2 provides inputs in most of the sea urchin embryonic “skeletogenic” differentiation genes (e.g. SM50, p19) as shown by GRN analysis (Oliveri et al. 2008; Rafiq et al. 2012); it is also present during sea urchin juvenile skeletogenesis; and it is evolutionary conserved in other echinoderm classes (Gao and Davidson, 2008).
Secondly, the optimized staging method to distinguish between proximal (differentiated) and distal (undifferentiated) segments used in QPCR experiments has provided informative data on the dynamics of expression along the whole length of the regenerating arm and complements spatial patterns. Further development of techniques for functional gene analysis will certainly enrich the molecular toolbox available for A. filiformis, although the results of this study already provide a good starting point for future research into deuterostome regeneration and increase our understanding of the evolution and mechanisms of this process.
Acknowledgments
We would like to thank Olga Ortega-Martinez, Sam Dupont and Michael Thorndyke along with the staff at the Sven Lovén Centre for Marine Sciences in Kristineberg, Sweden for support during this work. We also thank Michaela Egertová and Maurice Elphick for help with cryosections as well as Josefine Stångberg for making the Afi-αcoll in situ probe. We thank Anoop Kumar and the two anonymous reviewers for helpful commentary and discussion on the manuscript. The Amphiura filiformis cDNA library was done within the EU Network of Excellence, Marine Genomics project. Part of this study has been funded by the EU FP7 Research Infrastructure Initiative ASSEMBLE (Ref. 227799). A.C. was funded by a Wellcome Trust studentship. D.D. was funded by a Systems Biology UCL studentship.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.gep.2013.09.002.
Appendix A. Supplementary data
References
- Adoutte A., Balavoine G., Lartillot N., Lespinet O., Prud’homme B., Rosa R. The new animal phylogeny: reliability and implications. Proceedings of the National academy of Sciences of the United States of America. 2000;97:1–4. doi: 10.1073/pnas.97.9.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister R., McGonnell I., Graham A., Thorndyke M., Beesley P. Afuni, a novel transforming growth factor-beta gene is involved in arm regeneration by the brittle star Amphiura filiformis. Development Genes and Evolution. 2005;215:1–9. doi: 10.1007/s00427-005-0487-8. [DOI] [PubMed] [Google Scholar]
- Bannister R., McGonnell I., Graham A., Thorndyke M., Beesley P. Coelomic expression of a novel bone morphogenetic protein in regenerating arms of the brittle star Amphiura filiformis. Development Genes and Evolution. 2008;218:1–6. doi: 10.1007/s00427-007-0193-9. [DOI] [PubMed] [Google Scholar]
- Biressi A., Zou T., Dupont S., Dahlberg C., Benedetto C., Bonasoro F., Thorndyke M., Carnevali M. Wound healing and arm regeneration in Ophioderma longicaudum and Amphiura filiformis (Ophiuroidea, Echinodermata): comparative morphogenesis and histogenesis. Zoomorphology. 2010;129:1–19. [Google Scholar]
- Bowmer T., Keegan B. Field survey of the occurrence and significance of regeneration in Amphiura filiformis (Echinodermata: Ophiuroidea) from Galway Bay, west coast of Ireland. Marine Biology. 1983;74:1–7. [Google Scholar]
- Brusca R.C., Brusca G.J. Sinauer Associates; Sunderland, MA: 1990. Invertebrates. [Google Scholar]
- Burns G., Ortega-Martinez O., Dupont S., Thorndyke M., Peck L., Clark M. Intrinsic gene expression during regeneration in arm explants of Amphiura filiformis. Journal of Experimental Marine Biology and Ecology. 2012;413:1–7. [Google Scholar]
- Burns G., Ortega-Martinez O., Thorndyke M., Peck L., Dupont S., Clark M. Dynamic gene expression profiles during arm regeneration in the brittle star Amphiura filiformis. Journal of Experimental Marine Biology and Ecology. 2011;407:1–8. [Google Scholar]
- Cameron C., Garey J., Swalla B., Cameron C., Garey J., Swalla B., Cameron C., Garey J., Swalla B., Cameron C., Garey J., Swalla B. Special feature: evolution of the chordate body plan: new insights from phylogenetic analyses of deuterostome phyla. Proceedings of the National Academy of Sciences. 2000;97:1–6. doi: 10.1073/pnas.97.9.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevali M., Bonasoro F. Microscopic overview of crinoid regeneration. Microscopy Research and Technique. 2001;55:1–24. doi: 10.1002/jemt.1187. [DOI] [PubMed] [Google Scholar]
- Carnevali M.D. Regeneration in echinoderms: repair, regrowth, cloning. ISJ. 2006;55:403–426. [Google Scholar]
- Croce J., McClay D. The canonical Wnt pathway in embryonic axis polarity. Seminars in Cell & Developmental Biology. 2006;17:168–174. doi: 10.1016/j.semcdb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Davidson E., Rast J., Oliveri P., Ransick A., Calestani C., Yuh C.-H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., Otim O., Brown C., Livi C., Lee P., Revilla R., Schilstra M., Clarke P., Rust A., Pan Z., Arnone M., Rowen L., Cameron R., McClay D., Hood L., Bolouri H. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Developmental Biology. 2002;246:1–29. doi: 10.1006/dbio.2002.0635. [DOI] [PubMed] [Google Scholar]
- Dolmatov I., Ginanova T. Muscle regeneration in holothurians. Microscopy Research and Technique. 2001;55:1–12. doi: 10.1002/jemt.1190. [DOI] [PubMed] [Google Scholar]
- Dubois P., Ameye L. Regeneration of spines and pedicellariae in echinoderms: a review. Microscopy Research and Technique. 2001;55:1–11. doi: 10.1002/jemt.1188. [DOI] [PubMed] [Google Scholar]
- Dupont S., Thorndyke M. Growth or differentiation? Adaptive regeneration in the brittlestar Amphiura filiformis. Journal of Experimental Biology. 2006;209:1–9. doi: 10.1242/jeb.02445. [DOI] [PubMed] [Google Scholar]
- Edmondson C.H. Autonomy and regeneration of Hawaiian starfishes. Bishop Museum Occasional Papers. 1935;11:3–20. [Google Scholar]
- Ettensohn C.A., Illies M.R., Oliveri P., De Jong D.L. Alx1, a member of the Cart1/Alx3/Alx4 subfamily of Paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development. 2003;130:2917–2928. doi: 10.1242/dev.00511. [DOI] [PubMed] [Google Scholar]
- Gao F., Davidson E. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proceedings of the National academy of Sciences of the United States of America. 2008;105:1–6. doi: 10.1073/pnas.0801201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Arrarás J., Greenberg M. Visceral regeneration in holothurians. Microscopy Research and Technique. 2001;55:1–14. doi: 10.1002/jemt.1189. [DOI] [PubMed] [Google Scholar]
- Gorzelak P., Stolarski J., Dubois P., Kopp C., Meibom A. 26Mg labeling of the sea urchin regenerating spine: Insights into echinoderm biomineralization process. Journal of Structural Biology. 2011;176:1–8. doi: 10.1016/j.jsb.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Harkey M., Whiteley H., Whiteley A. Coordinate accumulation of five transcripts in the primary mesenchyme during skeletogenesis in the sea urchin embryo. Developmental Biology. 1988;125:381–395. doi: 10.1016/0012-1606(88)90219-9. [DOI] [PubMed] [Google Scholar]
- Koga H., Matsubara M., Fujitani H., Miyamoto N., Komatsu M., Kiyomoto M., Akasaka K., Wada H. Functional evolution of Ets in echinoderms with focus on the evolution of echinoderm larval skeletons. Development Genes and Evolution. 2010;220:107–115. doi: 10.1007/s00427-010-0333-5. [DOI] [PubMed] [Google Scholar]
- Lee J.-M., Im G.-I. PTHrP isoforms have differing effect on chondrogenic differentiation and hypertrophy of mesenchymal stem cells. Biochemical and Biophysical Research Communications. 2012;421:1–6. doi: 10.1016/j.bbrc.2012.04.096. [DOI] [PubMed] [Google Scholar]
- Livingston B., Killian C., Wilt F., Cameron A., Landrum M., Ermolaeva O., Sapojnikov V., Maglott D., Buchanan A., Ettensohn C. A genome-wide analysis of biomineralization-related proteins in the sea urchin Strongylocentrotus purpuratus. Developmental Biology. 2006;300:335–348. doi: 10.1016/j.ydbio.2006.07.047. [DOI] [PubMed] [Google Scholar]
- Mashanov V., Zueva O., Garcia-Arraras J. Expression of Wnt9, TCTP, and Bmp1/Tll in sea cucumber visceral regeneration. Gene expression patterns: GEP. 2012;12:1–12. doi: 10.1016/j.gep.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna S.C., Davidson E.H. A comprehensive analysis of Delta signaling in pre-gastrular sea urchin embryos. Developmental Biology. 2012;364:77–87. doi: 10.1016/j.ydbio.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minemura K., Yamaguchi M., Minokawa T. Evolutionary modification of T-brain (tbr) expression patterns in sand dollar. Gene Expression Patterns. 2009;9:468–474. doi: 10.1016/j.gep.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Minokawa T., Rast J., Arenas-Mena C., Franco C., Davidson E. Expression patterns of four different regulatory genes that function during sea urchin development. Gene expression patterns: GEP. 2004;4:449–456. doi: 10.1016/j.modgep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Nilsson H., Sköld M. Arm regeneration and spawning in the brittle star Amphiura filiformis (O.F. Müller) during hypoxia. Journal of Experimental Marine Biology and Ecology. 1996;199:193–206. [Google Scholar]
- Oliveri P., Carrick D.M., Davidson E.H. A regulatory gene network that directs micromere specification in the sea urchin embryo. Developmental Biology. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- Oliveri P., Tu Q., Davidson E. Global regulatory logic for specification of an embryonic cell lineage. Proceedings of the National academy of Sciences of the United States of America. 2008;105:1–8. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patruno, M., McGonnell, I., Graham, A., Beesley, P., Carnevali, M., Thorndyke, M., 2003. Anbmp2/4 is a new member of the transforming growth factor-beta superfamily isolated from a crinoid and involved in regeneration. Proceedings. Biological Sciences/The Royal Society 270, 1341-1347. [DOI] [PMC free article] [PubMed]
- Perry S. Vertebrate tropomyosin: distribution, properties and function. Journal of Muscle Research and Cell Motility. 2001;22:5–49. doi: 10.1023/a:1010303732441. [DOI] [PubMed] [Google Scholar]
- Philippe H., Lartillot N., Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Molecular Biology and Evolution. 2005;22:1246–1253. doi: 10.1093/molbev/msi111. [DOI] [PubMed] [Google Scholar]
- Rafiq K., Cheers M.S., Ettensohn C.A. The genomic regulatory control of skeletal morphogenesis in the sea urchin. Development. 2012;139:579–590. doi: 10.1242/dev.073049. [DOI] [PubMed] [Google Scholar]
- Rast J., Cameron R., Poustka A., Davidson E. Brachyury Target genes in the early sea urchin embryo isolated by differential macroarray screening. Developmental Biology. 2002;246:1–18. doi: 10.1006/dbio.2002.0654. [DOI] [PubMed] [Google Scholar]
- Rubilar T., Pastor C., Vivar E. Timing of fission in the starfish Allostichaster capensis (Echinodermata: Asteroidea) in laboratory. Revista de Biología Tropical. 2005;53(Suppl 3):299–303. [PubMed] [Google Scholar]
- Sodergren E. The genome of the sea urchin Strongylocentrotus purpuratus. Science (New York, NY) 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solek C.M., Oliveri P., Loza-Coll M., Schrankel C.S., Ho E.C.H., Wang G., Rast J.P. An ancient role for Gata-1/2/3 and Scl transcription factor homologs in specification of immunocytes Developmental Biology. 2013;382:280–292. doi: 10.1016/j.ydbio.2013.06.019. [DOI] [PubMed] [Google Scholar]
- Steinmetz P., Kraus J., Larroux C., Hammel J., Amon-Hassenzahl A., Houliston E., Wörheide G., Nickel M., Degnan B., Technau U. Independent evolution of striated muscles in cnidarians and bilaterians. Nature. 2012;487:231–234. doi: 10.1038/nature11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathmann R.R. Hypotheses on the origins of marine larvae. Annual Review of Ecology and Systematics. 1993;24:89–117. [Google Scholar]
- Tautz D., Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Torrele E. Regeneration in Holothuria. Zoologischer Anzeiger. 1909;35:15–22. [Google Scholar]
- Tu Q., Brown C., Davidson E., Oliveri P. Sea urchin Forkhead gene family: phylogeny and embryonic expression. Developmental biology. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Tu Q., Cameron R., Worley K., Gibbs R., Davidson E. Gene structure in the sea urchin Strongylocentrotus purpuratus based on transcriptome analysis. Genome Research. 2012;22:1–10. doi: 10.1101/gr.139170.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turbeville J., Schulz J., Raff R. Deuterostome phylogeny and the sister group of the chordates: evidence from molecules and morphology. Molecular Biology and Evolution. 1994;11:648–655. doi: 10.1093/oxfordjournals.molbev.a040143. [DOI] [PubMed] [Google Scholar]
- Vickery M., Vickery M., Amsler C., McClintock J. Regeneration in echinoderm larvae. Microscopy Research and Technique. 2001;55:464–473. doi: 10.1002/jemt.1191. [DOI] [PubMed] [Google Scholar]
- Winchell C., Sullivan J., Cameron C., Swalla B., Mallatt J. Evaluating hypotheses of deuterostome phylogeny and chordate evolution with new LSU and SSU ribosomal DNA data. Molecular Biology and Evolution. 2002;19:762–776. doi: 10.1093/oxfordjournals.molbev.a004134. [DOI] [PubMed] [Google Scholar]
- Zhu X., Mahairas G., Illies M., Cameron R., Davidson E., Ettensohn C. A large-scale analysis of mRNAs expressed by primary mesenchyme cells of the sea urchin embryo. Development (Cambridge, England) 2001;128:2615–2627. doi: 10.1242/dev.128.13.2615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.