Highlights
-
•
Spatial and temporal expression of nxph1 during zebrafish embryonic development.
-
•
High conservation of neurexophilins among vertebrates.
-
•
High homology of nxph1 between zebrafish and other vertebrates.
-
•
Expression of nxph1 in various clusters of post-mitotic neurons and in glia.
-
•
Zebrafish is a good model to understand the in vivo function of neurexophilin in vertebrates.
Keywords: Neurexophilins, Synapses, Neurexins, Epiphysis, Zebrafish, Interneurons, Neurons
Abstract
Neurexophilin 1 (Nxph1) is a specific endoligand of α-neurexins that is essential for trans-synaptic activation. Here, we report its dynamic expression during development in zebrafish. Our study revealed an early onset of expression of nxph1. RT-PCR on a series of embryonic stages showed that it is maternally deposited, although only readily detectable by whole mount in situ hybridization by 22 hpf. During embryogenesis and larval stages, the zygotic transcript is expressed dynamically in various clusters of post-mitotic neurons and in glia in the central nervous system.
Neurexophilin is a highly conserved secreted cysteine-rich glycoprotein in vertebrates. It was discovered as a 29-kDa protein that is co-purified with neurexin 1α on immobilized α-latrotoxin (Petrenko et al., 1993). The binding of the protein to neurexin is very tight, hence the name neurexophilin (Petrenko et al., 1996).
The protein neurexophilin comprises of four domains: a signal peptide, a non-conserved N-terminal region, a highly conserved central N-glycosylated domain, and an equally conserved C-terminal cysteine-rich domain. Expression and protein purification experiments revealed that neurexophilins are secreted glycoproteins that are proteolytically processed from the primary translation product, first by signal sequence cleavage and then by cleavage at the boundary between the N-terminal non-conserved and the central conserved domain. The proteolytic processing observed in neurexophilin, reminiscent of maturation processes in neuropeptides, together with their tight binding to neurexins indicates that they are endogenous ligands for α-neurexins (Missler and Sudhof, 1998). Neurexins (NRXNs) are neuronal cell surface proteins that are required for trans-synaptic activation of synaptic transmission but not synapse formation (Missler et al., 2003). They are polymorphic synaptic receptors encoded by at least three genes (NRXN1, NRXN2 and NRXN3) in mammals, each of which has two independent promoters directing transcription of long α-neurexins and short β-neurexins.
Molecular cloning revealed that there are at least four neurexophilins genes (nxph1, nxph2, nxph3 and nxph4) in mammals (Missler and Sudhof, 1998; Petrenko et al., 1996). Comparisons of neurexophilins show that they are closely related to each other in the C-terminus regions but diverge considerably in their N-terminal regions (Missler and Sudhof, 1998). Rodents have all four genes but only neurexophilins 1, 3, and 4 are expressed at detectable levels. Nxph2 is however expressed in bovine, strongly suggesting evolutionary changes in nxph gene expression. Nxph4 has a different linker region and doesn’t bind to α-neurexins, unlike other neurexophilins (Missler et al., 1998).
Although neurexophilin gene family has been studied in mammals, there is lack of data for these genes in lower vertebrates. Very little is known about the expression of neurexophilins in vivo. Zebrafish (Danio rerio) has a quite simple and well-characterized nervous system with broad developmental and physiological similarities to humans. Recent studies in zebrafish showed that nxph1 is expressed in discrete clusters in the habenula, pallium, and ventral thalamus at 72 hpf and is involved in Notch signaling (Hortopan and Baraban, 2011). We provide here a comprehensive description of nxph1 expression during zebrafish development.
1. Results and discussion
1.1. Phylogenetic study of vertebrate neurexophilins
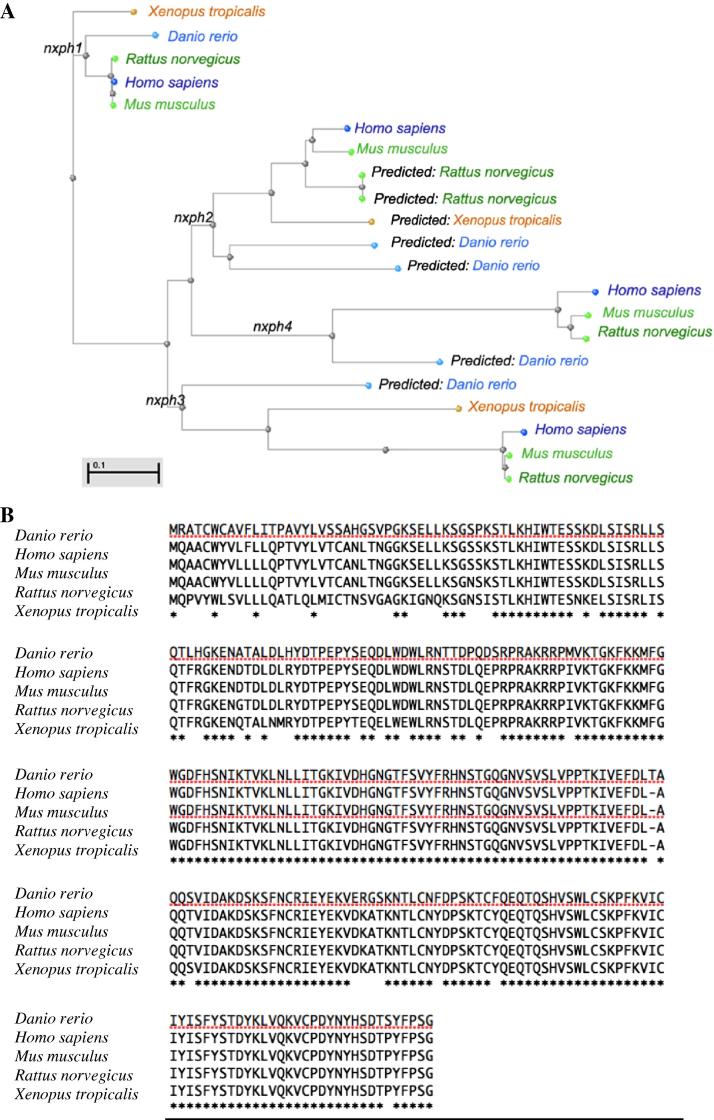
A phylogenetic tree was constructed (Fig. 1) based on the multiple sequence alignment of all neurexophilin orthologues of Homo sapiens, Rattus norvegicus, Mus musculus, Xenopus tropicalis and Danio rerio generated using COBALT alignment tool (Papadopoulos and Agarwala, 2007). The following orthologues were selected for analysis: human (NP_689958.1, NP_009157.1, NP_009156.2, NP_009155.1), mouse (NP_032777.3, NP_032778.1, NP_570928.1, NP_899120.2) rat (NP_037126.1, XP_001055001.1 XP_002726131.1, NP_067711.1, NP_067712.2), NP_570928.1, NP_899120.2), frog (NP_001120374.1, XP_002933980.1, NP_001096477.1) and zebrafish (NP_001002733.1, XP_698522.2, XP_001337769.1, XP_691473.2, XP_002667270.2). The phylogenetic analysis confirmed that these genes share a common ancestor.
Fig. 1.
Phylogenetic tree and amino acid alignment of neurexophilins. (A) Phylogenetic tree of the neurexophilin homologues of human (NP_689958.1, NP_009157.1, NP_009156.2, NP_009155.1), NP_067712.2), mouse (NP_032777.3, NP_032778.1, NP_570928.1, NP_899120.2), rat (NP_037126.1, XP_001055001.1, XP_002726131.1, NP_067711.1, frog (NP_001120374.1, XP_002933980.1, NP_001096477.1) and zebrafish (NP_001002733.1, XP_698522.2, XP_001337769.1, XP_691473.2, XP_002667270.2) was generated based on the full-length amino acid sequence available at NCBI. (B) Multiple sequence alignment of nxph1 in human, rat, mouse, frog and zebrafish. The nxph1 aminoacid sequences of Danio rerio, Homo sapiens, Mus musculus, Rattus norvegicus and Xenopus tropicalis were aligned based on Clustal W(2.1) algorithm. Conserved amino acids are indicated by an asterisk (identical in all cases), colon (conserved substitutions observed) or dot (semi-conserved substitutions observed).
The zebrafish genome contains one confirmed neurexophilin, nxph1 (ENSDARG00000070694) and four predicted paralogues, nxph2a (ENSDARG00000063224), nxph2b (ENSDARG00000079188), nxph3 (ENSDARG00000070694) and nxph4 (ENSDARG00000086716). Zebrafish nxph1 (NM_001002733.2; GI: 402692395) is located on chromosome 19 in the zebrafish genome ENSEMBL database. Comparison of z-nxph1 amino acid sequence with those of other vertebrate species of interest using Clustal W1.8 multiple sequence alignment (Fig. 1B) revealed a high conservation amongst vertebrates with a high amino acid homology of z-nxph1 with H. sapiens (87%), M. musculus (87%), R. norvegicus (86%) and X. tropicalis (80%) nxph1 proteins.
1.2. nxph1 transcript is maternally inherited in zebrafish
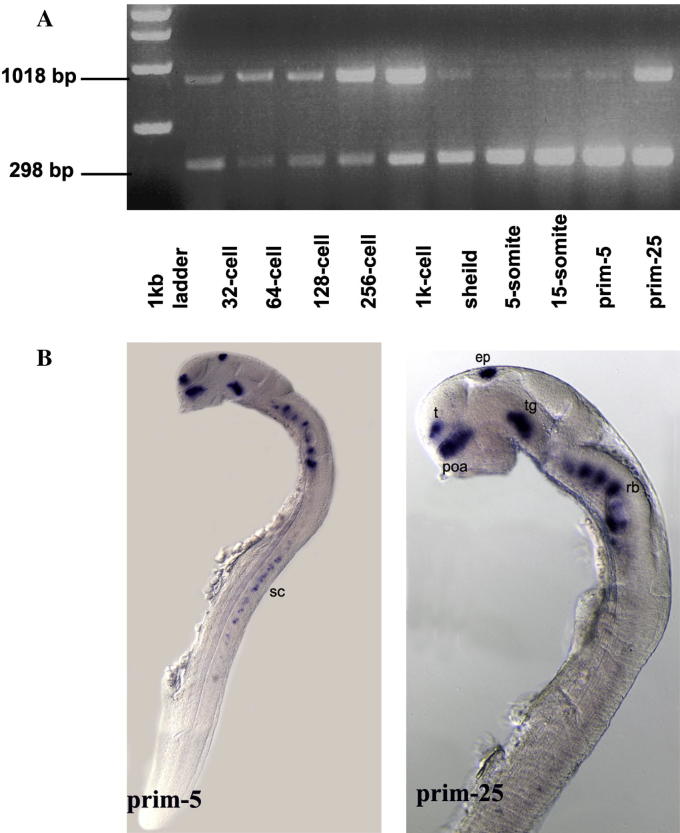
The presence of zebrafish nxph1 transcripts during embryonic development was shown by RT-PCR analysis of total RNA extracts at different embryonic stages: 32-cell (1.75 hpf), 64-cell (2 hpf), 128-cell (2.25 h), 256 cell (2.5 hpf), 1000-cell (3 hpf), shield (6 hpf), 5-somite (10.5 hpf), 15-somite (14 hpf), prim-5 (24 dpf), and prim-25 (36 dpf). Zebrafish nxph1 transcripts could be detected from as early as the 32-cell stage, indicating the presence of maternally deposited transcripts in the egg (Fig. 2A). Tissue-specific zygotic expression was detectable from 22 somite stage onwards in the pineal gland and soon after (prim-5) in post-mitotic neurons (data not shown and Fig. 2B).
Fig. 2.
Zebrafish nxph1 expression during embryonic development. (A) RT-PCR showing the presence of maternal nxph1 in zebrafish embryos. Lane 2–11: cDNA amplified from total RNA of 32-cell, 64-cell, 128-cell, 256 cell, 1000-cell, shield, 5-somite, 15-somite, prim-5 and prim-25 stage embryos, respectively. (B) Lateral view: anterior to the left of embryos at prim-5 and prim-25. nxph1 is expressed in discrete clusters in the forebrain, midbrain, hindbrain and spinal cord. Abbreviations: t, telencephalon; pOA, post-optic area; e, epiphysis; tg, tegmentum; rh, rhombomeres; sc, spinal cord. Scale bar = 100 μm.
1.3. Neuronal expression of nxph1 in zebrafish
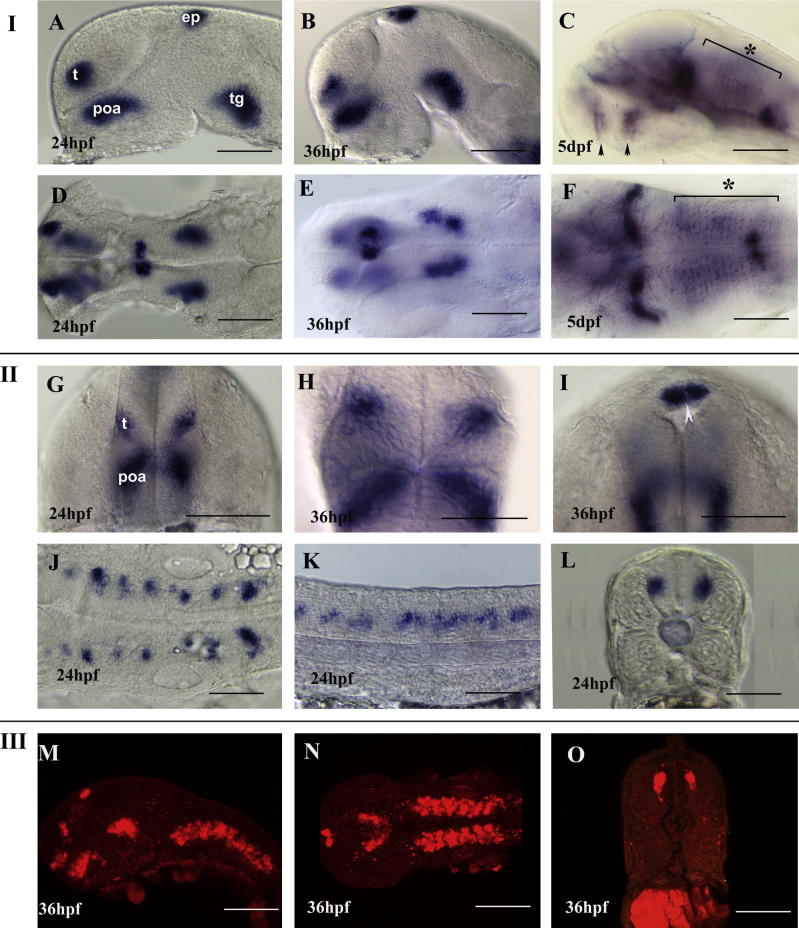
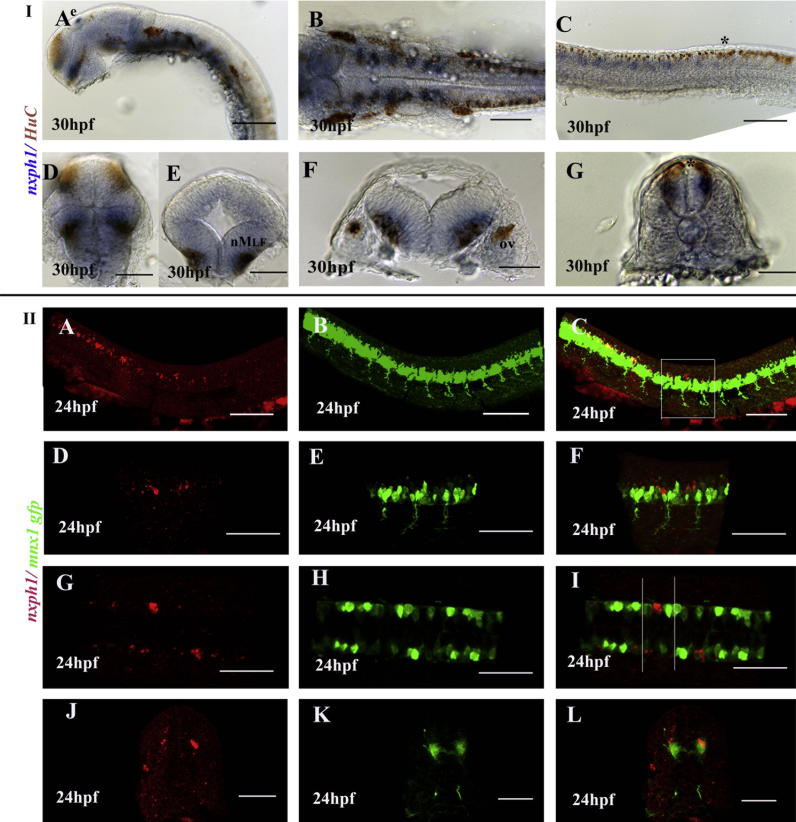
Embryos at Prim5 express nxph1 in discrete patch of cells in the ventral telencephalon (subpallium), pre-optic area, pineal gland/epiphysis, tegmentum, hindbrain rhombomeres and all along the spinal cord (Fig. 2B) The expression gets stronger and dynamic during the later stages of development except in the spinal cord where it gets down regulated from 48 hpf onwards. The pallial expression of nxph1 initially observed at 32 hpf by fast red staining gets stronger by 36 hpf (Fig. 3M, O; Fig. 5G, I), but not detected at the same stage by NBT/BCIP coloration (Fig. 3). In the 5 dpf larvae, the nxph1 expression around the anterior and posterior commissure (arrows in Fig. 3C) and in the hindbrain radial glia (asterisks in Fig. 3C, F) suggests a late expression in glia. The location of nxph1 in basal clusters inside the embryonic central nervous system (CNS) strongly suggested a neuronal expression, confirmed by co-localization with Hu-C (Fig. 4A–G).
Fig. 3.
Dynamic expression of nxph1 during zebrafish development. (I) Lateral (A–C) and dorsal (D–F) view of 24 hpf (A, D), 36 hpf (B, E) and 5 dpf (C, F) embryos showing nxph1 expression (blue). Black arrowhead indicates anterior and post-optic commissure and asterisk indicates hindbrain radial glia in C, F. Abbreviations: t, telencephalon; pOA, post-optic area; e; epiphysis; tg, tegmentum. (II) Transverse section showing nxph1 expression in telencephalon (t) and pre-optic area (poa) (G, H) at 24 hpf and 36 hpf. Transverse section through the diencephalon (I, white arrowhead indicates the pineal gland). Dorsal view of hindbrain (J), lateral view (K) and transverse section (L) of the spinal cord at 24 hpf. (III) Lateral (M) and dorsal (N) view of brain and transverse section of spinal cord (O) of nxph1 (red) at 36 hpf. Scale bar = 40 μm.
Fig. 5.
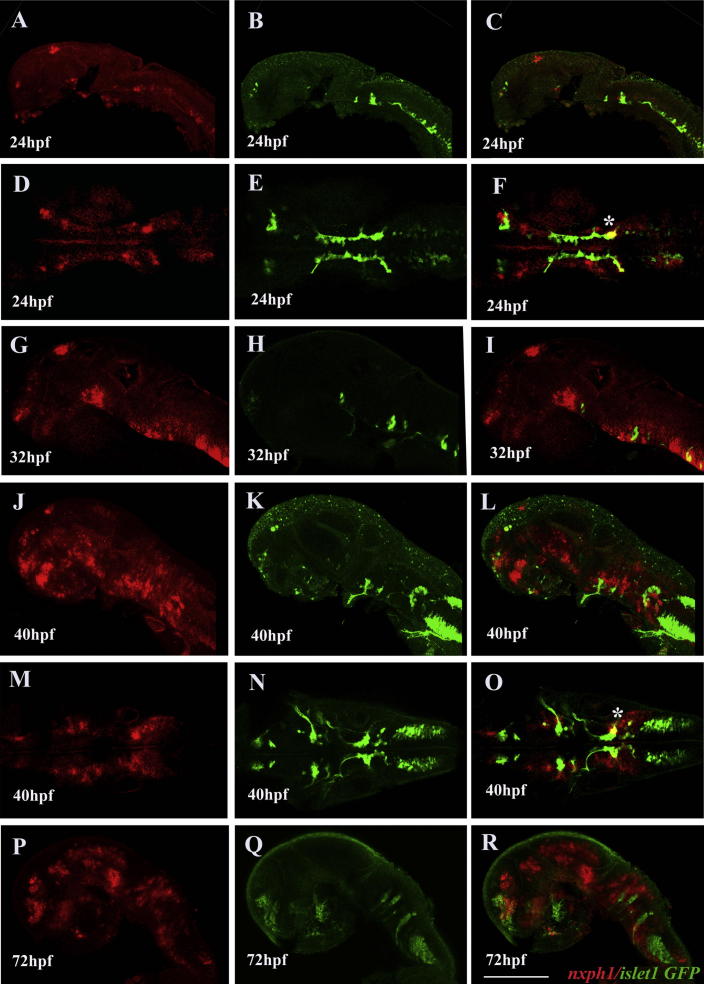
Motor neurons do not express nxph1 except for a small cluster of facial neurons. Lateral (A–C, G–L, P–R) and dorsal view (D–F, M–O) of zebrafish brain; anterior to the left. nxph1 expression in Tg(isl1: GFP) at 24 hpf (A–F), 32 hpf (G–I), 40 hpf (J–O) and 72 hpf (P–R). (∗) indicates co-expresssion in nVII facial motor neurons. Scale bar = 50 μm.
Fig. 4.
nxph1 is expressed in neurons, mostly interneurons. (I) Anti-HuC staining for post-mitotic neurons in 30 hpf embryos stained for nxph1 (A–G). Lateral view of zebrafish brain (A), frontal view of forebrain (D), transverse view of midbrain (E), dorsal (B) and transverse (F) view of hindbrain, lateral (C) and transverse (G) view of spinal cord at 36 hpf. Abbreviations: nMLF, nuclei of the media longitudinal fascicle); ov, otic vescicle; e, epiphysis. Scale bar: A–C (40 μm) and D–G (20 μm). (II) Lateral (A–F), dorsal view (G–I) and transverse section of spinal cord (J–L) of Tg(mnx1:GFP) zebrafish at 24 hpf, anterior to the left. Scale bar: A–C, J–L (40 μm) and D–I (20 μm). nxph1 is expressed ventral (blue) to the Rohon-Beard sensory neurons (∗) stained by HuC (I) and dorsal (red) to the motor neurons marked by Tg(mnx1:GFP)(II), thus indicating its expression in the interneurons.
1.4. nxph1 is mostly expressed in brain and spinal cord interneurons
Co-staining with Anti-HuC revealed that nxph1 is expressed ventral to the Rohon-Beard sensory neurons suggesting their expression in the interneurons in the spinal cord (Fig. 4C, G). At prim15 stage, all neurons labeled by the Hu-C express nxph1, but all regions of nxph1 expression overlap with neuronal area except in the pineal gland (Fig. 4A–G). Transgenic lines, Tg(isl1:GFP) and Tg(mnx1:GFP)ml2, expressing GFP in a subset or all motor neurons respectively, were used to test whether nxph1 is expressed in these neuronal populations. With the exception of a sub-domain of facial motor neuron (nVII), nxph1 populations do not overlap with GFP expressing motor neurons at prim5 (Fig. 5). The nxph1 labeling in the spinal cord did not co-localize, but appeared dorsal to the motor neurons marked by mnx1 at prim5, confirming its expression in spinal interneurons.
1.5. Conclusions and discussion
Expression patterns of different neurexophilin mRNAs showed tissue-specific variations between species (Missler and Sudhof, 1998). In humans, nxph1 expression is detected in spleen but not in brain unlike nxph2, nxph3 and nxph4 (Missler and Sudhof, 1998). In zebrafish, nxph1 is widely distributed in the central nervous system from late somite stage onwards, getting highly localized in distinct regions of the brain by 72 hpf and in more domains by 5 dpf (data not shown). Though nxph1 is maternally deposited in zebrafish, it can be detected by in situ hybridization by only 22 hpf (data not shown). In contrast to neurexins that are expressed in all neurons, we found that nxph1 is expressed at high levels only in scattered subpopulations of neurons that probably represent inhibitory interneurons. This finding support the localization in interneurons in adult rat brain (Petrenko et al., 1996), making the mouse a possible exception and indicating some level of conservation in the specificity of nxph1 expression in interneurons in vertebrates.
Previous studies demonstrated that in rats and mice, only Nxph1 and Nxph3 interact biochemically with α-neurexins (Missler et al., 1998). nxph3 is highly localized in mouse cortical and cerebellar regions and is functionally important for sensorimotor gating and motor coordination, but, the function of nxph1 in the CNS can’t be assessed in mouse where the gene is not expressed in the CNS (Beglopoulos et al., 2005). Due to the expression of most neurexophilins in the human brain, it is important to know the function of nxph gene family in a developing brain.
The function of NXPH is poorly understood; however, previous data suggest that they inhibit NRXN-mediated signalling (Beglopoulos et al., 2005; Zhang et al., 2005). Recent studies in animal models demonstrate the role of interneuron dysfunction in psychiatric disorders such as schizophrenia, autism spectrum disorder (ASD) and intellectual disabilities (ID) (Belforte et al., 2010; Chao et al., 2010; Fazzari et al., 2010). The haploinsufficiency of NRXN1α causes the ASD/ID and schizophrenia spectrum phenotypes in mice and involves synaptic dysfunction (Missler et al., 2003). It has become increasingly evident that the disruption of inhibitory circuits in these psychiatric disorders is linked to specific defects in the development and function of interneurons – the cells that are responsible for establishing inhibitory circuits in the brain. In zebrafish, nxph1 was found to be down regulated in the transcriptome analysis of mind bomb (mib) E3 ubiquitin ligase mutant that exhibits electrographic and behavioral seizure activity (Hortopan and Baraban, 2011). In humans, loss-of-function mutations in E3 ubiquitin ligase causes a neurogenetic disorder, Angelman syndrome (Kishino et al., 1997) which includes motor dysfunction leading to an ataxic gait, frequent severe seizures, profound learning disability, absent speech, and characteristic happy demeanor (Lossie et al., 2001). nxph1 being highly restricted to distinct neuronal clusters (mostly interneurons) in the zebrafish CNS, interfering with its function would give valuable insights about the role of interneurons and the function of NXPH genes in such neurogenetic or psychiatric disorders. Thus, zebrafish is a good model to study the in vivo function of neurexophilins and the current study provides the basis for future functional loss of function studies using TALEN-induced knockout technology to assess the role of neurexophilins in synapse development and dysfunction in vertebrates and explore its possible role in glia.
2. Experimental procedures
2.1. Zebrafish embryos maintenance
Zebrafish (Danio rerio) was maintained at 28 °C on a 14 h light/10 h dark cycle as described in the book (Brand et al., 2002). Embryos collected were cultured in fish water containing 0.003% 1-phenyl-2-thiouera to prevent pigmentation and 0.01% methylene blue to prevent fungal growth. Embryos were staged beginning from 4 hpf according to Kimmel (Kimmel et al., 1995) and fixed for further studies.
2.2. Cloning of zebrafish nxph1 gene
The IMAGE clone (IMAGE: 7155997) of nxph1 complete cDNA sequence was obtained (geneservice) in vector pDNR-LIB, which did not have any T7/sp6/T3 priming site that is necessary for in situ hybridisation. In order to reclone the insert into an appropriate vector, the insert was amplified by PCR using the oligonucleotides synthesized based on the information obtained on nxph1 gene sequence from the zebrafish genome database ENSEMBL available on the Sanger website (http://www.sanger.ac.uk/Projects/D_rerio/): nxph1 (GenBank Accession number: BC076424). The PCR product was recloned to PCR-4-TOPO vector using TOPO TA Cloning Kit for sequencing (Invitrogen) according to supplier’s protocol.
2.3. Whole mount in situ hybridisation and immunohistochemistry
Whole mount in situ hybridisation was performed on wild type embryos as previously described by (Macdonald et al., 1994) using Digoxigenin labeled antisense RNA probes synthesised from linearized plasmid nxph1 (Not1) and NBT/BCIP coloration (Roche). For neuronal cell type experiment, transgenic GFP embryos were labeled with digoxigenin, stained with Fast Red (Roche) and further processed for immunostaining. Antibody staining was performed (Shanmugalingam et al., 2000) using anti-GFP antibody (Torrey Pines Biolabs, USA) and secondary antibody Alexa 488 (Molecular Probes, Invitrogen, UK).
2.4. RT-PCR
Total RNA was extracted from WT embryos at various stages using TRIZOL® RNA isolation reagent (Invitrogen) and cDNA was synthesized (First strand CDNA synthesis kit, GE Healthcare) and amplified by PCR using the nxph1 specific primers, nxph1F (5′-TCTCATCACTCCGGCGGTCTAC-3′) and nxph1R (5′-ACGCATGGTATGGACAGTACAGG-3′). As an internal control for RNA quality and quantity, the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript expression was amplified using the primer pair GAPDH F (5′-TCAATGGATTTGGCCGTATT-3′) and GAPDH R (5′-GAGCTGAGGCCTTCTCAATG-3′) in the same reaction. PCR amplification of cDNA included 28 cycles with an initial denaturation at 94 °C (5 min) and final extension at 72 °C (10 min). Each reaction cycle consisted of incubations at 94 °C (60 s), 58 °C (60 s), and 72 °C (60 s) with Taq DNA Polymerase (GoTaq, Promega). The products were resolved on a 1.5% agarose gel with 1 μg/mL of ethidium bromide and analyzed in a gel-doc system.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Beglopoulos V., Montag-Sallaz M., Rohlmann A., Piechotta K., Ahmad M., Montag D., Missler M. Neurexophilin 3 is highly localized in cortical and cerebellar regions and is functionally important for sensorimotor gating and motor coordination. Molecular and Cellular Biology. 2005;25:7278–7288. doi: 10.1128/MCB.25.16.7278-7288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belforte J.E., Zsiros V., Sklar E.R., Jiang Z., Yu G., Li Y., Quinlan E.M., Nakazawa K. Postnatal NMDA receptor ablation in corticolimbic interneurons confers schizophrenia-like phenotypes. Nature Neuroscience. 2010;13:76–83. doi: 10.1038/nn.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, M., Granato, M., Nüsslein-Volhard, C., 2002. Keeping and raising zebrafish. Nüsslein-Volhard, C., Dahm, R., (Eds.), Oxford University Press, Oxford, pp. 7–37.
- Chao H.T., Chen H., Samaco R.C., Xue M., Chahrour M., Yoo J., Neul J.L., Gong S., Lu H.C., Heintz N., Ekker M., Rubenstein J.L., Noebels J.L., Rosenmund C., Zoghbi H.Y. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzari P., Paternain A.V., Valiente M., Pla R., Lujan R., Lloyd K., Lerma J., Marin O., Rico B. Control of cortical GABA circuitry development by Nrg1 and ErbB4 signalling. Nature. 2010;464:1376–1380. doi: 10.1038/nature08928. [DOI] [PubMed] [Google Scholar]
- Hortopan G.A., Baraban S.C. Aberrant expression of genes necessary for neuronal development and notch signaling in an epileptic mind bomb zebrafish. Developmental Dynamics. 2011;240:1964–1976. doi: 10.1002/dvdy.22680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Developmental Dynamics. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kishino T., Lalande M., Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nature Genetics. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Lossie A.C., Whitney M.M., Amidon D., Dong H.J., Chen P., Theriaque D., Hutson A., Nicholls R.D., Zori R.T., Williams C.A., Driscoll D.J. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. Journal of Medical Genetics. 2001;38:834–845. doi: 10.1136/jmg.38.12.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald R., Xu Q., Barth K.A., Mikkola I., Holder N., Fjose A., Krauss S., Wilson S.W. Regulatory gene expression boundaries demarcate sites of neuronal differentiation in the embryonic zebrafish forebrain. Neuron. 1994;13:1039–1053. doi: 10.1016/0896-6273(94)90044-2. [DOI] [PubMed] [Google Scholar]
- Missler M., Hammer R.E., Sudhof T.C. Neurexophilin binding to alpha-neurexins. A single LNS domain functions as an independently folding ligand-binding unit. The Journal of Biological Chemistry. 1998;273:34716–34723. doi: 10.1074/jbc.273.52.34716. [DOI] [PubMed] [Google Scholar]
- Missler M., Sudhof T.C. Neurexophilins form a conserved family of neuropeptide-like glycoproteins. The Journal of Neuroscience. 1998;18:3630–3638. doi: 10.1523/JNEUROSCI.18-10-03630.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M., Zhang W., Rohlmann A., Kattenstroth G., Hammer R.E., Gottmann K., Sudhof T.C. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Papadopoulos J.S., Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- Petrenko A.G., Lazaryeva V.D., Geppert M., Tarasyuk T.A., Moomaw C., Khokhlatchev A.V., Ushkaryov Y.A., Slaughter C., Nasimov I.V., Sudhof T.C. Polypeptide composition of the alpha-latrotoxin receptor. High affinity binding protein consists of a family of related high molecular weight polypeptides complexed to a low molecular weight protein. The Journal of Biological Chemistry. 1993;268:1860–1867. [PubMed] [Google Scholar]
- Petrenko A.G., Ullrich B., Missler M., Krasnoperov V., Rosahl T.W., Sudhof T.C. Structure and evolution of neurexophilin. The Journal of Neuroscience. 1996;16:4360–4369. doi: 10.1523/JNEUROSCI.16-14-04360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugalingam S., Houart C., Picker A., Reifers F., Macdonald R., Barth A., Griffin K., Brand M., Wilson S.W. Ace/Fgf8 is required for forebrain commissure formation and patterning of the telencephalon. Development (Cambridge, England) 2000;127:2549–2561. doi: 10.1242/dev.127.12.2549. [DOI] [PubMed] [Google Scholar]
- Zhang W., Rohlmann A., Sargsyan V., Aramuni G., Hammer R.E., Sudhof T.C., Missler M. Extracellular domains of alpha-neurexins participate in regulating synaptic transmission by selectively affecting N- and P/Q-type Ca2+ channels. The Journal of Neuroscience. 2005;25:4330–4342. doi: 10.1523/JNEUROSCI.0497-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]