Abstract
Low-density lipoprotein (LDL) cholesterol, high-density lipoprotein (HDL) cholesterol, triglycerides, and total cholesterol are heritable, modifiable, risk factors for coronary artery disease. To identify new loci and refine known loci influencing these lipids, we examined 188,578 individuals using genome-wide and custom genotyping arrays. We identify and annotate 157 loci associated with lipid levels at P < 5×10−8, including 62 loci not previously associated with lipid levels in humans. Using dense genotyping in individuals of European, East Asian, South Asian, and African ancestry, we narrow association signals in 12 loci. We find that loci associated with blood lipids are often associated with cardiovascular and metabolic traits including coronary artery disease, type 2 diabetes, blood pressure, waist-hip ratio, and body mass index. Our results illustrate the value of genetic data from individuals of diverse ancestries and provide insights into biological mechanisms regulating blood lipids to guide future genetic, biological, and therapeutic research.
Introduction
Blood lipids are heritable, modifiable, risk factors for coronary artery disease (CAD)1,2, a leading cause of death3. Human genetic studies of lipid levels can identify targets for new therapies for cholesterol management and prevention of heart disease, and can complement animal studies4,5. Studies of naturally occurring genetic variation can proceed through large-scale association analyses focused on unrelated individuals or through investigation of Mendelian forms of dyslipidemia in families6. We previously identified 95 loci associated with blood lipids, accounting for ~10-12% of the total trait variance4 and showed that variants with small effects can point to pathways and therapeutic targets that enable clinically-important changes in blood lipids4,7.
Here, we report on studies of naturally occurring variation in 188,578 European-ancestry individuals and 7,898 non-European ancestry individuals. Our analyses identify 157 loci associated with lipid levels at P < 5×10−8, including 62 new loci. Thirty of the 62 loci do not include genes implicated in lipid biology by previous literature. We tested lipid-associated SNPs for association with mRNA expression levels, carried out pathway analyses to uncover relationships between loci, and compared the locations of lipid-associated SNPs with those of genes and other functional elements in the genome. These results provide direction for biological and therapeutic research into risk factors for CAD.
Results
Novel loci associated with blood lipid levels
We examined subjects of European ancestry, including 94,595 individuals from 23 studies genotyped with GWAS arrays4 and 93,982 individuals from 37 studies genotyped with the Metabochip array8 (Supplementary Table 1 and Supplementary Fig. 1). The Metabochip includes variants representing promising loci from our previous GWAS (14,886 SNPs) and from GWAS of other CAD risk factors and related traits (50,459 SNPs), variants from the 1000 Genomes Project9 and focused resequencing10 efforts in 64 previously associated loci (28,923 SNPs), and fine-mapping variants in 181 loci associated with other traits (93,308 SNPs). In cases where Metabochip and GWAS array data were available for the same individuals, we used Metabochip data to ensure key variants were directly genotyped, rather than imputed.
We excluded individuals known to be on lipid lowering medications and evaluated the additive effects of each SNP on blood lipid levels after adjusting for age and sex. Genomic control values11 for the initial meta-analyses were 1.10 – 1.15, low for a sample of this size, indicating that population stratification should have only a minor impact on our results (Supplementary Fig. 2). After genomic control correction, 157 loci associated with blood lipid levels were identified (P < 5×10−8), including 62 new loci (Tables 1A-D, Figure 1, Supplementary Tables 2 and 3). Loci were >1 Mb apart and nearly independent (r2 < 0.10). Of the 62 novel loci, 24 demonstrated the strongest evidence of association with HDL cholesterol, 15 with LDL cholesterol, 8 with triglyceride levels, and 15 with total cholesterol (Supplementary Fig. 3). Several of these loci were validated by a similar extension based on GLGC GWAS results 12.
TABLE 1A. Novel Loci Primarily Associated with HDL Cholesterol Obtained from Joint GWAS and Metabochip Meta-analysis.
| Locus | MarkerName | Chr | hg19 Position (Mb) |
Associated trait(s) | MAF | Minor/major Allele |
Effect of A1 | Joint N (in 1000s) |
Joint P-value |
|---|---|---|---|---|---|---|---|---|---|
| PIGV-NR0B2 | rs12748152 | 1 | 27.14 | HDL, LDL, TG | .09 | T/C | −.051/.050/.037 | 187/173/178 | 1×10−15/3×10−12/1×10−9 |
| HDGF-PMVK | rs12145743 | 1 | 156.70 | HDL | .34 | G/T | .020 | 181 | 2×10−8 |
| ANGPTL1 | rs4650994 | 1 | 178.52 | HDL | .49 | G/A | .021 | 187 | 7×10−9 |
| CPS1 | rs1047891 | 2 | 211.54 | HDL | .33 | A/C | −.027 | 182 | 9×10−10 |
| ATG7 | rs2606736 | 3 | 11.40 | HDL | .39 | C/T | .025 | 129 | 5×10−8 |
| SETD2 | rs2290547 | 3 | 47.06 | HDL | .20 | A/G | −.030 | 187 | 4×10−9 |
| RBM5 | rs2013208 | 3 | 50.13 | HDL | .50 | T/C | .025 | 170 | 9×10−12 |
| STAB1 | rs13326165 | 3 | 52.53 | HDL | .21 | A/G | .029 | 187 | 9×10−11 |
| GSK3B | rs6805251 | 3 | 119.56 | HDL | .39 | T/C | .020 | 186 | 1×10−8 |
| C4orf52 | rs10019888 | 4 | 26.06 | HDL | .18 | G/A | −.027 | 187 | 5×10−8 |
| FAM13A | rs3822072 | 4 | 89.74 | HDL | .46 | A/G | −.025 | 187 | 4×10−12 |
| ADH5 | rs2602836 | 4 | 100.01 | HDL | .44 | A/G | .019 | 187 | 5×10−8 |
| RSPO3 | rs1936800 | 6 | 127.44 | HDL, TGa | .49 | C/T | .020/−.020 | 187/168 | 3×10−10/3×10−8 |
| DAGLB | rs702485 | 7 | 6.45 | HDL | .45 | G/A | .024 | 187 | 7×10−12 |
| SNX13 | rs4142995 | 7 | 17.92 | HDL | .38 | T/G | −.026 | 165 | 9×10−12 |
| IKZF1 | rs4917014 | 7 | 50.31 | HDL | .32 | G/T | .022 | 187 | 1×10−8 |
| TMEM176A | rs17173637 | 7 | 150.53 | HDL | .12 | C/T | −.036 | 184 | 2×10−8 |
| MARCH8-ALOX5 | rs970548 | 10 | 46.01 | HDL, TC | .26 | C/A | .026/−.026 | 187/187 | 2×10−10/8×10−9 |
| OR4C46 | rs11246602 | 11 | 51.51 | HDL | .15 | C/T | .034 | 176 | 2×10−10 |
| KAT5 | rs12801636 | 11 | 65.39 | HDL | .23 | A/G | .024 | 187 | 3×10−8 |
| MOGAT2-DGAT2 | rs499974 | 11 | 75.46 | HDL | .19 | A/C | −.026 | 187 | 1×10−8 |
| ZBTB42-AKT1 | rs4983559 | 14 | 105.28 | HDL | .40 | G/A | .020 | 184 | 1×10−8 |
| FTO | rs1121980 | 16 | 53.81 | HDL, TG | .43 | A/G | −.020/−.021 | 186/155 | 7×10−9/3×10−8 |
| HAS1 | rs17695224 | 19 | 52.32 | HDL | .26 | A/G | −.029 | 185 | 2×10−13 |
Chr, chromosome;MAF, minor allele frequency; A1, minor allele; A2, major allele.Effect sizes are given with respect to the minor allele (A1) in SD units. For loci associated with two or more traits at genome-wide significance, the trait corresponding to the strongest P-value is listed first. At one locus, the secondary trait was most strongly associated with a different SNP:
rs719726 (within 1Mb of rs1936800, r2 = 0.74).
TABLE 1D. Novel Loci Primarily Associated with Triglycerides Obtained from Joint GWAS and Metabochip Meta-analysis.
| Locus | MarkerName | Chr | hg19 Position (Mb) |
Associated trait(s) |
MAF | Minor/major Allele |
Effect of A1 | Joint N (in 1000s) |
Joint P-value |
|---|---|---|---|---|---|---|---|---|---|
| LRPAP1 | rs6831256 | 4 | 3.47 | TG, TCf,LDLf | .42 | G/A | 0.026/−0.022/− | 177/173/187 | 2×10−12/1×10−10/2×10−8 |
| 0.025 | |||||||||
| VEGFA | rs998584 | 6 | 43.76 | TG, HDL | .49 | A/C | 0.029/−0.026 | 175/184 | 3×10−15/2×10−11 |
| MET | rs38855 | 7 | 116.36 | TG | .47 | G/A | −0.019 | 178 | 2×10−8 |
| AKR1C4 | rs1832007 | 10 | 5.25 | TG | .18 | G/A | −0.033 | 178 | 2×10−12 |
| PDXDC1 | rs3198697 | 16 | 15.13 | TG | .43 | T/C | −0.020 | 176 | 2×10−8 |
| MPP3 | rs8077889 | 17 | 41.88 | TG | .22 | C/A | 0.025 | 176 | 1×10−8 |
| INSR | rs7248104 | 19 | 7.22 | TG | .42 | A/G | −0.022 | 176 | 5×10−10 |
| PEPD | rs731839 | 19 | 33.90 | TG, HDL | .35 | G/A | 0.022/−0.022 | 176/185 | 3×10−9/3×10−9 |
Chr, chromosome;MAF, minor allele frequency; A1, minor allele; A2, major allele.Effect sizes are given with respect to the minor allele (A1) in SD units. For loci associated with two or more traits at genome-wide significance, the trait corresponding to the strongest P-value is listed first. At one locus, secondary traits were most strongly associated with a different SNP:
rs6818397 (within 1 Mb of rs6831256, r2 = 0.18).
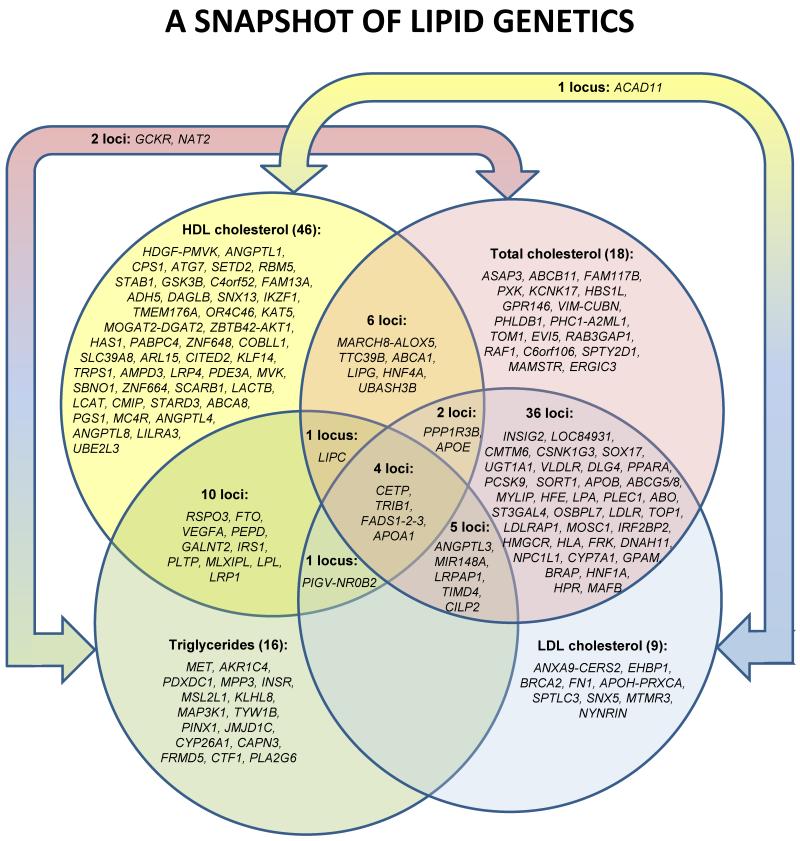
FIGURE 1. Overlap between loci associated with different lipid traits.
This Venn Diagram illustrates the number of loci that show association with multiple lipid traits. The number of loci primarily associated with only one trait is listed in parentheses after the trait name and the locus name is listed below in italics. Loci that show association with two or more traits are shown in the appropriate section.
The effects of newly identified loci were generally smaller than in earlier GWAS (Supplementary Fig. 4). For the 62 newly identified variants, trait variance explained in the Framingham offspring were 1.6% for HDL cholesterol, 2.1% for triglycerides, 2.4% for LDL cholesterol, and 2.6% for total cholesterol.
Overlap of genetic discoveries and prior knowledge
To investigate connections between our new loci and known lipid biology, we first catalogued genes within 100 kb of the peak associated SNPs and searched PubMed and OMIM for occurrences of these gene names and their aliases in the context of relevant keywords. After manual curation, we identified at least one strong candidate in 32 of the 62 loci (52%) (Supplementary Table 4). For the remaining 30 loci, we found no literature support for the role of a nearby gene on blood lipid levels. This search highlighted genes whose connections to lipid metabolism have been extensively documented in mouse models (such as VLDLR13 and LRPAP113) and human cell lines (such as VIM14), as well as candidates whose connection to lipid levels is more recent, such as VEGFA. For the latter, recent studies of VEGFB have suggested that vascular endothelial growth factors have an unexpected role in the targeting of lipids to peripheral tissues15, which we corroborate by associating variants near VEGFA with blood triglyceride and HDL levels.
Multiple types of evidence supported several literature candidates (Supplementary Table 2). For example, VLDLR is categorized by Gene Ontology16 in the retinoid × nuclear receptor (RXR) activation pathway, which also includes genes (APOB, APOE, CYP7A1, APOA1, HNF1A, HNF4A) in previously implicated loci4. However, since these additional sources of evidence build on overlapping knowledge they are not truly independent.
To estimate the probability of finding ≥32 literature supported candidates after automated search and manual review of results, we repeated our text-mining literature search using 100 permutations of SNPs matched for allele frequency, distance to the nearest gene, and number of linkage disequilibrium proxies. To approximate hand-curation of the text-mining results, we focused on genes implicated by 3 or more publications (25 in observed data, 8.7 on average in control SNP sets, P = 8×10−8).
Pathway analyses
We performed a gene-set enrichment analysis, using MAGENTA17, to evaluate over-representation of biological pathways among associated loci. Across the 157 loci, MAGENTA identified 71 enriched pathways. These pathways included at least one gene in 20 of our newly identified loci (Supplementary Table 5). Examples include DAGLB (connected to previously associated loci by genes in the triglyceride lipase activity pathway), INSIG2 (connected by the cholesterol and steroid metabolic process pathways), AKR1C4 (connected by the steroid metabolic process and bile acid biosynthesis pathways), VLDLR (connected by the retinoic × receptor activation and lipid transport pathways, among others), PPARA, ABCB11, and UGT1A1 (three genes assigned to pathways implicated in activation of nuclear hormone receptors, which play an important role in lipid metabolism through the transcriptional regulation of genes in sterol metabolic pathways18). Among the 16 loci where literature review and pathway analysis both suggested a candidate, the predictions overlapped 14 times (Supplementary Table 2; by chance, we expect 6.6 overlapping predictions, P = 1×10−5).
Protein-protein interactions
We assessed evidence for physical interactions between proteins encoded near our associated SNPs using DAPPLE19. We found an excess of direct protein-protein interactions for genes in loci associated with LDL (10 interactions, P = 0.0002), HDL (8 interactions, P = 0.002), and total cholesterol (6 interactions, P = 0.017), but not for triglycerides (2 interactions, P = 0.27) (Supplementary Fig. 5). Most of the interactions involved genes at known loci (such as the interaction network connecting PLTP, APOE, APOB, and LIPC) or highlighted the same genes as literature and pathway analyses (such as those connecting VLDLR, APOE, APOB, CETP, and LPL). Among novel loci, we identified a link between AKT1 and GSK3B. GSK3B has been shown to play a role in energy metabolism20 and its activity is regulated by AKT1 through phosphorylation21. Literature review also supported a role in blood lipid levels for these two genes.
Regulation of gene expression by associated variants
Many complex trait associated variants act through the regulation of gene expression. We examined whether our 62 novel variants were associated with expression levels of nearby genes in liver, omental fat, or subcutaneous fat. Fifteen were associated with expression of a nearby transcript with P < 5×10−8 (Supplementary Table 6) and, in seven, the lipid-associated variant was in strong disequilibrium with the strongest expression-quantitative trait locus (eQTL) for the region (r2 > 0.8). In three of these loci, literature search also prioritized candidate genes. In all three, eQTL analysis and literature review identified the same candidate (DAGLB, SPTLC3, and PXK, P = 0.05). For the remaining four loci (near RBM5, ADH5, TMEM176A, and GPR146), analysis of expression levels identified candidates that were not supported by literature or pathway analyses.
Coding variation
In some loci where previous coding variant association studies were inconclusive, we now find convincing evidence of association, demonstrating the benefits of the large sample sizes achievable by collaboration. For example, in the APOH locus22, our most strongly associated variant is rs1801689 (APOH C325G, P = 1×10−11 for LDL cholesterol). Overall, at 15 of the 62 new loci, there is at least one nonsynonymous variant within 100kb and in strong (r2>0.8) linkage disequilibrium with the index SNP (Supplementary Table 7)(18 loci with no restrictions on distance). This ~30% overlap between associated loci and coding variation is similar to that in other complex traits9. Unexpectedly, in the 11 loci where a candidate was suggested by literature review and by coding variation, the two coincided seven times (P = 0.03 compared to expected chance overlap of 3.8 times); thus, agreement between literature and coding variation was less significant than for eQTL and pathway analysis or protein-protein interactions.
Overlap between association signals and regulators of transcription in liver
Despite our efforts, 18 of the 62 new loci remain without prioritized candidate genes. The liver is an important hub of lipid biosynthesis and there is evidence that lipid loci might be associated with changes in gene regulation in liver cells23. Using ENCODE data23, we evaluated whether associated SNPs overlapped experimentally annotated functional elements identified in HepG2 cells, a commonly used model of human hepatocytes. To determine significance, we generated 100,000 lists of permuted SNPs, matched for minor allele frequency, distance to the nearest gene, and number of SNPs in r2 > 0.8 (described in Methods). In HepG2 cells, lipid-associated SNPs were enriched in eight of the 15 functional chromatin states defined by Ernst et al.24 (P < 1×10−5; Supplementary Table 8). The strongest enrichment was in regions with “strong enhancer activity” (3.7-fold enrichment, P = 2×10−25; Supplementary Table 9). In the other eight cell types examined by Ernst et al., no more than three functional chromatin states showed evidence for enrichment (and, when present, enrichment was weaker).
We proceeded to investigate the overlap between lipid loci and functional marks in HepG2 cells in more detail (Supplementary Table 9). Notable regulatory elements showing significant overlap with lipid loci included histone marks associated with active regulatory regions (H3K27ac, P = 3×10−20; H3K9ac, P = 3×10−22), promoters (H3K4me3, P = 2×10−15, H3K4me2, P = 8×10−12), transcribed regions (H3K36me3, P = 4×10−14), indicators of open chromatin (FAIRE, P = 5×10−9; DNase, P = 2×10−4), and regions that interact with transcription factors HNF4A (P = 6×10−10) and CEBP/B (P = 1×10−5). Overall, 56 of our 62 new loci contained at least one SNP that overlaps a functional mark24 and/or chromatin state23 highlighted in Supplementary Table 9, including all but 3 of the loci where no candidates were suggested by literature review or analyses of pathways, coding variation, or gene expression (Supplementary Table 10).
Initial fine-mapping of 65 lipid-associated loci
Previous fine-mapping of five LDL-associated lipid loci found that variants showing the strongest association were often substantially different in frequency and effect size from those identified in GWAS10. Metabochip genotypes enabled us to carry out an initial fine-mapping analysis for 65 loci: 60 selected for fine-mapping based on our previous study4 and 5 nominated for fine-mapping because of association to other traits.
For each of these loci, we identified the most strongly associated Metabochip variant and evaluated whether it (a) reached genome-wide significant evidence for association (to avoid chance fluctuations in regions where the signal was relatively weak) and (b) was different from the GWAS index SNP in terms of frequency and effect size (operationalized to r2 < 0.8 with the GWAS index SNP). In the European samples, fine-mapping identified eight loci where the fine-mapping signal was clearly different from the GWAS signal (Supplementary Table 11). The two largest differences were at the loci near PCSK9 (top GWAS variant with minor allele frequency f = 0.24 and P = 9×10−24; fine-mapping variant with f = 0.03, P = 2×10−136) and APOE (GWAS variant f = 0.20, P = 3×10−44, fine-mapping variant f = 0.07, P = 3×10−651), consistent with Sanna et al10. Large differences were also observed near LRP4 (GWAS f = 0.17, P = 8×10−14; fine-mapping f = 0.35, P = 1×10−26), IGF2R (GWAS f = 0.16, P = 7×10−9; fine-mapping f = 0.37, P = 2×10−13), NPC1L1 (GWAS f = 0.27, P = 2×10−5; fine-mapping f = 0.24, P = 1×10−12), ST3GAL4 (GWAS f = 0.26, P = 2×10−6; fine-mapping f = 0.07, P = 6×10−11), MED1 (GWAS f = 0.37, P = 3×10−5; fine-mapping f = 0.24, P = 2×10−10), and COBLL1 (GWAS f = 0.12, P = 2×10−6; fine-mapping f = 0.11, P = 6×10−9). Thus, although the large changes observed by Sanna et al10 after fine-mapping are by no means unique, they are not typical. Except for the R46L variant in PCSK9, the variants showing strongest association in fine-mapped loci all had minor allele frequency > .05.
We also attempted fine-mapping in African (N=3,263), East Asian (N=1,771), and South Asian (N=4,901) ancestry samples. Despite comparatively small samples, ancestry-specific analyses identified SNPs clearly distinct from the original GWAS variant in five loci (Supplementary Table 11). These were: APOE, consistent with European ancestry analyses above; three loci where differences in linkage disequilibrium between populations enabled fine-mapping in African (SORT1, LDLR) or East Asian (APOA5) ancestry samples; and CETP, where an African-specific variant was present. For CETP, SORT1, and APOA5, results are consistent with other fine-mapping and functional studies7,7,25,26.
Association of lipid loci with metabolic and cardiovascular traits
To evaluate the role of the 157 loci identified here on related traits, we evaluated the most strongly associated SNPs for each locus in genetic studies of coronary artery disease (CAD, N=114,590 including 37,653 cases)27,28, type 2 diabetes (T2D, N=47,117 including 8,130 cases)29, body mass index (BMI, N=123,865 individuals)30 and waist-hip ratio (WHR, N=77,167 individuals)31, systolic and diastolic blood pressure (SBP and DBP, N=69,395 individuals)32, and fasting glucose (N=46,186 non-diabetics)33. We observed an excess of SNPs nominally associated (P < 0.05) with all these traits: a 5.1 fold excess for CAD (40 nominally significant loci, P = 2×10−19), a 4.1 fold excess for BMI (32 loci, P = 1×10−11), 3.7 fold excesses for DBP (29 loci, P = 1×10−9), a 3.4 fold excess for WHR (27 loci, P = 1×10−9), a 2.5 fold excess for SBP (20 loci, P = 1×10−4), a 2.3 fold excess for T2D (18 loci, P = 0.001), and a 2.2 fold excess for fasting glucose (17 loci, P = 3×10−3) (Supplementary Table 12). Interestingly, among the novel loci, we observed greater overlap with BMI, SBP, and DBP (9 overlapping loci each) than with CAD (8 overlapping loci). Among new loci, the two SNPs showing strongest association to CAD map near RBM5 (rs2013208, PHDL = 9×10−12, PCAD = 7×10−5) and CMTM6 (rs7640978, PLDL = 1×10−8, PCAD = 4×10−4).
We tested whether the LDL-, total cholesterol- or triglyceride-increasing allele, or HDL-decreasing allele was associated with increased risk of cardiovascular disease or related metabolic outcomes; the direction of effect of each locus was categorized according to the primary association signal at the locus, as in Tables 1A-D. We observed association with increased CAD risk (104/149, P = 1×10−6), SBP (96/155, P = 2.7×10−3) and WHR adjusted for BMI (92/154, P = 0.019). There were many instances where a single locus was associated with many traits. These included variants near FTO, consistent with previous reports34; near VEGFA (associated with triglyceride levels, CAD, T2D, SBP, and DBP), near SLC39A8 (associated with HDL cholesterol, BMI, SBP, and DBP), and near MIR581 (associated with HDL cholesterol, BMI, T2D, and DBP). In some cases, like FTO, a strong association with BMI or another phenotype generates weaker association signals for other metabolic traits34. In other cases, like SORT1, a primary effect on lipid levels may mediate secondary association with other traits, like CAD7.
Association of individual lipids with coronary artery disease
Epidemiological studies consistently show high total cholesterol and LDL cholesterol levels are associated with increased risk of CAD, whereas high HDL cholesterol levels are associated with reduced risk of CAD35. In genetic studies, the connection between LDL cholesterol and CAD is clear, whereas the results for HDL cholesterol levels are more equivocal36-38. In our data, trait increasing alleles at the loci showing strongest association with LDL cholesterol (31 loci), triglycerides (30 loci), or total cholesterol (38 loci) were associated with increased risk of CAD (P = 2×10−12, P = 2×10−16, and P = 0.006). Conversely, trait decreasing alleles at loci showing the strongest association with HDL cholesterol (64 loci), were associated with increased CAD risk with P = 0.02. When we focused on loci uniquely associated with LDL cholesterol (12 loci where P > .05 for other lipids), triglycerides (6 loci), or HDL cholesterol (14 loci), only the LDL association remained significant (P = 0.03).
To better explore how associations with individual lipid levels related to CAD risk, we used linear regression to test whether association with lipid levels could predict impact on CAD risk. In this analysis, the effect on CAD of 149 lipid loci (CAD results were not available for 8 SNPs) was correlated with LDL (Pearson r=0.74, P = 7×10−6) and triglyceride (Pearson r=0.46, P = 0.02) effect sizes, but not HDL effect sizes (Pearson r=−9×10−4, P = 0.99; Supplementary Fig. 6). Since most variants affect multiple lipid fractions (Figure 1), dissecting the relationship between lipid level and CAD effects requires multivariate analysis. In a companion manuscript, we use multivariate analysis and detailed examination of triglyceride associated loci to show that increased LDL and triglyceride levels, but not HDL, appear causally related to CAD risk.
Evidence for additional loci, not yet reaching genome-wide significance
To evaluate evidence for loci not yet reaching genome-wide significance, we compared direction of effect in GWAS and Metabochip analyses of non-overlapping samples, outside the 157 genome-wide significant loci. Among independent variants (r2 < 0.1) with P < 0.1 in the GWAS-only analysis, a significant excess were concordant in direction of effect for HDL (62.9% in 1,847 SNPs, P < 10−16), LDL (58.6% of 1,730 SNPs, P < 10−16), triglyceride levels (59.1% of 1,783 SNPs, P < 10−16), and total cholesterol (61.0% of 1,904 SNPs, P < 10−16), suggesting many additional loci to be discovered in future studies.
Discussion
Molecular understanding of the genes and pathways that modify blood lipid levels in humans will facilitate the design of new therapies for cardiovascular and metabolic disease. This understanding can be gained from studies of model organisms, in vitro experiments, bioinformatic analyses, and human genetic studies. Here, we demonstrate association between blood lipid levels and 62 new loci, bringing the total number of lipid-associated loci to 157 (See Tables 1A-D and Figure 1). All but one of the loci identified here include protein-coding genes within 100 kb of the SNP showing strongest association. While 38 of the 62 new loci include genes whose role in blood lipid levels is supported by literature review or analysis of curated pathway databases, the remainder includes only genes whose role on blood lipid levels has not been documented.
In total, there are 240 genes within 100 kb of one of our 62 new lipid-associated loci – providing a daunting challenge for future functional studies. Prioritizing on the basis of literature review, pathway analysis, regulation of mRNA expression levels, and protein altering variants suggests that 70 genes in 44 of the 62 new loci might be the focus of the first round of functional studies (summarized in Supplementary Table 2). While we found significant overlap, different sources of prioritization sometimes disagreed. This result suggests that truly understanding causality will be very challenging. The Supplementary Note includes an interpreted digest of genes highlighted by our study. Clearly, a range of approaches will be needed to follow-up these findings. To illustrate possibilities, consider U. S. Patent Application #20,090,036,394 disclosing that, in the mouse, knockout of Gpr146 modifies blood lipid levels. Here, we show that variants near the human homologue of this gene, GPR146, are associated with levels of total cholesterol – providing an added incentive for studies of GPR146 inhibitors in humans. GPR146 encodes a G-protein coupled receptor – an attractive pharmaceutical target – so it is tempting to speculate that, one day, pharmaceutical inhibition of GPR146 may modify cholesterol levels and reduce risk of heart disease.
Each locus typically includes many strongly associated (and potentially causal) variants. Our fine-mapping results illustrate how genetic analysis of large samples and individuals of diverse ancestry can help focus the search for causal variants. In our fine-mapping analysis of 65 lipid-associated loci, we were able to separate the strongest signal in a region from the prior GWAS signal in 12 instances. In three of these 12 instances, fine-mapping was enabled by analysis of a few thousand African or East Asian ancestry individuals, whereas in the remaining instances, fine-mapping was possible through examination of nearly 100,000 European ancestry samples. A more detailed fine-mapping exercise, including imputation of variants from emerging very large reference panels, may help refine the location of additional signals.
Lipid-associated loci were strongly associated with CAD, T2D, BMI, SBP, and DBP. In univariate analyses, we found that impact on LDL and triglycerides all predicted association with CAD, but HDL did not. In a more detailed multivariate investigation, a companion manuscript shows that our data is consistent with the hypothesis that both LDL and triglycerides, but not HDL, are causally related to CAD risk. HDL, LDL, and triglycerides levels summarize aggregate levels of different lipid particles, each with potentially distinct consequences for CAD risk. We evaluated association of our loci with lipid subfractions in 2,900 individuals from the Framingham Heart Study (Supplementary Table 13, Supplementary Fig. 7) and with sphingolipids, which are components of lipid membranes in cells, in 4,034 individuals from five samples of European ancestry39 (Supplementary Table 14). The results suggest HDL-associated variants can have a markedly different impact on these sub-phenotypes. For example, among HDL loci, variants near LIPC were strongly associated with plasmalogen levels (P < 10−40), variants near ABCA1 were associated with sphingomyelin levels (P < 10−5), and variants near CETP – which show the strongest association with HDL cholesterol overall – were associated with neither of these. Detailed genetic dissection of these sub-phenotypes in larger samples, could lead to functional groupings of HDL-associated variants that reconcile the results of genetic studies (which show no clear connection between HDL cholesterol-associated variants and CAD risk) and epidemiologic studies (which show clear association between plasma HDL levels and CAD risk).
In summary, we report the largest genetic association study of blood lipid levels yet conducted. The large number of loci identified, the many candidate genes they contain, and the diverse proteins they encode generate new leads and insights into lipid biology. It is our hope that the next round of genetic studies will build on these results, using new sequencing, genotyping, and imputation technologies to examine rare loss-of-function alleles and other variants of clear functional impact to accelerate the translation of these leads into mechanistic insights and improved treatments for CAD.
Online Methods
Samples studied
We collected summary statistics for Metabochip SNPs from 45 studies. Among these, 37 studies consisted primarily of individuals of European ancestry (see Supplementary Table 1 and Supplementary Note for details), including both population-based studies and case-control studies of CAD and T2D. Another 8 studies consisted primarily of individuals with non-European ancestry: two studies of South Asian descent, AIDHS/SDS (N=1,516) and PROMIS (N=3,385); two studies of East Asian descent, CLHNS (N=1,771) and TAI-CHI (N=7044); and five studies of recent African ancestry, MRC/UVRI GPC (N=1,687) from Uganda, SEY (N=426) from the Caribbean, and FBPP (N=1,614, TG results unavailable), GXE (N=397), and SPT (N=838) from the United States (more details in Supplementary Table 1 and Supplementary Note).
Genotyping
We genotyped 196,710 genetic variants prioritized on the basis of prior GWAS for cardiovascular and metabolic phenotypes using the Illumina iSelect Metabochip8 genotyping array. To design the Metabochip, we used our previous GWAS of ~100,000 individuals4 to prioritize 5,023 SNPs for HDL cholesterol, 5,055 for LDL cholesterol, 5,056 for triglycerides, and 938 for total cholesterol. These independent SNPs represent most loci with P < .005 in our original GWAS for HDL cholesterol, LDL cholesterol and triglycerides and with P < .0005 for total cholesterol. An additional 28,923 SNPs were selected for fine-mapping of 65 previously identified lipid loci. The Metabochip also included 50,459 SNPs prioritized based on GWAS of non-lipid traits and 93,308 SNPs selected for fine-mapping of loci associated with non-lipid traits (5 of these loci were associated with blood lipids by the analyses described here).
Phenotypes
Blood lipid levels were typically measured after > 8 hours of fasting. Individuals known to be on lipid-lowering medication were excluded when possible. LDL cholesterol levels were directly measured in 10 studies (24% of total study individuals) and estimated using the Friedewald formula40 in the remaining studies. Trait residuals within each study cohort were adjusted for age, age2, and sex, and then quantile normalized. Explicit adjustments for population structure using principal component41 or mixed model approaches42 were carried out in 24 studies (35% of individuals); all studies were adjusted using genomic control prior to meta-analysis11. In studies ascertained on diabetes or CVD status, cases and controls were analyzed separately (Supplementary Table 1). All meta-analyses were limited to a single ancestral group (e.g. European only).
Primary statistical analysis
Individual SNP association tests were performed using linear regression with the inverse normal transformed trait values as the dependent variable and the expected allele count for each individual as the independent variable. These analyses were performed using PLINK (26 samples, 53% of the total number of individuals), SNPTEST (4 samples, 20% of individuals), EMMAX (9 samples, 14% of individuals), Merlin (4 samples, 9% of individuals), GENABEL (1 sample, 3% of individuals), and MMAP (1 sample, 1% of individuals) (Supplementary Table 1).
Meta-analysis
Meta-analysis was performed using the Stouffer method43,44, with weights proportional to the square root of the sample size for each sample. To correct for inflated test statistics due to potential population stratification, we first applied genomic control to each sample and then repeated the procedure with initial meta-analysis results. For GWAS samples, we used all available SNPs when estimating the median test statistic and inflation factor λ. For Metabochip samples, we used a subset of SNPs (N = 7,168) that had P-values > 0.50 for all lipid traits in the original GWAS, expecting that the majority of these would not be associated with lipids and would behave as null variants in the Metabochip samples. Signals were considered to be novel if they reached a P-value < 5×10−8 in the combined GWAS and Metabochip meta-analysis and were >1 Mb away from the nearest previously described lipid locus and other novel loci. We used only European samples for the discovery of novel genome-wide significant loci. The non-European samples were meta-analyzed and examined only for fine-mapping analyses.
Quality control
To flag potentially erroneous analyses, we carried out a series of quality control steps. Average standard errors for association statistics from each study were plotted against study sample size to identify outlier studies. We inspected allele frequencies to ensure all analyses used the same strand assignment of alleles. We evaluated whether reported statistics and allelic effects were consistent with published findings for known loci. Genomic control values for study specific analyses were inspected, and all were <1.20. Finally, within each study, we excluded variants for which the minor allele was observed <7 times.
Proportion of trait variance explained
We estimated the increase in trait variance explained by novel loci in the Framingham cohort (N=7,132) using three models for each trait-residual: 1) lead and secondary SNPs from the previously published loci4 and 2) previously published lipid loci plus newly reported loci; and 3) newly reported loci. We regressed lipid residuals on these sets of SNPs using the lme kinship package in R.
Initial automated review of the published literature
An initial list of candidates within each locus was generated with Snipper (http://csg.sph.umich.edu/boehnke/snipper/) and then subjected to manual review. For each locus, Snipper first generates a list of nearby genes and then checks for the co-occurrence of the corresponding gene names and selected search terms (“cholesterol”, “lipids”, “HDL”, “LDL”, or “triglycerides”) in published literature and OMIM. We supplemented this approach with traditional literature searches using PubMed and Google.
Generating permuted sets of non-associated SNPs
To estimate the expected chance overlap between literature searches and our loci, we generated lists of permuted SNPs. To generate these lists, we first identified all non-associated lipid SNPs (P > 0.10 for any of the 4 lipid traits) and created bins based on 3 statistics: minor allele frequency, distance to the nearest gene, and number of SNPs with r2 > 0.8. For each index SNP, we identified 500 non lipid-associated SNPs that fell within the same 3 bins and randomly selected one SNP for each permuted list.
Pathway analyses
To investigate if lipid-associated variants overlapped previously annotated pathways, we used gene set enrichment analysis (GSEA), as implemented in MAGENTA17 using the meta-analysis of all studies, including GWAS and Metabochip SNPs. Briefly, MAGENTA first assigns SNPs to a given gene when within 110 kb upstream or 40 kb downstream of transcript boundaries. The most significant SNP P-value within this interval is then adjusted for confounders (gene size, marker density, LD) to create a gene association score. When the same SNP is assigned to multiple genes, only the gene with the lowest score is kept for downstream analyses. Subsequently, MAGENTA attaches pathway terms to each gene using several annotation resources, including GO, PANTHER, Ingenuity, and KEGG. Finally, the genes are ranked on their gene association score, and a modified GSEA test is used to test the null hypothesis that all gene score ranks above a given rank cutoff are randomly distributed with regard to a given pathway term (and compared to multiple randomly sampled gene sets of identical size). We evaluated enrichment by using a rank cutoff of 5% of the total number of genes. A minimum of 10,000 gene set permutations were performed, and up to 1,000,000 permutations for GSEA P-values below 1×10−4.
We used the Disease Association Protein–Protein Link Evaluator package (DAPPLE; http://www.broadinstitute.org/mpg/dapple/dapple.php) to examine evidence for protein-protein interaction networks connecting genes across different lipid loci. This analysis included the 62 novel loci as well as the 95 previously known loci; we focus our discussion on pathways that included one or more genes from novel loci.
Cis-expression quantitative trait locus analysis
To determine whether lipid-associated SNPs might act as cis-regulators of nearby genes, we examined association with expression levels of 39,280 transcripts in 960 human liver samples, 741 human omental fat samples, and 609 human subcutaneous fat samples. Tissue samples were collected postmortem or during surgical resection from donors; tissue collection, DNA and RNA isolation, expression profiling, and genotyping were performed as described45. MACH was used to obtain imputed genotypes for ~2.6 million SNPs in the HapMap release 22 for each of the samples. We examined the correlation between each of the 62 new index SNPs and all transcripts within 500 kb of the SNP position, performing association analyses as previously described46.
Functional annotation of associated variants
We attempted to identify lipid-associated SNPs that fall in important regulatory domains. We initially created a list of all potentially causal variants by selecting index SNPs at loci identified in this study or in Teslovich et al4. We then selected any variant in strong linkage disequilibrium (r2 > 0.8 from 1000 Genomes or HapMap) with each index SNP. We compared the position of the index SNPs and their proxies to previously described functional marks23,24. To assess the expected overlap with functional marks, we created 100,000 permuted sets of non-associated SNPs (see above) and evaluated permuted SNP lists for overlap with functional domains. We estimated a P-value for each functional domain as the proportion of permuted sets with an equal or greater number of loci overlapping functional domains (for large P-values). For small P-values we used a normal approximation to the empirical overlap distribution to estimate P-values.
Association with lipid subfractions
Lipoprotein fractions for Women’s Genome Health Study (WGHS) samples (N = 23170) were measured using the LipoProtein-II assay (Liposcience Inc. Raleigh, NC) and Framingham Heart Study Offspring samples (N = 2900) were measured with the LipoProtein-I assay (Liposcience Inc. Raleigh, NC)47. Additional information on sub-fraction measurements can be found in Supplementary Fig. 7. Log transformations were used for non-normalized traits. All models were adjusted for age, sex, and PCs. The genetic association analysis of WGHS used SNP genotypes imputed from the HapMap r22 CEU reference panel using MACH. 16,730 out of 23,170 WGHS participants were fasting for 8 hours prior to blood draw (72.2%).
Supplementary Material
TABLE 1B. Novel Loci Primarily Associated with LDL Cholesterol Obtained from Joint GWAS and Metabochip Meta-analysis.
| Locus | MarkerName | Chr | hg19 Position (Mb) |
Associated trait(s) |
MAF | Minor/major Allele |
Effect of A1 | Joint N (in 1000s) |
Joint P-value |
|---|---|---|---|---|---|---|---|---|---|
| ANXA9-CERS2 | rs267733 | 1 | 150.96 | LDL | .16 | G/A | −.033 | 165 | 5×10−9 |
| EHBP1 | rs2710642 | 2 | 63.15 | LDL | .35 | G/A | −.024 | 173 | 6×10−9 |
| INSIG2 | rs10490626 | 2 | 118.84 | LDL, TCb | .08 | A/G | −.051/.042 | 173/184 | 2×10−12/6×10−9 |
| LOC84931 | rs2030746 | 2 | 121.31 | LDL, TC | .40 | T/C | .021/.020 | 173/187 | 9×10−9/4×10−8 |
| FN1 | rs1250229 | 2 | 216.30 | LDL | .27 | T/C | −.024 | 173 | 3×10−8 |
| CMTM6 | rs7640978 | 3 | 32.53 | LDL, TC | .09 | T/C | −.039/−.038 | 172/186 | 1×10−8 |
| ACAD11 | rs17404153 | 3 | 132.16 | LDL, HDLc | .14 | T/G | −.034/.028 | 172/187 | 2×10−9/5×10−9 |
| CSNK1G3 | rs4530754 | 5 | 122.86 | LDL, TC | .46 | G/A | −.028/−.023 | 173/187 | 4×10−12/2×10−9 |
| MIR148A | rs4722551 | 7 | 25.99 | LDL, TGd, TC | .20 | C/T | .039/.029/.023 | 173/187/178 | 4×10−14/9×10−11/7.0×10−9 |
| SOX17 | rs10102164 | 8 | 55.42 | LDL, TC | .21 | A/G | .032/.030 | 173/187 | 4×10−11/5×10−11 |
| BRCA2 | rs4942486 | 13 | 32.95 | LDL | .48 | T/C | .024 | 172 | 2×10−11 |
| APOH-PRXCA | rs1801689 | 17 | 64.21 | LDL | .04 | C/A | .103 | 111 | 1×10−11 |
| SPTLC3 | rs364585 | 20 | 12.96 | LDL | .38 | A/G | −.025 | 172 | 4×10−10 |
| SNX5 | rs2328223 | 20 | 17.85 | LDL | .21 | C/A | .03 | 171 | 6×10−9 |
| MTMR3 | rs5763662 | 22 | 30.38 | LDL | .04 | T/C | .077 | 163 | 1×10−8 |
Chr, chromosome;MAF, minor allele frequency; A1, minor allele; A2, major allele.Effect sizes are given with respect to the minor allele (A1) in SD units. For loci associated with two or more traits at genome-wide significance, the trait corresponding to the strongest P-value is listed first. At three loci, secondary traits were most strongly associated with different SNPs.
rs17526895 (within 1Mb of rs10490626, r2 = 0.98);
rs13076253 (within 1Mb of rs17404153, r2 = 0.00);
rs4719841 (within 1Mb of rs4722551, r2 = 0.10).
TABLE 1C. Novel Loci Primarily Associated with Total Cholesterol Obtained from Joint GWAS and Metabochip Meta-analysis.
| Locus | MarkerName | Chr | hg19 Position (Mb) |
Associated trait(s) |
MAF | Minor/major Allele |
Effect of A1 | Joint N (in 1000s) |
Joint P-value |
|---|---|---|---|---|---|---|---|---|---|
| ASAP3 | rs1077514 | 1 | 23.77 | TC | .15 | C/T | −0.03 | 184 | 6×10−9 |
| ABCB11 | rs2287623 | 2 | 169.83 | TC | .41 | G/A | 0.027 | 184 | 4×10−12 |
| FAM117B | rs11694172 | 2 | 203.53 | TC | .25 | G/A | 0.028 | 187 | 2×10−9 |
| UGT1A1 | rs11563251 | 2 | 234.68 | TC, LDL | .12 | T/C | 0.037/0.034 | 187/173 | 1×10−9/5×10−8 |
| PXK | rs13315871 | 3 | 58.38 | TC | .10 | A/G | −0.036 | 187 | 4×10−8 |
| KCNK17 | rs2758886 | 6 | 39.25 | TC | .30 | A/G | 0.023 | 187 | 3×10−8 |
| HBS1L | rs9376090 | 6 | 135.41 | TC | .28 | T/C | −0.025 | 187 | 3×10−9 |
| GPR146 | rs1997243 | 7 | 1.08 | TC | .16 | G/A | 0.033 | 183 | 3×10−10 |
| VLDLR | rs3780181 | 9 | 2.64 | TC, LDL | .08 | G/A | −0.044/−0.044 | 186/172 | 7×10−10/2×10−9 |
| VIM-CUBN | rs10904908 | 10 | 17.26 | TC | .43 | G/A | 0.025 | 187 | 3×10−11 |
| PHLDB1 | rs11603023 | 11 | 118.49 | TC | .42 | T/C | 0.022 | 187 | 1×10−8 |
| PHC1-A2ML1 | rs4883201 | 12 | 9.08 | TC | .12 | G/A | −0.035 | 187 | 2×10−9 |
| DLG4 | rs314253 | 17 | 7.09 | TC, LDL | .37 | C/T | −0.023/−0.024 | 184/170 | 3×10−10/3×10−10 |
| TOM1 | rs138777 | 22 | 35.71 | TC | .36 | A/G | 0.021 | 185 | 5×10−8 |
| PPARA | rs4253772 | 22 | 46.63 | TC, LDLe | .11 | T/C | 0.032/−0.031 | 185/171 | 1×10−8/3×10−8 |
Chr, chromosome;MAF, minor allele frequency; A1, minor allele; A2, major allele.Effect sizes are given with respect to the minor allele (A1) in SD units. For loci associated with two or more traits at genome-wide significance, the trait corresponding to the strongest P-value is listed first. At one locus, the secondary trait was most strongly associated with a different SNP:
rs4253776 (within 1Mb of rs4253772, r2 = 0.95).
ACKNOWLEDGEMENTS
We especially thank the >196,000 volunteers who participated in our study. Detailed acknowledgement of funding sources is provided in the supplementary online material.
Footnotes
URLs
Summary results for our studies are available. We hope that they will facilitate continued research into the genetics of blood lipid levels and, eventually, help identify improved treatments for CAD. To browse the full result set, go to http:/www.sph.umich.edu/csg/abecasis/lipids2013/
Disclosures
CHS
Bruce Psaty serves on the DSBM of a clinical trial funded by the manufacturer (Zoll), and he serves on the Steering Committee of the Yale Open-Data Project funded by the Medtronic.
CoLaus
Peter Vollenweider received an unrestricted grant from GSK to build the CoLaus study.
deCODE
Authors affiliated with deCODE Genetics/Amgen, a biotechnology company, are employees of deCODE Genetics/Amgen.
GLACIER
Inês Barroso and spouse own stock in GlaxoSmithKline and Incyte Ltd.
AUTHOR CONTRIBUTIONS
Writing and Analysis Group
G.R.A., M.B., L.A.C., P.D., P.W.F., S.K., K.L.M., E.I., G.M.P., S.S.R., S.R., M.S.S., E.M.S., S.S., C.J.W. (Lead). E.M.S. and S.S. performed meta-analysis and E.M.S., S.S., G.M.P., M.B., J.C., S.G., A.G., and S. K. performed bioinformatics analyses. E.M.S. and S.S. prepared the tables, figures and supplementary material. C.J.W. led the analysis and bioinformatics efforts. E.I. and K.M. led the biological interpretation of results. C.J.W. and G.R.A. wrote the manuscript. All analysis and writing group authors extensively discussed the analysis, results, interpretation and presentation of results.
All authors contributed to the research and reviewed the manuscript.
Design, management and coordination of contributing cohorts
(ADVANCE) T.L.A.; (AGES Reykjavik study) T.B.H., V.G.; (AIDHS/SDS) D.K.S.; (AMC-PAS) P.D., G.K.H.; (Amish GLGC) A.R.S.; (ARIC) E.B.; (B58C-WTCCC & B58C-T1DGC) D.P.S.; (B58C-Metabochip) C.M.L., C.P., M.I.M.; (BLSA) L.F.; (BRIGHT) P.B.M.; (CHS) B.M.P., J.I.R.; (CLHNS) A.B.F., K.L.M., L.S.A.; (CoLaus) P.V.; (deCODE) K.S., U.T.; (DIAGEN) P.E.S., S.R.B.; (DILGOM) S.R.; (DPS) M.U.; (DR’s EXTRA) R.R.; (EAS) J.F.P.; (EGCUT (Estonian Genome Center of University of Tartu)) A.M.; (ELY) N.W.; (EPIC) N.W., K.K.; (EPIC_N_OBSET GWAS) E.H.Young; (ERF) C.M.V.; (ESS (Erasmus Stroke Study)) P.J.K.; (Family Heart Study FHS) I.B.B.; (FBPP) A.C., R.S.C., S.C.H.; (FENLAND) R.L., N.W.; (FIN-D2D 2007) A.K., L.M.; (FINCAVAS) M.K.; (Framingham) L.A.C., S.K., J.M.O.; (FRISCII) A.S., L.W.; (FUSION GWAS) K.L.M., M.B.; (FUSION stage 2) F.S.C., J.T., J.S.; (GenomEUTwin) J.B.W., N.G.M., K.O.K., V.S., J.K., A.J., D.I.B., N.P., T.D.S.; (GLACIER) P.W.F.; (Go-DARTS) A.D.M., C.N.P.; (GxE/Spanish Town) B.O.T., C.A.M., F.B., J.N.H., R.S.C.; (HUNT2) K.H.; (IMPROVE) U.D., A.H., E.T., S.E.H.; (InCHIANTI) S.B.; (KORAF4) C.G.;(LifeLines) B.H.W.; (LOLIPOP) J.S.K., J.C.C.; (LURIC) B.O.B.; W.M.; (MDC) L.C.G., S.K.; (METSIM) J.K., M.L.; (MICROS) P.P.P.; (MORGAM) D.A., J.F.; (MRC/UVRI GPC GWAS) P.K., G.A., J.S., E.H.Y.; (MRC National Survey of Health & Development) D.K.; (NFBC1986) M-R.J.; (NSPHS) U.G.; (ORCADES) H.C.; (PARC) Y.I.C., R.M.K., J.I.R.; (PIVUS) E.I., L.L.; (PROMIS) J.D., P.D., D.S.; (Rotterdam Study) A.H., A.G.U.; (SardiNIA) G.R.A.; (SCARFSHEEP) A.H., U.D.; (SEYCHELLES) M.B., M.B.; P.B.; (SUVIMAX) P.M.; (Swedish Twin Reg.) E.I., N.L.P.; (TAICHI) T.L.A., Y.I.C., C.A.H., T.Q., J.I.R., W.H.S.; (THISEAS) G.D., P.D.; (Tromsø) I.N.; (TWINGENE) U.D., E.I.; (ULSAM) E.I.; (Whitehall II) A.H., M.K.
Genotyping of contributing cohorts
(ADVANCE) D.A.; (AIDHS/SDS) L.F.B., M.L.G.; (AMC-PAS) P.D., G.K.H.; (B58C-WTCCC & B58C-T1DGC) W.L.M.; (B58C-Metabochip) M.I.M.; (BLSA) D.H.; (BRIGHT) P.B.M.; (CHS) J.I.R.; (DIAGEN) N.N., G.M.; (DILGOM) A.P.; (DR’s EXTRA) T.A.L.; (EAS) J.F.W.; (EGCUT (Estonian Genome Center of University of Tartu)) T.E.; (EPIC) P.D.; (EPIC_N_SUBCOH GWAS) I.B.; (ERF) C.M.V.; (ESS (Erasmus Stroke Study)) C.M.V.; (FBPP) A.C., G.B.E.; (FENLAND) M.S.S.; (FIN-D2D 2007) A.J.S.; (FINCAVAS) T.L.; (Framingham) J.M.O.; (FUSION stage 2) L.L.B.; (GLACIER) I.B.; (Go-DARTS) C.G., C.N.P., M.I.M.; (IMPROVE) A.H.; (KORAF3) H.G., T.I.; (KORAF4) N.K.; (LifeLines) C.W.; (LOLIPOP) J.S.K., J.C.C.; (LURIC) M.E.K.; (MDC) B.F.V., R.D.; (MICROS) A.A.H.; (MORGAM) L.T., P.B.; (MRC/UVRI GPC GWAS) M.S.S.; (MRC National Survey of Health & Development) A.W., D.K., K.K.O.; (NFBC1986) A-L.H., M.J, M.M., P.E., S.V.; (NSPHS and FRISCII) Å.J.; (ORCADES) H.C.; (PARC) M.O.G., M.R.J., J.I.R.; (PIVUS) E.I., L.L.; (PROMIS) P.D., K.S.; (Rotterdam Study) A.G.U., F.R.; (SardiNIA) R.N.; (SCARFSHEEP) B.G., R.J.S.; (SEYCHELLES) F.M., G.B.E.; (Swedish Twin Reg.) E.I., N.L.P.; (TAICHI) D.A., T.L.A., E.K., T.Q., L.L.W.; (THISEAS) P.D.; (TWINGENE) A.H., E.I.; (ULSAM) E.I.; (WGHS) D.I.C., P.M.R.; (Whitehall II) A.H., C.L., M.K., M.K.
Phenotype definition of contributing cohorts
(ADVANCE) C.I.; (AGES Reykjavik study) T.B.H., V.G.; (AIDHS/SDS) L.F.B.; (AMC-PAS) J.J.K.; (Amish GLGC) A.R.S., B.D.M.; (B58C-WTCCC & B58C-T1DGC) D.P.S.; (B58C-Metabochip) C.P.; E.H.; (BRIGHT) P.B.M.; (CHS) B.M.P.; (CoLaus) P.V.; (deCODE) G.I.E., H.H., I.O.; (DIAGEN) G.M.; (DILGOM) K.S.; (DPS) J.L.; (DR’s EXTRA) P.K.; (EAS) J.L.B.; (EGCUT (Estonian Genome Center of University of Tartu)) A.M.; (EGCUT (Estonian Genome Center of University of Tartu)) K.F.; (ERF and Rotterdam Study) A.H.; (ERF) C.M.V; (ESS (Erasmus Stroke Study)) E.G.V., H.M.D., P.J.K.; (FBPP) A.C., R.S.C., S.C.H.; (FINCAVAS) T.V.N.; (Framingham) S.K., J.M.O.; (GenomEUTwin: MZGWA) J.B.W.; (GenomEUTwin-FINRISK) V.S.; (GenomEUTwin-FINTWIN) J.K., K.H.; (GenomEUTwin-GENMETS) A.J.; (GenomEUTwin-NLDTWIN) G.W.; (Go-DARTS) A.S.D., A.D.M., C.N.P., L.A.D.; (GxE/Spanish Town) C.A.M., F.B.; (IMPROVE) U.D.; A.H., E.T.; (KORAF3) C.M.; (KORAF4) A.D.; (LifeLines) L.J.; (LOLIPOP) J.S.K., J.C.C.; (LURIC) H.S.; (MDC) L.C.G.; (METSIM) A.S.; (MORGAM) G.C.; (MRC/UVRI GPC GWAS) R.N.N.; (MRC National Survey of Health & Development) D.K.; (NFBC1986) A.R., A-L.H., A.P., M-R.J.; (NSPHS and FRISCII) Å.J.; (NSPHS) U.G.; (ORCADES) S.H.W.; (PARC) Y.I.C., R.M.K.; (PIVUS) E.I., L.L.; (PROMIS) D.F.F.; (Rotterdam Study) A.H.; (SCARFSHEEP) U.D., B.G.; (SEYCHELLES) M.B., M.B., P.B.; (Swedish Twin Reg.) E.I., N.L.P.; (TAICHI) H.C., C.A.H., Y.H., E.K., S.L., W.H.S.; (THISEAS) G.D., M.D.; (Tromsø) T.W.; (TWINGENE) U.D., E.I.; (ULSAM) E.I.; (WGHS) P.M.R.; (Whitehall II) M.K.
Primary analysis from contributing cohorts
(ADVANCE) L.L.W.; (AIDHS/SDS) R.S.; (AMC-PAS) S.K.; (Amish GLGC) J.R.O., M.E.M.; (ARIC) K.A.V.; (B58C-Metabochip) C.M.L., E.H., T.F.; (B58C-WTCCC & B58C-T1DGC) D.P.S.; (BLSA) T.T.; (BRIGHT) T.J.; (CLHNS) Y.W.; (CoLaus) J.S.B.; (deCODE) G.T.; (DIAGEN) A.U.J.; (DILGOM) M.P.; (EAS) R.M.F.; (DPS) A.U.J.; (DR ’S EXTRA) A.U.J.; (EGCUT (Estonian Genome Center of University of Tartu)) E.M., K.F., T.E.; (ELY) D.G.; (EPIC) K.S., D.G.; (EPIC_N_OBSET GWAS) E.Y., C.L.; (EPIC_N_SUBCOH GWAS) N.W.; (ERF) A.I.; (ESS (Erasmus Stroke Study)) C.M.V., E.G.V.; (EUROSPAN) A.D.; (Family Heart Study FHS) I.B.B., M.F.F.; (FBPP) A.C., G.B.E.; (FENLAND) T.P., C.P.; (FENLAND GWAS) J.H.Z., J.L.; (FIN-D2D 2007) A.U.J.; (FINCAVAS) L.L.; (Framingham) L.A.C., G.M.P.; (FRISCII and NSPHS) Å.J.; (FUSION stage 2) T.M.T.; (GenomEUTwin-FINRISK) J.K.; (GenomEUTwin-FINTWIN) K.H.; (GenomEUTwin-GENMETS) I.S.; (GenomEUTwin-SWETWIN) P.K.M.; (GenomEUTwin-UK-TWINS) M.M.; (GLACIER) D.S.; (GLACIER) P.W.F.; (Go-DARTS) C.N.P., L.A.D.; (GxE/Spanish Town) C.D.P.; (HUNT) A.U.J.; (IMPROVE) R.J.S.; (InCHIANTI) T.T.; (KORAF3) M.M.; (KORAF4) A.P.; (LifeLines) I.M.N.; (LOLIPOP) W.Z.; (LURIC) M.E.K.; (MDC) B.F.V.; (MDC) P.F., R.D.; (METSIM) A.U.J.; (MRC/UVRI GPC GWAS) R.N.N.; (MRC National Survey of Health & Development) A.W., J.L.; (NFBC1986) M.K., I.S., S.K.S.; (NSPHS and FRISCII) Å.J.; (PARC) X.L.; (PIVUS) C.S., E.I.; (PROMIS) J.D., D.F.F., K.S.; (Rotterdam Study) A.I.; (SardiNIA) C.S., J.L.B., S.S.; (SCARFSHEEP) R.J.S.; (SEYCHELLES) G.B.E., M.B.; (SUVIMAX) T.J.; (Swedish Twin Reg.) C.S., E.I.; (TAICHI) D.A., T.L.A., H.C., M.G., C.A.H., T.Q., L.L.W; (THISEAS) S.K.; (Tromsø) A.U.J.; (TWINGENE) A.G., E.I.; (ULSAM) C.S., E.I., S.G.; (WGHS) D.I.C.; (Whitehall II) S.S.
REFERENCES
- 1.Kannel WB, Dawber TR, Kagan A, Revotskie N, Stokes J., 3rd Factors of risk in the development of coronary heart disease--six year follow-up experience. The Framingham Study. Annals of Internal Medicine. 1961;55:33–50. doi: 10.7326/0003-4819-55-1-33. [DOI] [PubMed] [Google Scholar]
- 2.Castelli WP. Cholesterol and lipids in the risk of coronary artery disease--the Framingham Heart Study. Canadian Journal of Cardiology. 1988;4(Suppl A):5A–10A. [PubMed] [Google Scholar]
- 3.Lloyd-Jones D, et al. Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation. 2010;121:e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 4.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barter PJ, Rye KA. Cholesteryl ester transfer protein (CETP) inhibition as a strategy to reduce cardiovascular isk. Journal of Lipid Research. 2012 doi: 10.1194/jlr.R024075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahalkar AR, Hegele RA. Monogenic pediatric dyslipidemias: classification, genetics and clinical spectrum. Molecular Genetics and Metabolism. 2008;93:282–94. doi: 10.1016/j.ymgme.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Musunuru K, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–9. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voight BF, et al. The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, nd Anthropometric Traits. PLoS Genetics. 2012 doi: 10.1371/journal.pgen.1002793. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The 1000 Genomes Project A map of human genome variation from population scale sequencing. Nature. 2010;467 doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanna S, et al. Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet. 2011;7:e1002198. doi: 10.1371/journal.pgen.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 12.Asselbergs FW, et al. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am J Hum Genet. 2012;91:823–38. doi: 10.1016/j.ajhg.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welch CL, et al. Genetic regulation of cholesterol homeostasis: chromosomal organization of candidate genes. Journal of Lipid Research. 1996;37:1406–21. [PubMed] [Google Scholar]
- 14.Sarria AJ, Panini SR, Evans RM. A functional role for vimentin intermediate filaments in the metabolism of lipoprotein-derived cholesterol in human SW-13 cells. Journal of Biological Chemistry. 1992;267:19455–63. [PubMed] [Google Scholar]
- 15.Hagberg CE, et al. Vascular endothelial growth factor B controls endothelial fatty acid uptake. Nature. 2010;464:917–21. doi: 10.1038/nature08945. [DOI] [PubMed] [Google Scholar]
- 16.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald ML, Moore KJ, Freeman MW. Nuclear hormone receptors and cholesterol trafficking: the orphans find a new home. J Mol Med (Berl) 2002;80:271–81. doi: 10.1007/s00109-001-0318-y. [DOI] [PubMed] [Google Scholar]
- 19.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plyte SE, Hughes K, Nikolakaki E, Pulverer BJ, Woodgett JR. Glycogen synthase kinase-3: functions in oncogenesis and development. Biochimica et Biophysica Acta. 1992;1114:147–62. doi: 10.1016/0304-419x(92)90012-n. [DOI] [PubMed] [Google Scholar]
- 21.Toker A, Cantley LC. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387:673–6. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 22.Kaprio J, Ferrell RE, Kottke BA, Kamboh MI, Sing CF. Effects of polymorphisms in apolipoproteins E, A-IV, and H on quantitative traits related to risk for cardiovascular disease. Arteriosclerosis and Thrombosis. 1991;11:1330–48. doi: 10.1161/01.atv.11.5.1330. [DOI] [PubMed] [Google Scholar]
- 23.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The ENCODE Project Consortium A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buyske S, et al. Evaluation of the metabochip genotyping array in African Americans and implications for fine mapping of GWAS-identified loci: the PAGE study. PLoS ONE. 2012;7:e35651. doi: 10.1371/journal.pone.0035651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmen J, et al. The functional interaction on in vitro gene expression of APOA5 SNPs, defining haplotype APOA52, and their paradoxical association with plasma triglyceride but not plasma apoAV levels. Biochimica et Biophysica Acta. 2008;1782:447–52. doi: 10.1016/j.bbadis.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nature Genetics. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Coronary Artery Disease (C4D) Consortium A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nature Genetics. 2011;43:339–44. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 29.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genetics. 2010;42:579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass ndex. Nature Genetics. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heid IM, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nature Genetics. 2010;42:949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehret GB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature Genetics. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freathy RM, et al. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes. 2008;57:1419–26. doi: 10.2337/db07-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clarke R, et al. Cholesterol fractions and apolipoproteins as risk factors for heart disease mortality in older men. Archives of Internal Medicine. 2007;167:1373–8. doi: 10.1001/archinte.167.13.1373. [DOI] [PubMed] [Google Scholar]
- 36.Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nature Genetics. 2008;40:161–9. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voight BF, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012 doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frikke-Schmidt R, et al. Association of loss-of-function mutations in the ABCA1 gene with high-density lipoprotein cholesterol levels and risk of ischemic heart disease. JAMA. 2008;299:2524–32. doi: 10.1001/jama.299.21.2524. [DOI] [PubMed] [Google Scholar]
- 39.Demirkan A, et al. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 2012;8:e1002490. doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein holesterol in plasma, without use of the preparative ultracentrifuge. Clinical Chemistry. 1972;18:499–502. [PubMed] [Google Scholar]
- 41.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 42.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nature Genetics. 2010;42:348–54. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stouffer SA, Suchman EA, DeVinney LC, Star SA, Williams RMJ. Adjustment During Army Life. Princeton University Press.; Princeton, NJ.: 1949. [Google Scholar]
- 44.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keating BJ, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chasman DI, et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.