Summary
Hubs integrate and distribute information in powerful ways due to the number and positioning of their contacts in a network. Several resting state functional connectivity MRI reports have implicated regions of the default mode system as brain hubs; we demonstrate that previous degree-based approaches to hub identification may have identified portions of large brain systems rather than critical nodes of brain networks. We utilize two methods to identify hub-like brain regions: 1) finding network nodes that participate in multiple sub-networks of the brain, and 2) finding spatial locations where several systems are represented within a small volume. These methods converge on a distributed set of regions that differ from previous reports on hubs. This work identifies regions that support multiple systems, leading to spatially constrained predictions about brain function that may be tested in terms of lesions, evoked responses, and dynamic patterns of activity.
Introduction
Hubs are intuitively important features of networks: high-volume airports are more important than smaller airfields in facilitating air travel, and people with many acquaintances are more powerful distributors of information than people with few acquaintances. Hubs, in an intuitive sense, are nodes with special importance in a network by virtue of their many, often diverse, connections.
The quantitative importance of hubs has been demonstrated in a series of graph theoretic studies (Albert et al., 2000; Albert et al., 1999; Barabasi and Albert, 1999; Jeong et al., 2001; Jeong et al., 2000). Graphs are mathematical models of complex systems (e.g., air traffic) in which the items in a system become a set of nodes (e.g., airports) and the relationships in the system become a set of edges (e.g., flights). Hubs are defined as nodes with many edges or with edges that place them in central positions for facilitating traffic over a network. The number of edges on a node is called the node’s degree, and degree is the simplest and most commonly used means of identifying hubs in graphs. Over the past decade it has become clear that many real-world networks contain nodes that vary by many orders of magnitude in their degree such that a handful of nodes have very powerful roles in networks (e.g., Google.com in the World Wide Web) (Albert et al., 1999; Barabasi and Albert, 1999; Jeong et al., 2000). The loss of such well-connected hubs can be particularly devastating to network function (Albert et al., 2000; Jeong et al., 2001; Jeong et al., 2000). Given the role of hubs and their importance to networks, the locations and functions of hubs in the brain are of clear interest to neuroscientists.
Over the past 15 years, advances in MRI techniques have enabled comprehensive estimates of structural and functional connectivity in the living human brain, leading to the first estimates of hub locations in human brain networks. In an influential study, Buckner and colleagues (Buckner et al., 2009) examined voxelwise resting state functional connectivity MRI (RSFC) networks, identifying hubs (high-degree nodes) in portions of the default mode system, as well as some regions of the anterior cingulate, anterior insula, and frontal and parietal cortex. Other investigations targeting “globally connected” regions in RSFC data have converged on similar sets of regions (Cole et al., 2010; Tomasi and Volkow, 2011). These “hubs” have garnered much interest because they are principally located in the default mode system, a collection of brain regions that are implicated in various “high-level” cognitive processes and that often degenerate in Alzheimer disease, thereby seeming to fit ideas about information integration and vulnerability to attack.
In this article we outline reasons to suspect that degree-based hubs reported in functional connectivity networks may not be hubs in the interesting and intuitive sense outlined at the beginning of this article, but rather that they might simply be members of the largest sub-network(s) (systems) of the brain. We follow two separate lines of argumentation to this conclusion. The first argument demonstrates that in networks formed using Pearson correlations (RSFC graphs are commonly formed using Pearson correlations between BOLD timeseries, and unless otherwise specified, “correlation” signifies Pearson correlation in this paper), node degree is substantially explained by sub-network size. The second argument is concerned with amplifications of the first argument that can occur when systems are not modeled at their inherent levels of organization, such as when brains (cortically organized at levels of columns, areas, and systems (Churchland and Sejnowski, 1988; Felleman and Van Essen, 1991)) are modeled as voxels (an arbitrary volumetric element).
Since some classic methods of hub identification are confounded in correlation networks, we develop two alternative methods for identifying hubs that are more suited to RSFC correlation networks. Both methods aim to identify regions of the brain that are well-situated to support and/or integrate multiple types of information. Both methods leverage the correspondence between functional brain systems (e.g., dorsal attention system) and graph sub-networks observed in recently described RSFC graphs (Power et al., 2011); see also (Yeo et al., 2011)). First, using a model of the brain at the level of functional areas we identify nodes that participate in many sub-networks of the brain (e.g., a node that has relationships with members of multiple brain systems, such as visual, default mode, or fronto-parietal control systems). These nodes are candidate brain hubs. We identify these candidate hubs using the established measure of participation coefficients (Guimerà and Nunes Amaral, 2005). Second, we examine a high-resolution brain network to identify spatial locations where many sub-networks are present within a small volume (e.g., finding, within a small sphere, voxels representing the dorsal attention, visual, fronto-parietal control, and default mode systems). We call these locations articulation points – they are not hubs in the traditional graph theoretic sense but they are locations where such hubs might be situated. Both methods identify similar sets of brain regions in the anterior insula, anterior, middle and superior frontal cortex, medial superior frontal cortex, medial parietal cortex, inferior parietal, and temporo-occipital cortex. Notably, these regions do not emphasize the default mode system.
Results
Argument 1: Degree is a confounded measure for identifying hubs in Pearson correlation networks
Several influential reports have identified brain hubs in RSFC networks using (variations of) a measure called degree (or degree centrality), which is the number of edges on a node (Buckner et al., 2009; Cole et al., 2010; Fransson et al., 2011; Tomasi and Volkow, 2010, 2011; van den Heuvel et al., 2008). Hubs, when identified by high degree, are nodes with many edges. In weighted networks the analogous measure, strength, is defined as the sum of the weights of the edges on a node.
Degree (or strength) is usually an appropriate measure for identifying hubs (e.g., an airport with 200 connections is almost certainly more important than an airport with 20 connections). In the computer network shown in Figure 1A, degree is an accurate means of identifying hubs.
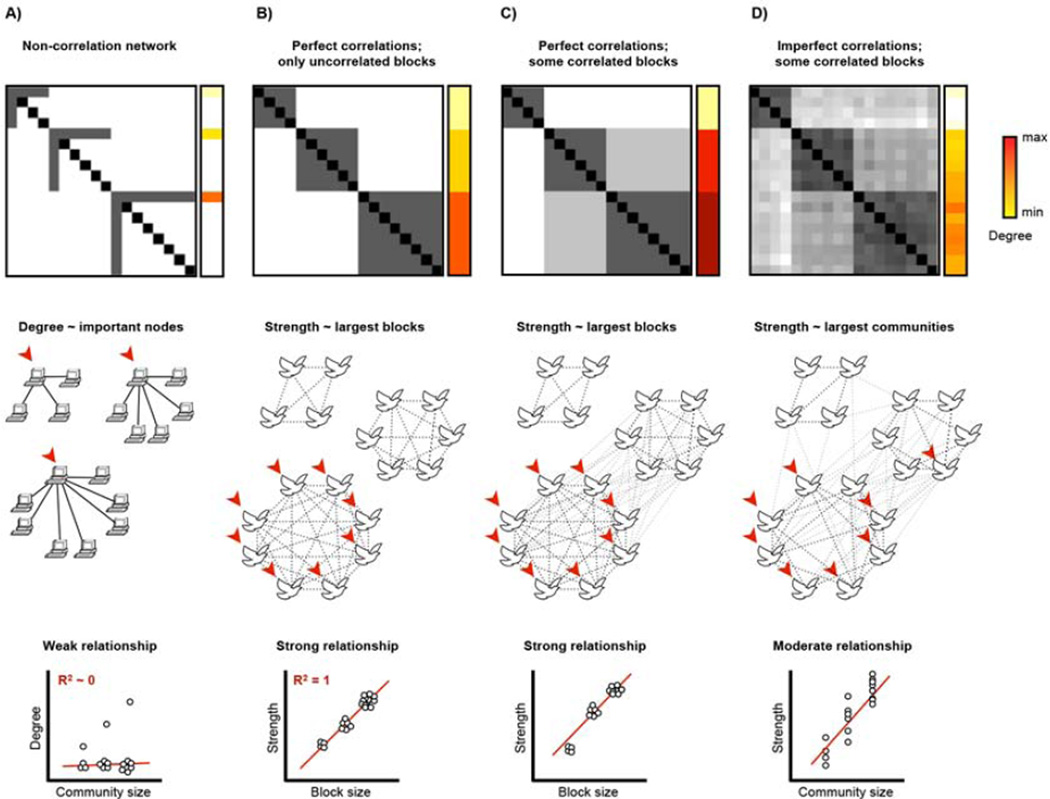
Figure 1. Degree is a problematic measure of node importance in Pearson correlation networks.
A) A computer network with 3 communities is shown. Degree identifies uniquely important nodes in the graph and there is a weak relationship between degree and community size. B) A block model corresponding to a birdsong correlation network. Three flocks are present, each singing a song uncorrelated with the other flock. C) As in (B) except that blocks are now allowed to correlate. This could correspond to a situation where there was similarity in the birdsong of different flocks, or where flocks sang the same song for limited periods of time. D) As in (C) except that the perfect correlations within blocks are relaxed to imperfect correlations. This could correspond to individual imperfections in birdsong, or individual birds switching songs occasionally.
In correlation networks, however, degree is a problematic means of identifying hubs. We argue this point using conceptual networks and real RSFC data. Two comments preface the data. First, the conceptual correlation networks in Figure 1 are presented to illustrate how the meaning of degree can change in various situations; they are not intended to be full-fledged models of RSFC signal. Second, our argument is intended to apply to networks formed using Pearson correlations; our argument may be less relevant to other types of correlation networks. We return to this topic in the Discussion.
Our argument is first demonstrated using networks of perfect correlations and then relaxed into a form that is more relevant to the imperfect correlations found in RSFC networks. Suppose there is a system composed of groups of nodes with perfectly covarying timecourses. An example is shown Figure 1B, where a system of songbirds segregates into 3 flocks singing different songs. In this example each flock sings a song with no similarity to the song of the other flock. Such a system is called a “block model” (see the matrix), and nodes within the blocks (here, flocks), are structurally equivalent, meaning they have identical sets of connections and are therefore interchangeable (Newman, 2010). All nodes within a block have identical degree, and this degree is directly related to the size of the block. Thus, degree will identify hubs in the largest blocks of the graph. If blocks correlate to any extent, then degree will depend not only on the size of a node’s block, but also on the sizes of related blocks (Figure 1C). If one relaxes “perfectly correlated” to “more correlated than average“, blocks become groups of nodes called communities and degree will tend to identify hubs in the largest communities of a correlation network (Figure 1D).
Degree thus has different meanings in different types of network. In many graphs, such as the computers of Figure 1A, high degree means that an individual node has many connections and is probably important. In others, such as the block model in Figure 1B, high degree means nothing more than that a node is part of a large block. In networks like RSFC networks, which are noisy and in which nodes may display individual temporal dynamics (Chang and Glover, 2010), degree is probably somewhat driven by unique properties of individual nodes as in Figure 1A, but also somewhat driven by community size as in Figure 1B. The meaning of degree is thus ambiguous in RSFC networks. This ambiguity has critical implications for studies that have identified hubs in RSFC on the basis of degree, since such hubs may be identified due to community size rather than important roles in information processing.
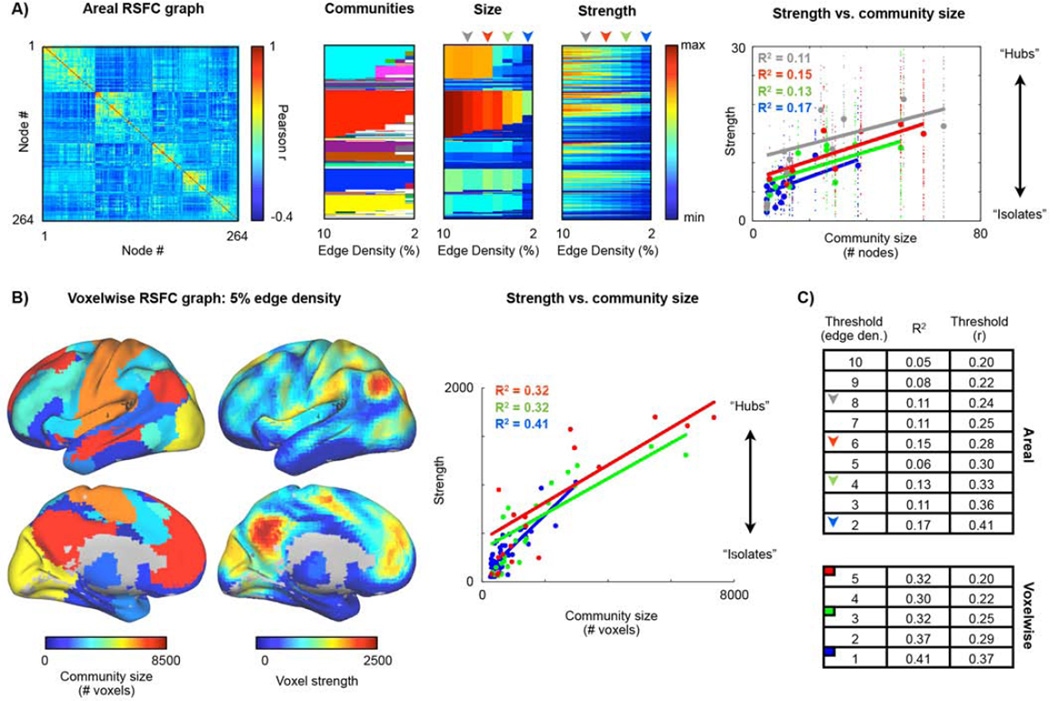
To determine whether these theoretical concerns are evident in reality, two versions of RSFC graphs were formed using data from 120 healthy young adults (60F, 24.7 ± 2.4 years old). The graphs were formed using methods consistent with the previous literature, and the relationship between community size and node strength was quantified for both graphs. Figure 2A shows the correlation matrix that defines a graph formed of 264 putative areas (Power et al., 2011), the communities found within this graph, the sizes of these communities, and node strength at multiple thresholds. Linear fits of strength to community size are plotted. There is an evident relation between community size and node strength. Similar analyses performed in a voxelwise network in the same dataset are shown in Figure 2B. In the voxelwise network the relationship between community size and node strength is considerably stronger. Because there is no “correct” threshold at which to analyze a graph these analyses were performed at many thresholds (those used in (Power et al., 2011)). Across thresholds, community size explained 11% ± 4% of the variance in strength in the areal network and 34 ± 5% of the variance in strength in the voxelwise network.
Figure 2. Degree is influenced by community size in RSFC graphs.
A) The RSFC correlation matrix of a 264-node graph in 120 young adults. Communities over a range of thresholds are shown as colors in the second panel. The number of nodes in the communities and node strengths at every threshold are shown in the third and fourth panels. A linear fit of node strength to community size is plotted for the 2, 4, 6, and 8% edge density analyses. Small dots indicate individual datapoints, large dots indicate average values in a community. Fits excluded communities with fewer than 5 nodes. The threshold range used corresponds to that used in (Power et al., 2011) and spans thresholds where many communities are present (higher edge densities such as 20% or 15% yield coarse structure with 2 or 4 communities) down to thresholds where the graph begins to fragment due to edge removal. B) Communities were identified in a voxelwise graph formed in the same subjects. For the 5% edge density analysis, for every voxel, the size of its community and its strength is shown on a brain surface. The default mode system is the largest community and contains the voxels with highest degree. Linear fits of node strength to community size at several thresholds are shown. Fits excluded communities with less than 250 nodes. These thresholds correspond to those used in (Power et al., 2011). C) R2 of linear fits of node strength to community size at several thresholds in each network (thresholds are reported in terms of edge density and the threshold used on the correlation matrix to produce the desired edge density).
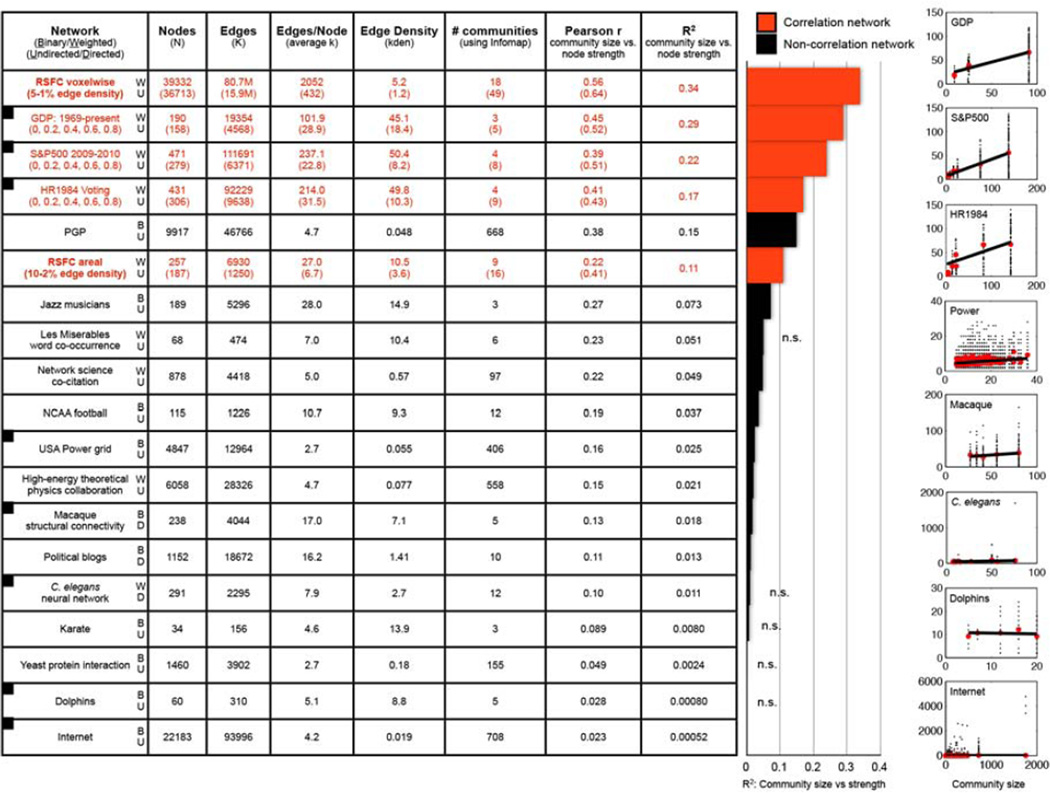
It is possible that strong relationships between strength and community size are actually typical of real-world networks. To investigate this possibility, seventeen other real-world datasets (3 correlation, 14 non-correlation) were analyzed in the manner just described (see Methods and Figures 3 and S1 for sources and details of the networks). Strong relationships between strength and community size were observed in real-world correlation networks, but were generally absent in real-world non-correlation networks, consistent with the theoretical considerations outlined above.
Figure 3. Degree is influenced by community size in Pearson correlation networks.
The table lists the properties of 19 real-world networks, 5 of which are correlation networks (red text). For each network Infomap was used to identify communities, and the the r and R2 values for linear fits of community size vs. node strength are shown. The bar graph plots the R2 values. The plots at right depict several of the linear fits (depicted networks have squares in the first column). For the correlation networks, several thresholds were analyzed (the edge densities from Figure 2 for the RSFC graphs, and r > 0, 0.2, 0.4, 0.6, and 0.8 for the 3 other correlation networks). For correlation networks, the top numbers are for the lowest threshold and the bottom numbers (in parentheses) are for the highest threshold, conveying the range of values the networks displayed; the R2 values are the mean over all analyses. As in Figure 2, fits for all graphs excluded communities with fewer than 5 nodes (and fewer than 250 nodes for the voxelwise graph). Reported graph properties reflect the properties of the nodes qualifying for the fits (small communities excluded). See Figure S1 for further details and graph properties.
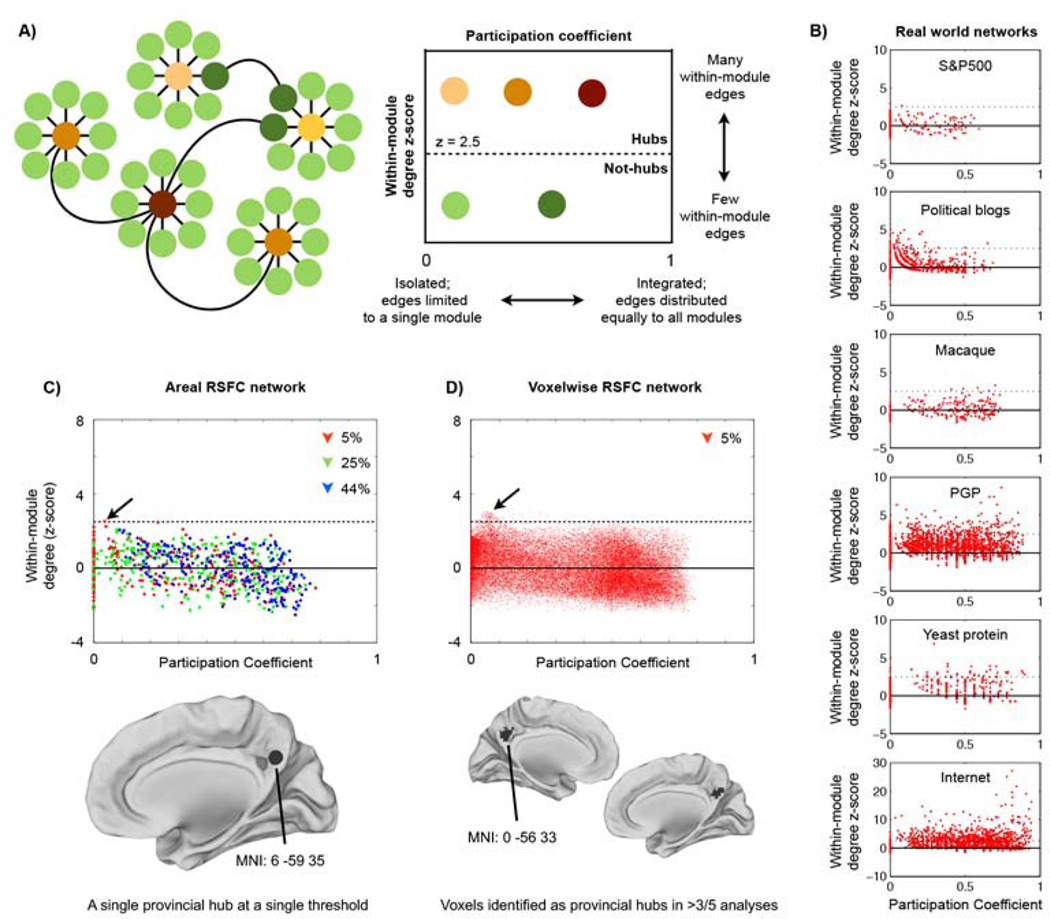
If the meaning of degree is confounded by community size in correlation networks, one might wonder whether important nodes could still be identified as nodes with high degree relative to other nodes within their community. Guimera and Amaral have proposed a widely-used classification scheme to identify node roles based on such a framework (Guimerà and Nunes Amaral, 2005). Their approach uses two measures to characterize nodes: within-module degree z-score and participation coefficient (Figure 4A). Within-module degree z-score is the z-score of a node’s within-module degree; z-scores greater than 2.5 denote hub status. Participation coefficients measure the distribution of a node’s edges among the communities of a graph. If a node’s edges are entirely restricted to its community, its participation coefficient is 0. If the node’s edges are evenly distributed among all communities, the participation coefficient is a maximal value that approaches 1 (the maximal value depends on the number of communities present). Hubs with low participation coefficients are called “provincial” hubs because their edges are not distributed widely among communities, whereas hubs with higher participation coefficients are called “connector” hubs.
Figure 4. Degree-based hubs are weak and provincial in RSFC graphs but not in other real-world graphs.
A) A model network depicting how (Guimerà and Nunes Amaral, 2005) define node roles. B) Node role plots for several real-world networks. C) Node roles were calculated in the areal RSFC graph for each threshold in the 10-2% threshold range. Only a single hub ROI was found (this is true across all positive thresholds: 44%-1% edge density). This node, in the precuneus, is a provincial hub (the black sphere). One other node immediately anterior to this node approaches but does not meet hub classification criteria (the gray sphere). D) Node roles were calculated in the voxelwise RSFC graph for each threshold in the 5-1% threshold range. Node roles at 5% edge density are plotted; this plot is typical of the other thresholds. The surfaces show locations of voxels that, across thresholds, are identified as hubs in at least 3 of 5 analyses. These voxels are provincial hubs located in the precuneus.
The node role approach indicates that the RSFC networks of Figure 2, relative to other networks such as communication or metabolic networks, are structured in ways such that they contain a very small number of hubs, all of which are quite weak by graph theoretic standards (Figure 4). In the areal network a single node at a single threshold meets criteria for being a hub. This node, in the precuneus, is a provincial hub with few strong correlations outside of its community (the default mode system). In the voxelwise network, 90–199 voxels are identified as hubs across thresholds, mainly as part of a large cluster in the precuneus. Like the single areal node, these voxels are also provincial hubs - part of the largest community in the network (the default mode system), with few strong correlations to nodes outside of their community. This provincial quality of RSFC hubs is similar to the pattern found in other real-world correlation networks (e.g., the S&P500 network in Figure 4B), but stands in contrast to the patterning of hubs found in many real-world non-correlation networks, where hubs display a wide range of participation coefficients (Figure 4B). These findings are echoed in Figure S1, where node strength correlates negatively with participation coefficients in the 3 real-world correlation networks (such that nodes with many edges are often isolated from other communities) but positively in most real-world non-correlation networks (such that nodes with many edges often contact many communities). The RSFC networks have intermediate findings: weakly negative correlations of node strength and participation coefficients, consistent with our conceptual arguments above. Importantly, though the node role approach does identify a small number of provincial hubs in RSFC networks, it still uses degree as the basis of hub identification and does not address the fundamental uncertainty about what degree signifies in correlation networks.
The essential points from this section are that 1) degree is normally a good indicator of a node’s importance in a non-correlation network, 2) degree has an unclear meaning in Pearson correlation networks due to the influence of community size, and 3) degree-based RSFC hubs may, to a substantial extent, reflect community size rather than a privileged role in information processing.
Argument 2: Volume-based graphs distort functional brain properties
Having established that RSFC correlation networks entail strong (confounding) relationships between community size and node degree, we now discuss a second problem that can amplify this relationship. Estimates of degree-based hubs in functional connectivity networks have often used voxel-based networks or approximations of them (Buckner et al., 2009; Cole et al., 2010; Fransson et al., 2011; Tomasi and Volkow, 2010, 2011; van den Heuvel et al., 2008). On the face of it, these approaches are sensible because they maximize the resolution of the analysis and minimize the possibility of conflating unique signals in a single node (Fornito et al., 2010). However, there are reasons to believe that such approaches introduce inaccuracies into representations of brain properties. Our second argument concerns the distortions that accompany volume-based models of brain organization.
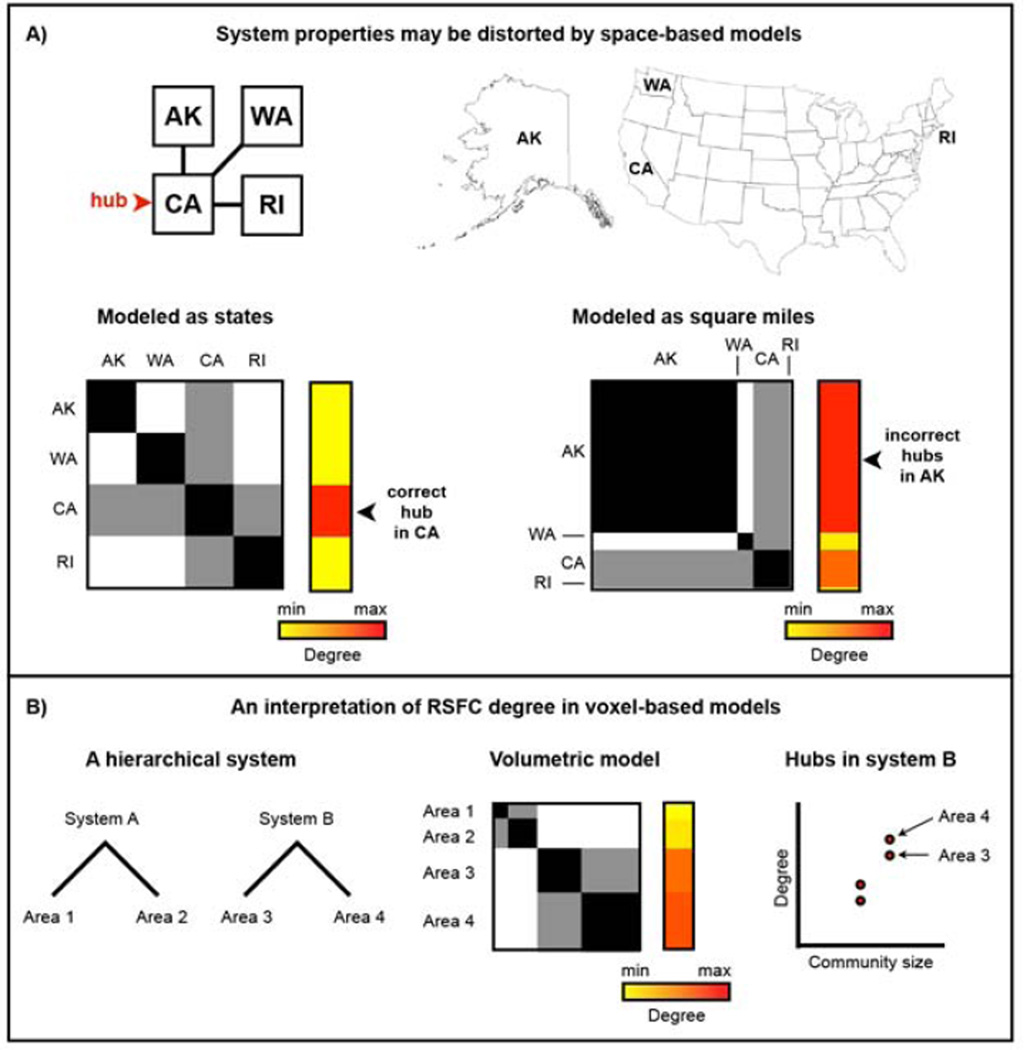
Complex systems, composed of items and their interrelationhips, are modeled as nodes and edges in graphs. For the properties of a graph to accurately reflect properties of the system it models, the nodes in the graph need to correspond to the items of the system (Butts, 2009; Power et al., 2011; Smith et al., 2011; Wig et al., 2011). Consider, for example, the set of interstate relationships shown in Figure 5A, in which California has relationships to Alaska, Washington, and Rhode Island. This spatially embedded system, organized at the level of states, can be represented using nodes of states or nodes of space. An item-based model (node = state) accurately represents this system, and identifies California as the hub of this simple network. If the same set of relationships is preserved but this system is instead represented by land area (node = square mile) the graph acquires a very different structure and hubs are identified in Alaska.
Figure 5. Volume-based models of brain organization may distort information processing properties of the brain.
A) Assume a spatially embedded economic system in which California (CA) is a hub of interstate commerce and Alaska (AK), Washington (WA), and Rhode Island (RI) play more peripheral roles. A graph in which nodes represent states correctly identifies CA as a hub. However, if states are represented by their areas (e.g., nodes of square miles), Alaska dominates the graph structure and is identified as the seat of hubs in the network simply by being the largest physical entity in the system. B) The parallels to RSFC are straightforward: areas contain voxels in proportion to their volume, and nodes within larger areas (and by extension members of larger systems) will tend be identified as hubs by degree simply because they are part of a large physical entity. Self-connections are allowed in the state graphs to emulate how voxels can and will correlate strongly to other voxels within the same area or system.
Analogous arguments apply to RSFC networks. The brain is a spatially embedded functional network: billions of neurons (in the cortex, at least) are spatially and functionally organized into columns, areas (e.g., primary visual cortex), and systems (e.g., visual system) (Churchland and Sejnowski, 1988). Areas have different sizes (Carmichael and Price, 1994), as do systems (e.g., visual vs. auditory systems). By representing the brain with voxels, a space-based model rather than an item-based model is adopted such that different areas (and systems) are represented by variable numbers of voxels. Since voxels within areas tend to have similar signals, and areas within systems have similar signals, nodes within large areas will tend to have many high correlations to other nodes within their area, and nodes within large systems will tend to have many moderate-to-high correlations to other nodes within their system.
These considerations suggest that voxel degree is driven in substantial part by the physical size of a voxel’s area and system (Figure 5B). For example, V1 may comprise hundreds of voxels whereas A1 may comprise only a few dozen voxels. The large number of strong within-area correlations in V1 will confer higher degree to voxels in this region than to voxels in A1. Similarly, the visual system spans many thousands of voxels, whereas the auditory system only includes a few hundred voxels. Voxels in the visual system will display more within-system correlations and therefore higher degree than voxels in the auditory system.
Because the locations and sizes of areas in humans are presently unknown, this argument cannot be fully demonstrated. Note, however, that the variance in node strength explained by community size rose from 11% in the areal network to 34% in the voxelwise network and also note that many communities correspond roughly to brain systems (Figure 2), all consistent with this line of argumentation. The essential point of this section is that using volumetric elements to form graphs results in network properties that may more closely represent volumetric properties of brain organization rather than the organization of information processing.
A renewed search for brain hubs
We have highlighted two difficulties with degree-based hub identification in RSFC data: the influence of community size on degree in Pearson correlation networks and the susceptibility of degree to distortion in volume-based brain networks. The latter problem can be ameliorated by proper network definition but the former problem suggests that degree has a fundamentally ambiguous interpretation in RSFC correlation networks. If degree-based methods of hub identification are confounded, can other methods identify hubs in RSFC correlation networks?
Many other centrality measures based upon combinations of degree and path length exist to characterize hubs (e.g., betweenness, closeness, eigenvector, and PageRank centralities). Some of these measures have been used to identify RSFC hubs (Achard et al., 2006; He et al., 2009; Joyce et al., 2010; Lohmann et al., 2010; Zuo et al., 2011). In many systems, such as transit networks, these centrality measures, which combine information about path length and node degree, are appropriate and interpretable. However, in correlation networks, where degree is a problematic measure, and where path lengths are often created from thresholded correlation matrices (despite ‘distances’ being already defined by the correlation coefficient), it is less clear how to interpret these measures. Other authors have used the node role approach, wherein centrality measures identify hubs (e.g., using within-module degree z-score or betweenness centrality), and then participation coefficients classify hub type (He et al., 2009; Meunier et al., 2009; Meunier et al., 2010). Possibly due to the variety of parcellation strategies employed (AAL atlas parcels, random parcellations), these studies have produced divergent descriptions of hub locations.
Due to the reservations we have expressed about degree-based measures and our lack of confidence in interpreting path-based measures in Pearson correlation networks, we have pursued different ways of identifying hubs.
Recall that hubs are parts of networks that are critical for integrating and distributing information. In graph theory, such nodes are often identified by the number of edges a node has and by the importance of a node’s edges for facilitating network traffic (Newman, 2010). In other words, it is not just the number, but also the qualities of a node’s edges that establish its importance in a network. Since the brain is composed of systems, we reason that nodes that are well-positioned to communicate among multiple systems are good candidates for being brain hubs and we utilize two methods to identify such regions.
Method 1: Hubs as nodes that participate in many functional systems in an areal network
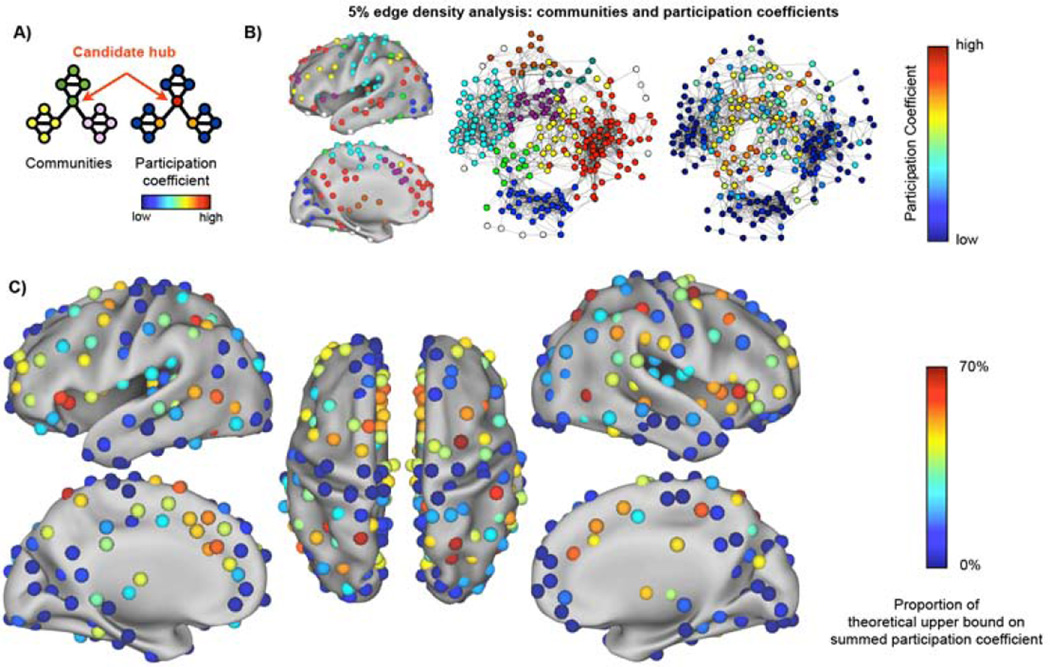
We have recently defined and described a network of 264 putative functional areas (Power et al., 2011). This graph is a first-draft model of area-level relationships in the brain, and communities in this network correspond well to functional systems (Power et al., 2011). In this areal graph, nodes that participate in multiple systems could potentially support or integrate different types of information. Our first method therefore identifies putative hubs as nodes in this areal network that have edges to many different communities. To find such nodes, we alter the node role approach of Guimera and Amaral: we discard the traditional measure of centrality due to the reservations expressed above, and instead use the participation coefficient as the sole measure of node importance. Figure 6A shows a network with 3 communities (yellow, green, and pink) and the participation coefficient of each node. Nodes in blue have no relationships outside their community and low participation coefficients whereas the red node has relationships to every community and the highest participation coefficient in the network. Our approach searches the areal network for nodes like the red node.
Figure 6. Putative hubs in the areal network identied by high participation coefficients.
A) A graph with several communities (yellow, green, pink) illustrates the meaning of participation coefficient. B) Surface and spring-embedded plots of communities in the areal graph at 5% edge density, with nodes colored by participation coefficient at right. C) Summed participation coefficients across thresholds. See Figures S2–S3 for replicability over sub-cohorts and the robustness of these calculations to data smoothing and global signal regression.
In the first half of this paper, in order to replicate and expand on previous findings related to degree-based hubs, graphs were formed in ways corresponding to the previous literature. In the second half of the paper, graphs will be formed using our preferred methodology (Power et al., 2011), which excludes short-distance relationships (less than 20 mm apart). This exclusion is performed because short-distance correlations are inflated by unavoidable steps in image processing (realigning, registration, reslicing), partial voluming, and head motion (Power et al., 2012). Additionally, short-distance correlations are virtually always high (the bloom around any seed in a seed map), thus acting as a spatial lattice of high short-range correlations that provide little distinguishing information between nodes. Eliminating correlations spanning less than 20 mm removes 4% of the edges in both the areal and voxelwise graph, and does not alter our observations about the confounding relationship between community size and degree in RSFC graphs (Figure S1).
An areal network was formed in 120 healthy young adults and community assignments were obtained over many thresholds (10% to 2% edge density in 1% steps) as in (Power et al., 2011). Figure 6B shows the participation coefficients in the average network at a single threshold. The participation coefficients were summed over thresholds to identify nodes that routinely participate in multiple communities, and the summed participation coefficients are plotted in Figure 6C.
Several control analyses were performed to establish the robustness of these results. Identical analyses performed in matched 40-subject sub-cohorts of the main cohort yielded very similar results (Table S1 and Figure S2; correlations between sub-cohorts = 0.87 ± 0.04). Identical analyses performed without global signal regression also produced very similar results (Figure S3; r = 0.83). Because short-distance relationships were excluded, nodes identified by high participation coefficients are not identified simply for being proximal to nodes belonging to other communities. Because nodes within relatively small communities tend to have higher participation coefficients (Figure S1), we also counted the number of communities contacted by each node, and this index, which is not biased by community size, identified a similar set of nodes as participation coefficients (Figure S3; r = 0.85).
Method 2: Spatial locations that contain many functional systems in a modified voxelwise network
We have also proposed a high-resolution modification of voxelwise networks (Power et al., 2011). Communities in this graph are in good agreement with functional systems and with communities in the areal network (Power et al., 2011). This graph has excellent spatial resolution but also some distorted network properties (see Argument 2). We therefore focus on the spatial properties of this model. Our second method examines the spatial topography of this graph to identify locations where many communities are present within a small volume. Such locations, which we call articulation points, would be well-suited for integrating (or distributing) a variety of types of information represented in different systems.
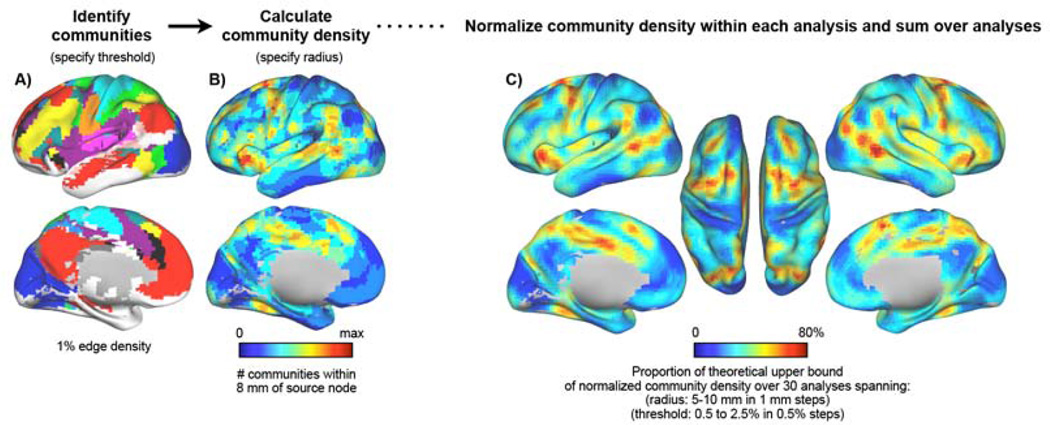
Figure 7 outlines our methodology. A modified voxelwise network was formed in the 120-subject cohort and community assignments were obtained for all voxels in the AAL atlas (cortical and subcortical) over multiple thresholds (2.5% to 0.5% edge density in 0.5% steps) as in (Power et al., 2011). Community density was then calculated for each voxel as the number of unique communities found within some radius of that voxel (see Methods). Radii of 5 to 10 mm in 1 mm steps were sampled. We use high thresholds because more communities are detected at high thresholds, yielding more focal community density maps (often articulations of 4–7 commumities); at lower thresholds, fewer communities are found (often 4–6), yielding non-focal maps of community density. A representative analysis at threshold 1% and radius 8 mm is shown in Figure 7A and 7B. To identify peaks in community density that are reliable across thresholds and sampling distances, results were summed from analyses performed across these parameter spaces after normalizing the values within each analysis (Figure 7C). The topography of community density is very similar in 40-subject sub-cohorts of the main cohort and across parameter spaces (Figure S4; correlations between sub-cohorts = 0.85 ± 0.04). When calculating community density, each hemisphere was analyzed separately to avoid contributions from tissue across the midline, and subcortical structures were excluded from calculations to avoid inflated estimates in the insula (Figure S5).
Figure 7. Articulation points: brain locations that are densely populated by functional systems.
A) Communities in the 1% edge density analysis is shown; colors represent communities. All communities with fewer than 125 voxels are colored white (and are treated as a single community in community density calculations). B) Community density is calculated as the number of unique communities present within some distance of a source node (here, within 8 mm of a source voxel, in the 1% edge density analysis). C) Summed community density. See Figures S4–S5 for analyses of the influence of subcortical and contralateral tissue on these calculations, replicability in sub-cohorts, and stability over the parameter spaces of thresholds and sampling radii. The data in this figure and subsequent figures are derived from single-hemisphere community analyses of all voxels within the AAL atlas, followed by community density calculation excluding subcortical structures.
Integration and generalizability of these findings
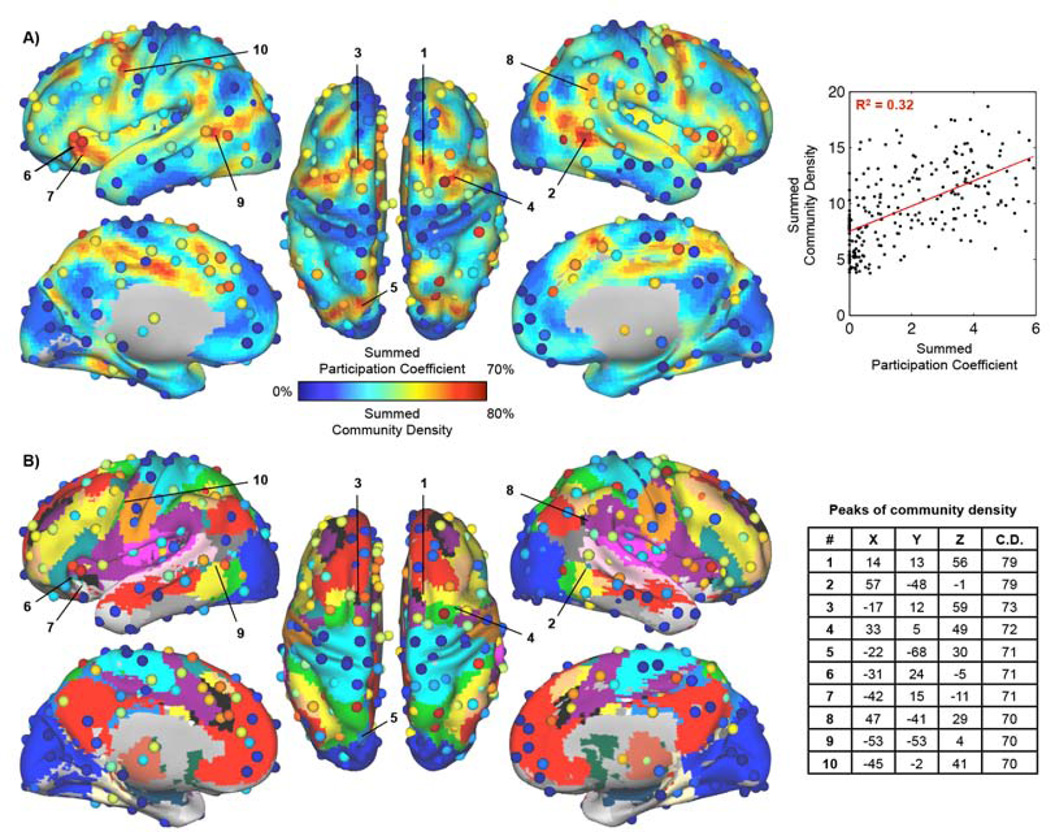
We have developed two methods aimed at identifying brain regions that support or integrate multiple functional systems. Figure 8 plots the results of both methods on a single surface and with reference to the consensus community assignments from (Power et al., 2011) (see Figure S6 for flat maps). Nodes with high participation coefficients tend to be adjacent to regions of high community density, though this is not always the case (e.g., left intraparietal sulcus). This proximity is consonant with our reasoning that brain regions where multiple functional systems are represented would be good locations for hubs.
Figure 8. Areal participation coefficients plotted over community density, with consensus communities for reference.
A) Overlaid data from Figures 6 and 7. The correlation between the two measures is r = 0.57 (calculated in ROIs where at least 10/19 voxels were defined in the community density analysis, 245/264 ROIs). B) The consensus community assignments from (Power et al., 2011) are provided as a reference to illustrate the communities present near areas of high community density. Positions and MNI coordinates for peaks in community density are shown. See Table S2 for ROI locations and summed measures. See Figure S6–S8 for flat-map illustrations of the same data, similar findings in a separate cohort, and plots of the interdependence of participation coefficient, community density, and degree.
This proximity is also consistent with an argument that high participation coefficients arise from signal blurring due to proximity to several distinct signals. The data were therefore reanalyzed without spatial blurring as part of functional connectivity processing, yielding results very similar to those with blurring (r = 0.94, Figure S3).
The subjects studied thus far are mainly university students who met strict inclusion criteria. To determine whether our results generalize to more typical populations, a 40-subject cohort (40F, 30.0 ± 3.2 years old) from a prospective twin study in the general population was also examined, including subjects with psychiatric and neurologic disease and psychotropic medication use. Analyses identical to those shown in Figure 8 were performed on this cohort. Results in the main 120-subject cohort explained 74% of the variance in summed participation coefficients and 77% of the variance in summed community density in this accessory cohort (Figure S7).
Discussion
Hubs exist in many real-world networks and they often play critical roles in facilitating network traffic and maintaining network integrity (Albert et al., 2000; Albert et al., 1999; Jeong et al., 2001). In this report, we aimed to advance the study of brain hubs by clarifying some important issues and by providing some conceptually straightforward methods to identify putative hubs in RSFC correlation networks. We now discuss our findings and their implications for previous and future work.
Interpreting degree-based hubs
Several points are worth noting when considering how to interpret degree-based hubs. First, unlike many real-world networks, RSFC networks formed using Pearson correlations do not tend to contain nodes that are convincing outliers in strength, meaning that any degree-based hubs in RSFC networks are rather weak hubs from a graph theoretic perspective (Figure 4). Second, given the block-like structure of correlation networks, these hubs tend to be provincial, meaning their connections are largely restricted to their community (Figure 4). This provincial quality stands in contrast to hubs found in many real-world networks, which often connect strongly to a wide variety of other communities (Figure 4 and Figure S1). Third, strength in RSFC graphs strongly reflects community size, which is indirectly related to the physical sizes of areas and systems (Figures 1–5). Fourth, degree can represent the dynamic and unique couplings of a node to other nodes, a property of great interest that is clouded by the considerations just discussed.
It is possible that some degree-based hubs (like those in the precuneus) are provincial hubs that play central roles in particular systems. It is also possible that these hubs do not have hub-like roles in information processing and that their “hubness” arises from the factors discussed above. We shall return to this topic.
Interpreting our areal hubs and articulation points
In the areal network, nodes represent our current best estimate of the centers of brain areas (Power et al., 2011). If a node has a high participation index, it has modest-to-high correlations with multiple communities. Since these communities correspond reasonably well to systems (Power et al., 2011), we infer that such nodes likely have access to a variety of types of different information processing represented among different systems.
In the modified voxelwise network, nodes do not correspond to any “unit” of brain organization. Here, the peaks in community density represent points of spatial articulation between multiple brain systems. These peaks do not represent areas but rather locations where areas from multiple systems exist in close proximity to one another. Cortex in such regions does not necessarily integrate different types of information but would be well-situated to perform such integration.
On relationships between community density, participation coefficients, and degree
Regions with high community density tend to have high participation coefficients (Figure 8A). Convergence between measures is especially prominent at some regions in the anterior insula, dorsal medial prefrontal cortex, dorsal prefrontal cortex, lateral occipito-temporal cortex, and superior parietal cortex. There are also some regions where the measures diverge, such as the inferior parietal sulcus (high participation coefficient, low community density) or the mid-cingulate (low participation coefficient, high community density). Differences between the measures in these latter regions may be of eventual interest, but our present focus is on regions where both measures are congruent.
The methods advocated in this report generally highlight different parts of the brain than degree-based methods. Indeed, community density and node strength (normalized and summed across thresholds) are negatively correlated (r = −0.37, Figure S8), as are participation coefficient and node strength (r = − 0.12, Figure S8). No analogue of community density exists in the real-world graphs, but the relationship between participation coefficient and node strength seen across networks in Figure S1 is instructive: it is strongly negative in the 3 real-world correlation networks, mildly negative in the RSFC networks and in a few real-world non-correlation networks, but usually positive in real-world non-correlation networks. This is consistent with the idea that RSFC networks occupy a conceptual space somewhere between the computer and birdsong networks of Figure 1. The negative relationship between node strength and our measures of node importance indicates a spatial complementarity that may be leveraged when trying to discern if and how various measures (e.g., degree vs. participation coefficient) denote hub-like roles in cognition, as discussed below.
Evaluating the functional importance of hubs
A challenging topic is how to characterize the functional role of hubs. One approach might be to study the various systems involved with a hub. However, the functions performed by systems are often unclear. For example, what functions are performed by the default mode system or cingulo-opercular systems? There are many ideas but little consensus. Another approach is to examine the proposed functions of individual hub regions. However, the brain is everywhere “integrative” in some sense and the “functions” of much non-sensory cortex are contested or unknown. Defensible conclusions about hub-like processing seem unlikely to emerge from this approach.
Another approach would be to study the hodology of hub regions and to infer the function and importance of a hub from the physical projections it sends and receives. This approach may prove quite fruitful. However, it also has important limitations. First, because detailed anatomical information is mainly available in non-human primates, inferences in humans would depend on the similarity between human and non-human primate anatomy (and function). Our hub regions and degree-based hubs largely avoid unimodal sensory or motor cortex, making such inferences tenuous. Second, the relationship between the structural and functional properties of a network are not simple or clear. For example, it is not obvious that hubs in a structural network should correspond to a degree-based hub in a functional network, or even a hub of the sort we are advocating (Honey et al., 2009). There is no doubt that anatomical connections, chemoarchitecture, and cytoarchitecture will eventually inform our understanding of hub location and function, but they may not be the most fruitful starting point for creating functional descriptions of hubs at present.
We suggest a lesion-based approach to characterizing hub function. Hubs are interesting because they are single nodes that exert disproportionate influence over network structure and dynamics due to of the number and placement of their edges. As such, their elimination can produce profound effects in a network (Albert et al., 2000; Jeong et al., 2001; Jeong et al., 2000). Our observations lead to several predictions in the brain. The removal of a provincial hub should produce effects mainly within a single community, with limited impact on global network function. The removal of the sort of hubs identified in this report should produce effects within multiple communities, producing more global effects in the network. The removal of non-hub nodes should minimally alter community and global network function. These predictions can be tested by studying spontaneous activity, evoked activity, and behavior in the context of transient or permanent inactivation of nodes. Recently, RSFC results consistent with these predictions were reported by Gratton and colleagues (Gratton et al., 2012): lesions to nodes with high participation coefficients decreased network modularity, but lesions to nodes with high within-module degree did not produce such effects.
Future Directions
Our methods targeted brain regions that may play roles in multiple brain systems. Lesion studies could offer strong support for this characterization. The large nature of most lesions makes it difficult to draw firm conclusions along such lines from the literature, but inroads may be possible using voxel-based lesion symptom studies (e.g. (Bates et al., 2003)). Studies that target hubs using TMS combined with comprehensive investigations of cognitive function (e.g., (Pitcher et al., 2009)) may also possess sufficient precision to test this hypothesis. Alternatively, investigation of temporal dynamics at hub locations using RSFC, EEG, or MEG could test and refine our observations. We are actively pursuing the lesion-based and dynamic implications of this work.
This study has outlined some difficulties in using graph theoretic techniques in RSFC data. Measures like degree, and probably path length, have unclear significance in Pearson correlation networks. Other properties, like community structure or participation coefficients, remain relatively interpretable. The Pearson correlation is widely used in RSFC due to its familarity, its simplicity of interpretation (the linear dependence between timeseries), and the ability to study large sets of nodes (264 and 40,100 in this study). Future studies that elaborate on the significance of existing graph theoretic measures in Pearson correlation networks will improve the ability of the field to utilize and interpret such networks, as will studies that propose measures designed for use in such networks. Alternative methods of RSFC edge definition, perhaps based on partial correlations or generative models, may enable more standard interpretations of graph theoretic measures. However, experience with such techniques is at present mainly limited to small networks (of a few dozen nodes or less) and it is not clear how well such approaches can scale to networks of the size explored in this report. Despite these complexities, the validation of methods that expand the utility of graph theoretic approaches in RSFC networks will be a valuable step forward for the field.
Limitations
The present work is based on analyses of RSFC data and shares the general limitations of this technique. Two limitations are especially worth noting.
First, RSFC is focused on low-frequency fluctuations in BOLD signal that only indirectly reflect neuronal activity via blood oxygenation. Our characterization of a node’s “participation” with different systems is inferential, based on correlations in these spontaneous fluctuations, not demonstrations of causal interactions. However, because temporal coherence in BOLD activity (in fMRI or RSFC) is typically interpreted to represent functional relationships, and because our study is essentially exploratory, this limitation may also be viewed as a strength. By studying spontaneous correlations, we placed no particular limitations on the types of information processing that might occur, thereby obtaining a less constrained, more “natural” sampling of interactions between brain regions than a task-based experiment would provide.
The second principal limitation of this work is spatial resolution. In our RSFC analyses, BOLD activity is sampled in voxels 3–4 mm on each side. Blurring of data is unavoidable in the process of data realignment, resampling, registration, and subject averaging. As such, nearby voxels share signal for non-biological reasons, hampering accurate estimation of BOLD correlations between brain regions. In network analyses, this means that spatially proximal relationships contain artifactual influence, but also that distant relationships (from node X to node Y) could be influenced (if voxels similar to voxel Y are present near node X and are blurred into X’s signal). We have made every effort to discount these effects, including ignoring relationships between voxels or ROIs less than 20 mm apart, reanalyzing data without blurring, and analyzing hemispheres separately in the modified voxelwise graphs to avoid the particularly high homotopic correlations that might also reflect local blurring (though dual-and single-hemisphere results were very similar, Figure S5). However some blurring of data is unavoidable and one could argue that participation coefficients are increased near regions of high community density due to blurring of signals.
Although this effect is likely present, several lines of evidence suggest that its impact is modest and did not drive the present results. First, because we only examined strong correlations (within the top few percentiles of positive correlations), blurring would have to induce very large changes in correlations to create edges that would enter our analyses for spurious reasons (unlike if we had examined thresholdfree graphs). Second, the fact that nodes with higher participation indices did not have high degree, despite being in the vicinity of many functional systems, also suggests that blurring did not spuriously induce widespread correlations to distal nodes in multiple communities at nodes proximal to multiple systems. Finally, even if high participation coefficients were due to proximity to multiple community representations, it would not detract from the observation that certain parts of the brain are densely populated with systems, and the predictions this observation entails.
Conclusions
In this report we demonstrated that brain regions previously identified as degree-based hubs in RSFC graphs may have been identified because they are members of large areas or systems rather than because of special roles in information processing. Guided by the intuitive notion of what makes hubs important, we developed approaches to search for nodes that link different communities of areal brain networks and to identify brain locations where multiple systems exist in close proximity. By re-contextualizing the nature of previously reported hubs, and by identifying a new set of hub regions with conceptually different properties, this work generates new, spatially constrained predictions about brain function that may be tested in a variety of experimental settings.
Experimental Procedures
Subjects
For the main analyses, 120 healthy young adults (60M/60F; 25.0 ± 2.4 years old) were recruited from the Washington University campus and the surrounding community. All subjects were native English speakers, right-handed, reported no history of neurological or psychiatric disease, and were not on psychotropic medications. For sub-cohort analyses, sub-cohorts were matched on sex, age, and all QC-related measures (see Table S1). For the generalizability analyses an accessory cohort of 40 subjects from a twin study in the general population (40F; 30.0 ± 3.2 years old) was examined. These subjects were recruited with relaxed restrictions on handedness (4 left-handed, 4 ambidextrous), psychotropic medication use (8 subjects), and reported psychiatric or neurological history (6 subjects). Only one twin from each twin pair was examined. All subjects gave informed consent and were compensated for their participation.
Data Collection and Processing
All subjects were scanned in a Siemens MAGNETOM Tim Trio 3.0T scanner with a Siemens 12 channel Head Matrix Coil (Erlangen, Germany). A T1-weighted sagittal MP-RAGE was obtained (TE = 3.06 ms, TR-partition = 2.4 s, TI = 1000ms, flip angle = 8°, 127 slices with 1×1×1 mm voxels). A T2-weighted turbo spin echo structural image (TE = 84 ms, TR = 6.8 s, 32 slices with 2×1×4 mm voxels) in the same anatomical plane as the BOLD images was also obtained to improve alignment to an atlas. Functional images were obtained using a BOLD contrast sensitive gradient echo echo-planar sequence (TE = 27 ms, flip angle = 90°, in-plane resolution= 4×4 mm; volume TR = 2.5 s). Whole brain coverage for the functional data was obtained using 32 contiguous interleaved 4 mm axial slices. The number of volumes obtained in the main cohort was 336 ± 121 (range 184–724). In the accessory cohort: 386 ± 35 (range 264–396).
Functional images underwent standard fMRI preprocessing to reduce artifacts (Shulman et al., 2010). These steps included: (i) sinc interpolation of all slices to the temporal midpoint of the first slice, accounting for differences in the acquisition time of each individual slice, (ii) correction for head movement within and across runs and (iii) within-run intensity normalization to a whole brain mode value (across voxels and TRs) of 1000. Atlas transformation of the functional data was computed for each individual via the MP-RAGE scan. Each run was then resampled in atlas space on an isotropic 3 mm grid combining movement correction and atlas transformation in a single interpolation (Shulman et al., 2010).
Functional Connectivity (RSFC) Processing
BOLD runs were obtained from subjects fixating a white crosshair on a black background for RSFC data. When preparing this data, standard processing steps were utilized to reduce spurious variance unlikely to reflect neuronal activity (Fox et al., 2009). These steps included: (i) a multiple regression of nuisance variables from the BOLD data, (ii) a frequency filter (f < 0.08 Hz) using a 1st order Butterworth filter in forward and reverse directions, and (iii) spatial smoothing (6 mm full width at half maximum). Nuisance regressions included ventricular signal averaged from ventricular regions of interest (ROIs), white matter signal averaged from white matter ROIs, whole brain signal averaged across the whole brain, six detrended head realignment parameters obtained by rigid body head motion correction, and the derivatives of these signals and parameters.
Motion scrubbing
Head motion can cause spurious but spatially structured changes in RSFC correlations (Power et al., 2012; Van Dijk et al., 2012). The data in this report underwent a “scrubbing” procedure (see (Power et al., 2012; Power et al., 2013)) to minimize motion-related effects. This procedure uses temporal masks to remove motion-contaminated data from regression and correlation calculations by excising unwanted data and concatenating the remaining data. For this report, the data were first processed without temporal masks. Then volume-to-volume head displacement (FD) was calculated from realignment parameters and volume-to-volume signal change (DVARS) was calculated from the functional connectivity image. A temporal mask was formed by flagging any volume with FD > 0.2 mm, as well as volumes 2 forward and 2 back from these FD-flagged volumes to account for modeled temporal spread of artifactual signal during temporal filtering. Any volume with DVARS > 0.25% change in BOLD signal was also flagged. The data were then re-processed using temporal masks that excluded all flagged volumes. Because regressions precede temporal filtering, the betas generated from the censored regressions were applied to the entire uncensored dataset to generate residuals, which were temporally filtered, followed by re-censoring for correlation calculations. In this way, motion-contaminated data contributed to neither regressions nor correlations, and temporal spread of artifactual signal during temporal filtering was minimized by augmenting temporal masks. This procedure removed 26 ± 18% (range 1 – 74%) of the data from the 120-subject cohort, leaving 245 ± 107 (range 126 – 715) volumes of usable data per subject. In the accessory cohort, 22 ± 16% (range 4 – 68%) of the data were removed, leaving 300 ± 70 (range 125 – 379) volumes of data per subject.
Node Definitions
For the areal network, a collection of 264 ROIs defined in (Power et al., 2011) were used as network nodes (Table S2). These ROIs represent the centers of putative functional areas defined by task fMRI meta-analyses and by the fc-Mapping technique (Cohen et al., 2008). All ROIs are modeled as 10 mm diameter spheres centered upon ROI coordinates.
For the voxelwise and modified voxelwise networks, all voxels (N = 40,100) within the AAL atlas (Tzourio-Mazoyer et al., 2002) were used as in (Power et al., 2011). All voxels are cubes with sides of 3 mm.
Edge Definitions
The subject-specific temporal masks formed from Motion Scrubbing were applied to each subject’s reprocessed data, and a correlation matrix was calculated from node RSFC timecourses (e.g., 264 nodes yields a 264 × 264 correlation matrix in each subject). For the main analyses, 120-subject average matrices were used. All averages and comparisons of correlations use Fisher z(r) transformations for calculations, followed by reconversion to Pearson r values for reporting.
In Figure 2, all correlations were used regardless of the distances between nodes for consistency with the previous literature. Short-distance correlations can arise from shared patterns of local neuronal activity, but they can also arise from data processing (e.g., blurring, reslicing) and from head motion (Power et al., 2012). To minimize the effects of questionable correlations on network structure, as in (Power et al., 2011), short-distance correlations (Euclidean distance < 20 mm) were excluded from graph analyses in Figures 6–8.
Graph Analyses
Graphs were formed using the nodes and edges described above. Traditionally, analyses of weighted graphs must ignore negative edges and explore a range of thresholds to characterize the properties of a network (Rubinov and Sporns, 2010). Proposals have been made to modify some graph theoretic measures for unthresholded matrices (Rubinov and Sporns, 2011), but here we follow the traditional approach. Many real-world networks have edge densities of a few percent or less (see Figure 3) and the graph measures used in this paper are developed in such networks. Accordingly, we applied thresholds to graphs to bring them to similar levels of sparseness (~10%−2% for the areal graph, 5%−0.5% for the voxel-based graphs) as in (Power et al., 2011). In general, results are presented over a range of thresholds to give the reader a sense of the (lack of) dependence of a property upon thresholds, and no formal definition of threshold ranges is proposed since it is essentially arbitrary. Our thresholds matched the ranges used in (Power et al., 2011), which were chosen to 1) yield complex and interesting community structures (> 4 communities), and 2) occupy a range of edge densities often seen in the real-world networks in which techniques like Infomap and measures like participation coefficients were originally developed.
Community detection was accomplished by subjecting thresholded (weights retained) graphs to the InfoMap algorithm (Rosvall and Bergstrom, 2008), one of the best-performing community detection algorithms currently available (Fortunato, 2010), as in (Power et al., 2011).
Degree (strength) was calculated as the sum of binary (weighted) edges on a node at a given threshold. Participation coefficients and within-module z-scores were calculated after (Guimerà and Nunes Amaral, 2005) on thresholded graphs. Relevant formulas are provided below.
Degree for node i is defined as ki = Σj Aij, where Aij is the adjacency matrix of the graph. Within-module z-score for node i is defined as , where κi is the number of edges of node i to other nodes in its module si, κ̄si is the average of κ over all the nodes in si, and σsi is the standard deviation of κ in si. Participation index for node i is defined as , where κis is the number of edges of node i to nodes in module s, ki, is the degree of node i, and NMis the total number of modules in the graph.
In Figure 6, the areal graph was analyzed at 9 thresholds (10% to 2% edge density in 1% steps) and the participation coefficients arising from InfoMap community assignments were summed and plotted as the proportion of the theoretical upper bound attainable over thresholds.
In Figure 7, the modified voxelwise network was analyzed at 5 thresholds (2.5% to 0.5% edge density in 0.5% steps; these thresholds all displayed complex community structure and focal articulation points, see Figure S4) and the number of unique communities present within a certain radius of the center of a source voxel were calculated using InfoMap community assignments. Radii of 5–10 mm in 1 mm steps were sampled. Thus Figure 7 shows the results pooled from 30 analyses (5 thresholds × 6 radii; each analysis normalized to its maximal value).
Computations and Visualizations
MRI preprocessing and RSFC processing was performed with in-house software. Network calculations were performed in Matlab (2007a, The Mathworks, Natick, MA). Brain visualizations were created with Caret software and the PALS surface (Van Essen, 2005; Van Essen et al., 2001). Consensus assignments from (Power et al., 2011) are available at: http://sumsdb.wustl.edu/sums/directory.do?id=8293343&dir_name=power_Neuron11.
Real-world Graphs
The real-world graphs presented in Figures 3, 4, and S1 are publicly available datasets (www-personal.umich.edu/~mejn/netdata/). The citations for the networks are:
Yeast protein: (Jeong et al., 2000); Network science co-citation: (Newman, 2006); Political blogs: (Adamic and Glance, 2005); Les Miserables word co-occurrence: (Knuth, 1993); High-energy theory collaborations: (Newman, 2001); NCAA football: (Girvan and Newman, 2002); USA power grid: (Watts and Strogatz, 1998); C. elegans neural network: (Watts and Strogatz, 1998); Karate club: (Zachary, 1977); Dolphins: (Lusseau et al., 2003); Internet: Mark Newman, unpublished; Macaque: (Harriger et al., 2012); Jazz musicians: (Gleiser and Danon, 2003); PGP: (Boguñá et al., 2004); GDP: (Frank and Asuncion, 2010)
GDP by country in present day dollars, 1969-present: http://www.ers.usda.gov/data-products/international-macroeconomic-data-set.aspx
S&P500 2009–2010: http://pages.swcp.com/stocks/#historical%;20data
US House of Representatives voting patterns, 1984: http://archive.ics.uci.edu/ml/datasets/Congressional+Voting+Records
For the GDP dataset, one year was removed due to corrupted data. In the HR1984 dataset, one representative was removed who abstained from every vote. For the S&P500 dataset, if a stock was off of the S&P for more than 5 of the possible 245 days, it was removed from the analysis. All other missing days were replaced with within-stock mean values. Real-world correlation networks were analyzed with and without global signal regression. For congruence with RSFC results, results with global signal regression are presented. Results without global signal regression were similar, with even stronger relationships between community size and node strength.
Supplementary Material
Highlights.
-
-
Reveals confounds in degree-based hub detection techniques in correlation networks
-
-
Utilizes multiple methods to convergently identify hubs in correlation networks
-
-
Identifies regions and nodes that support and link different parts of brain networks
-
-
Generates differential, testable, and spatially constrained hypotheses regarding hubs
Acknowledgments
The impetus to write this paper came from discussions during the 2011 Summer Institute for Cognitive Neuroscience. We thank Tom Pearce, Steve Nelson, Chris Fetch, and Brad Miller for comments on an earlier version of the manuscript, and Jessica Church, Joe Dubis, Eric Feczko, Katie Ihnen, Maital Neta, and Alecia Vogel for data contribution. This work was funded by NIH F30 MH940322 (J.D.P.), NIH R21NS061144 (S.E.P.), a McDonnell Foundation Collaborative Action Award (S.E.P.), Simons Foundation Award 95177 (S.E.P.), NIH 5R01 HD057076-03-S1 (B.L.S.), NIH R01HD057076 (B.L.S.), and NSF IGERT DGE-0548890 (Kurt Thoroughman). Data were acquired with the support of NIH K12 EY16336 (John Pruett), NIH K01DA027046 (C.N.L.-S.), the Barnes-Jewish Hospital Foundation (C.N.L.-S.), the McDonnell Center for Systems Neuroscience at Washington University (C.N.L.-S.), and the Alvin J. Siteman Cancer Center (via NCI Cancer Center Support Grand P30 CA91842) (C.N.L.-S.). This project was supported by the Intellectual and Developmental Disabilities Research Center at Washington University (NIH/NICHD P30 HD062171).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jonathan D Power, Email: powerj@wusm.wustl.edu.
Bradley L Schlaggar, Email: schlaggarb@neuro.wustl.edu.
Christina N Lessov-Schlaggar, Email: schlaggc@psychiatry.wustl.edu.
Steven E Petersen, Email: sep@npg.wustl.edu.
References
- Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2006;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamic LA, Glance N. The political blogosphere and the 2004 U.S. election: divided they blog. Proceedings of the 3rd international workshop on Link discovery; New York, NY, USA, ACM. 2005. pp. 36–43. [Google Scholar]
- Albert, Jeong, Barabasi Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Albert R, Jeong H, Barabasi A-L. Internet: Diameter of the World-Wide Web. Nature. 1999;401:130–131. [Google Scholar]
- Barabasi, Albert Emergence of scaling in random networks. Science (New York, N.Y.) 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Bates E, Wilson SM, Saygin AP, Dick F, Sereno MI, Knight RT, Dronkers NF. Voxelbased lesion-symptom mapping. Nat Neurosci. 2003;6:448–450. doi: 10.1038/nn1050. [DOI] [PubMed] [Google Scholar]
- Boguñá M, Pastor-Satorras R, Díaz-Guilera A, Arenas A. Models of social networks based on social distance attachment. Physical Review E. 2004;70:056122. doi: 10.1103/PhysRevE.70.056122. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butts CT. Revisiting the Foundations of Network Analysis. Science. 2009;325:414–416. doi: 10.1126/science.1171022. [DOI] [PubMed] [Google Scholar]
- Carmichael ST, Price JL. Architectonic subdivision of the orbital and medial prefrontal cortex in the macaque monkey. The Journal of Comparative Neurology. 1994;346:366–402. doi: 10.1002/cne.903460305. [DOI] [PubMed] [Google Scholar]
- Chang C, Glover GH. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. NeuroImage. 2010;50:81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchland PS, Sejnowski TJ. Perspectives on cognitive neuroscience. Science (New York, N.Y.) 1988;242:741–745. doi: 10.1126/science.3055294. [DOI] [PubMed] [Google Scholar]
- Cohen AL, Fair DA, Dosenbach NUF, Miezin FM, Dierker D, Van Essen DC, Schlaggar BL, Petersen SE. Defining functional areas in individual human brains using resting functional connectivity MRI. NeuroImage. 2008;41:45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Pathak S, Schneider W. Identifying the brain's most globally connected regions. NeuroImage. 2010;49:3132–3148. doi: 10.1016/j.neuroimage.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cerebral Cortex (New York, N.Y.:1991) 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Bullmore ET. Network scaling effects in graph analytic studies of human resting-state FMRI data. Frontiers in Systems Neuroscience. 2010;4:22. doi: 10.3389/fnsys.2010.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato S. Community detection in graphs. Physics Reports. 2010;486:75–174. [Google Scholar]
- Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. Journal of Neurophysiology. 2009;101:3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank A, Asuncion A. UCI Machine Learning Repository. University of California, Irvine, School of Information and Computer Sciences; 2010. [Google Scholar]
- Fransson P, Aden U, Blennow M, Lagercrantz H. The functional architecture of the infant brain as revealed by resting-state FMRI. Cerebral Cortex (New York, N.Y.:1991) 2011;21:145–154. doi: 10.1093/cercor/bhq071. [DOI] [PubMed] [Google Scholar]
- Girvan M, Newman MEJ. Community structure in social and biological networks. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleiser P, Danon L. Community Structure in Jazz arXiv:cond-mat/0307434. 2003 [Google Scholar]
- Gratton C, Nomura EM, Perez F, D'Esposito M. Focal brain lesions to critical locations cause widespread disruption of the modular organization of the brain. Journal of cognitive neuroscience. 2012;24:1275–1285. doi: 10.1162/jocn_a_00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimerà R, Nunes Amaral LA. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriger L, van den Heuvel MP, Sporns O. Rich Club Organization of Macaque Cerebral Cortex and Its Role in Network Communication. PloS One. 2012;7:e46497. doi: 10.1371/journal.pone.0046497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wang J, Wang L, Chen ZJ, Yan C, Yang H, Tang H, Zhu C, Gong Q, Zang Y, Evans AC. Uncovering intrinsic modular organization of spontaneous brain activity in humans. PloS One. 2009;4:e5226. doi: 10.1371/journal.pone.0005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- Joyce KE, Laurienti PJ, Burdette JH, Hayasaka S. A new measure of centrality for brain networks. PloS One. 2010;5:e12200. doi: 10.1371/journal.pone.0012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuth D. The Stanford GraphBase : a platform for combinatorial computing. New York N.Y: Reading Mass.: ACM Press ;;Addison-Wesley; 1993. [Google Scholar]
- Lohmann G, Margulies DS, Horstmann A, Pleger B, Lepsien J, Goldhahn D, Schloegl H, Stumvoll M, Villringer A, Turner R. Eigenvector centrality mapping for analyzing connectivity patterns in FMRI data of the human brain. PloS One. 2010;5:e10232. doi: 10.1371/journal.pone.0010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusseau D, Schneider K, Boisseau OJ, Haase P, Slooten E, Dawson SM. The bottlenose dolphin community of Doubtful Sound features a large proportion of long-lasting associations. Behavioral Ecology and Sociobiology. 2003;54:396–405. [Google Scholar]
- Meunier D, Achard S, Morcom A, Bullmore E. Age-related changes in modular organization of human brain functional networks. NeuroImage. 2009;44:715–723. doi: 10.1016/j.neuroimage.2008.09.062. [DOI] [PubMed] [Google Scholar]
- Meunier D, Lambiotte R, Bullmore ET. Modular and hierarchically modular organization of brain networks. Frontiers in Neuroscience. 2010;4:200. doi: 10.3389/fnins.2010.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M. Networks: An Introduction. Oxford University Press; 2010. [Google Scholar]
- Newman ME. The structure of scientific collaboration networks. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:404–409. doi: 10.1073/pnas.021544898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. Modularity and community structure in networks. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher D, Charles L, Devlin JT, Walsh V, Duchaine B. Triple dissociation of faces, bodies, and objects in extrastriate cortex. Current Biology: CB. 2009;19:319–324. doi: 10.1016/j.cub.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Steps toward optimizing motion artifact removal in functional connectivity MRI; a reply to Carp. NeuroImage. 2013;76:439–441. doi: 10.1016/j.neuroimage.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, Petersen SE. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. NeuroImage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Weight-conserving characterization of complex functional brain networks. NeuroImage. 2011;56:2068–2079. doi: 10.1016/j.neuroimage.2011.03.069. [DOI] [PubMed] [Google Scholar]
- Shulman GL, Pope DLW, Astafiev SV, McAvoy MP, Snyder AZ, Corbetta M. Right hemisphere dominance during spatial selective attention and target detection occurs outside the dorsal frontoparietal network. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2010;30:3640–3651. doi: 10.1523/JNEUROSCI.4085-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, Woolrich MW. Network modelling methods for FMRI. NeuroImage. 2011;54:875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND. Functional connectivity density mapping. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9885–9890. doi: 10.1073/pnas.1001414107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND. Functional connectivity hubs in the human brain. NeuroImage. 2011;57:908–917. doi: 10.1016/j.neuroimage.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Stam CJ, Boersma M, Hulshoff Pol HE. Small-world and scale-free organization of voxel-based resting-state functional connectivity in the human brain. NeuroImage. 2008;43:528–539. doi: 10.1016/j.neuroimage.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Van Dijk KRA, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. NeuroImage. 2012;59:431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC. A Population-Average, Landmark- and Surface-based (PALS) atlas of human cerebral cortex. NeuroImage. 2005;28:635–662. doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA, Dickson J, Harwell J, Hanlon D, Anderson CH. An integrated software suite for surface-based analyses of cerebral cortex. Journal of the American Medical Informatics Association: JAMIA. 2001;8:443–459. doi: 10.1136/jamia.2001.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of 'small-world' networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Wig GS, Schlaggar BL, Petersen SE. Concepts and principles in the analysis of brain networks. Annals of the New York Academy of Sciences. 2011;1224:126–146. doi: 10.1111/j.1749-6632.2010.05947.x. [DOI] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of neurophysiology. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachary WW. An Information Flow Model for Conflict and Fission in Small Groups. Journal of Anthropological Research. 1977;33:452–473. [Google Scholar]
- Zuo X-N, Ehmke R, Mennes M, Imperati D, Castellanos FX, Sporns O, Milham MP. Network Centrality in the Human Functional Connectome. Cerebral Cortex (New York, N.Y.:1991) 2011 doi: 10.1093/cercor/bhr269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.