Abstract
Nuclear structure and mechanics play a critical role in diverse cellular functions, such as organizing direct access of chromatin to transcriptional regulators. Here, we use a new, to our knowledge, hybrid method, based on microscopy and hyperelastic warping, to determine three-dimensional strain distributions inside the nuclei of single living cells embedded within their native extracellular matrix. During physiologically relevant mechanical loading to tissue samples, strain was transferred to individual nuclei, resulting in submicron distributions of displacements, with compressive and tensile strain patterns approaching a fivefold magnitude increase in some locations compared to tissue-scale stimuli. Moreover, nascent RNA synthesis was observed in the interchromatin regions of the cells studied and spatially corresponded to strain patterns. Our ability to measure large strains in the interchromatin space, which reveals that movement of chromatin in the nucleus may not be due to random or biochemical mechanisms alone, but may result from the transfer of mechanical force applied at a distant tissue surface.
Introduction
The nucleus is a membrane-bound organelle and regulation center for gene expression in the cell (1). The position of a gene in the interior of the nucleus changes when it becomes highly expressed, and is often found to extend out of its chromosome territory into the interchromatin space (2). The accessibility of DNA regions by transcription factors may be driven by a variety of mechanisms, including diffusive or thermal conformational changes (3,4), or through biochemical processes (5), which affects the chromatin structure and the complex local binding affinities of the chromatin and RNA molecules surrounding a gene. Less clear is the role of mechanical force transfer as a directed movement mechanism for DNA accessibility, due perhaps to the technical challenges in measuring small-scale mechanics inside the nuclei of cells embedded in their native extracellular environment.
Mechanical forces transfer to the nucleus directly and indirectly through specific cellular pathways and cytoskeletal structures (6,7). There is increasing evidence that mechanical forces are transferred to the nucleus to orchestrate transcriptional activity (8). Protein dynamics inside the nucleus are additionally important for maintaining the nuclear structure and in facilitating gene expression at the transcription level (9). Probing spatiotemporal relationships between distributed mechanical forces and localized gene expression (i.e., biophysical and biochemical interactions) in the nuclei of individual cells is necessary because the individual cells experience different mechanical stimuli resulting from variations in local cell and extracellular matrix interactions. Studies that provide average measures over cells in a given tissue would oversimplify the heterogeneity intrinsic to the population. To understand the inherent variability of large cell populations, innovative methods are therefore required for combined measurements of single nuclei biophysical and biochemical interactions in cells maintained in their three-dimensional (3D) extracellular matrix microenvironment.
Current methods used to simultaneously probe biophysical or biochemical interactions in small subcellular structures like the nucleus are lacking. Methods to characterize nuclear mechanics typically study isolated cells or cells embedded in 3D gel matrices, and often report aspect ratio and volume change measures (10–12) that do not easily reveal the inherent complexity of internal strain patterns. Additionally, such methods lack the spatial resolution necessary for the correlation of intranuclear biomechanics and simultaneous internal biochemical activity. Recent approaches to link nuclear mechanics to biochemical responses have explored unique microscopy-based experimental designs, including the use of photobleaching and fluorescence resonance energy transfer pairs (13,14).
We simulated physiologically -relevant shear loading to tissues while simultaneously measuring nuclear mechanics and nascent RNA synthesis. Applied dynamic or static tissue shear loading mimics routine activities of cartilage-to-cartilage contact in the body that may be seen during walking or standing activities. Here, we describe detailed patterns of intranuclear strains and newly synthesized RNA in the nuclei of single cells in situ during tissue-scale loading. The use of a new, to our knowledge, hybrid imaging technique enabled us to measure biomechanical and biochemical activity in the nuclei of single cells, and contributes to our understanding of whether mechanical force applied directly to tissue surface transfers to the nuclei of embedded cells to possibly influence gene expression. Measured subcellular displacements and strains suggest that the nucleus is a complex structure that is actively deformed during mechanical loading at the tissue scale, with large motions and deformations that may regulate DNA accessibility in part by direct physical interactions.
Materials and Methods
Tissue harvesting and sample preparations
Articular cartilage explants with embedded cells (chondrocytes) were chosen as a model system due to their spatially heterogeneous mechanics and ultrastructure (15), with mechanically linked and significant disease relevance (16). This model system was additionally useful to closely mimic physical forces in a common daily activity, e.g., tissue contact during the walking cycle or while standing. Briefly, articular cartilage explants were harvested from juvenile bovines within 36 h of slaughter. Using a cork borer and custom cutting jig, explants (diameter = 5 mm, thickness = 2 mm) were obtained under standard sterile conditions for tissue/organ culture. Explants and embedded chondrocytes were maintained in DMEM/F-12, supplemented with 0.1% bovine serum albumin, 100 units/ml penicillin, 100 μg/ml streptomycin, 50 μg/ml ascorbate-2-phosphate, and 10% fetal bovine serum (Invitrogen, Carlsbad, CA). After harvesting, the explants were cut along the depth direction to produce hemi-cylinder pairs that were incubated and equilibrated for 24 h before testing.
Mechanical loading and imaging of DNA and newly synthesized RNA
A custom load application device, built for biaxial (compression and shear) loading of tissue samples, was used to apply a 15% simple shear strain at the surface of the cartilage explant, whereas confocal (z-slice) images were captured before and at multiple time points during deformation (Fig. 1). The loading apparatus included two piezoelectric motors with mounted magnetic encoders, and computerized displacement control in small (±61 nm) increments (Nanos Instruments, GmbH; Hamburg, Germany). The device was mounted over a confocal microscope (Olympus Fluoview) to allow for simultaneous loading of the cartilage tissue and imaging of the chondrocyte nuclei in their native extracellular matrix (Figs. 1 and 2).
Figure 1.
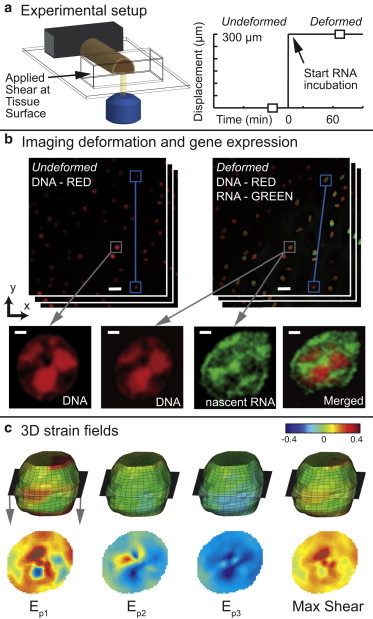
Combined confocal microscopy and hyperelastic warping reveals 3D strain fields and nascent RNA expression in the nuclei of single cells. (a) A custom tissue deformation device and microscope objective was used to image a cartilage explant during shear loading. (b) Projection images were constructed from undeformed and deformed confocal z-stack images (Red = DNA, Green = nascent RNA at 60 min, scale bar = 20 μm). Connected blue boxes track two nuclei to show tissue-scale shear deformation. Gray boxes point to a magnified middle slice of the nucleus volume (scale bar = 1 μm). (c) The selected nucleus from (b) is shown as a warped 3D volume with color maps representing principal strain fields and max shear, with the middle slice cross section detailed below. Of importance, the nuclei aspect ratios (undeformed = 1.08 and deformed = 1.15) do not capture the complexity of the intranuclear deformations. To see this figure in color, go online.
Figure 2.
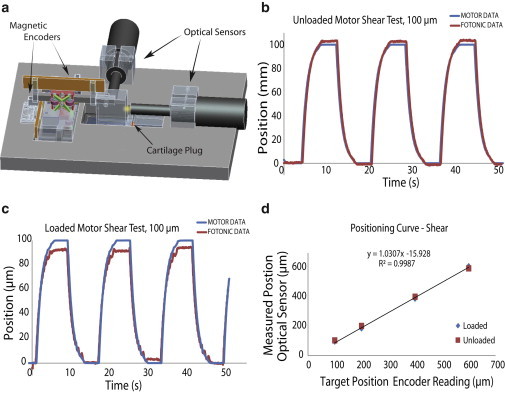
The tissue deformation system was validated using a fiber optic displacement measurement system. Motion was provided by two magnetic linear encoders (Nanos Instruments) mounted on Piezo LEGS LL1011A motors (Micromo; Clearwater, FL), and verified using a MTI-2000 Fotonic Sensor (MTI Instruments; Albany, NY). (a) CAD representation of the loading device with encoders as well as optical sensor in their respective positions. (b) Plot of a representative square-wave (100 μm) displacement profile in control experiments (i.e., actuator motion without a sample present). There was a small, micronscale offset between the encoder positioning and the Fotonic Sensor readout. (c) Plot of a square-wave (100 μm) displacement profile with a sample present. (d) Three square wave cycles at each position were averaged to produce the linear calibration curve loading a sample (diamonds) and without a sample (squares). To see this figure in color, go online.
At the time of testing, a pair of hemi cylinders from the same explant was selected, with one randomly assigned for treatment (mechanical loading) and the other one used as the nonloaded control. The explants were exposed for 8 min to a DNA stain (DRAQ5, Cell Signaling Technology,). The treated sample was affixed at both ends with cyanoacrylate to the loading apparatus with the cut surface positioned next to the coverslip and near the objective, noting that cell viability was maintained throughout the duration of testing (Fig. S1 in the Supporting Material). Both samples were placed in respective cell culture dishes and covered with phosphate buffered saline (PBS).
For imaging before and following deformation, an area of interest near the articular surface was visualized using a confocal microscope with a 60× water objective (NA = 1.20). 3D (z-stack) images (matrix = 1600 × 1600 pixel2; number of slices = 19) were sequentially captured with a z step of 0.5 μm/slice and a calculated in-plane resolution of 0.132 × 0.132 μm2. DNA was visualized by DRAQ5 staining (633 nm) before loading (Fig. 1 a). DNA was imaged again 10 and 60 min following shear loading using the same imaging parameters. A Click-iT RNA Alexa Fluor 488 Imaging Kit (Invitrogen) was used to detect RNA synthesis during the 60 min deformation period (described subsequently). Two channels (i.e., DNA at 633 nm and RNA at 488 nm) were acquired in sequential mode to eliminate cross talk. To detect the newly synthesized RNA, a filter bandwidth (520–600 nm) was additionally selected to exclude background from the autofluorescence of collagen in the extracellular matrix (Fig. 3). Although the peak of the collagen autofluorescence was around 488 nm, there was still a very weak signal that was detected from the tail of the collagen emissions spectrum at 520 nm, and which was easily removed in image postprocessing. The untreated control sample underwent the same protocol, except that shear loading was not performed.
Figure 3.
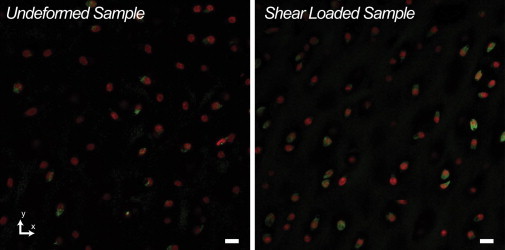
Control (nondeformed) tissue explants exhibited a reduce number of cells with nascent RNA expression compared to explants exposed to mechanical shear. The images shown are z projections of 20 z-slices, and the nascent RNA (green channel) was filtered to eliminate uneven background intensities so that a threshold could be applied to facilitate cell counting. The image postprocessing was performed with ImageJ (NIH, Bethesda, MD). The control sample showed 14 out of 54 cells (26%) with nascent RNA expression, whereas mechanical shear of the deformed (treated) sample resulted in 44 out of 75 (59%) cells with nascent RNA expression. Nuclei in Figs. 4 and 7–9 were analyzed from this deformed image field of view (Scale bar = 10 μm). To see this figure in color, go online.
Calculating nucleus 3D strain maps by hyperelastic warping
Hyperelastic warping was used to find the displacement field of the deformed nucleus. To measure internal nuclear deformation, a 3D finite element mesh was created from the confocal images depicting DNA, and a hyperelastic warping algorithm (nike3d) was used to calculate displacement and strain patterns throughout the nuclear volume (Figs. 1 and 4) (17,18). The algorithm deformed a 3D mesh of the z-stack image of the nucleus in the reference configuration until it matched the target image of the nucleus in a deformed configuration based upon the minimization of the differences in image intensities between the reference image and the deformed image (19,20). Nodal displacements were used to compute finite Lagrangian 3D strain fields, and principal strains and directions.
Figure 4.
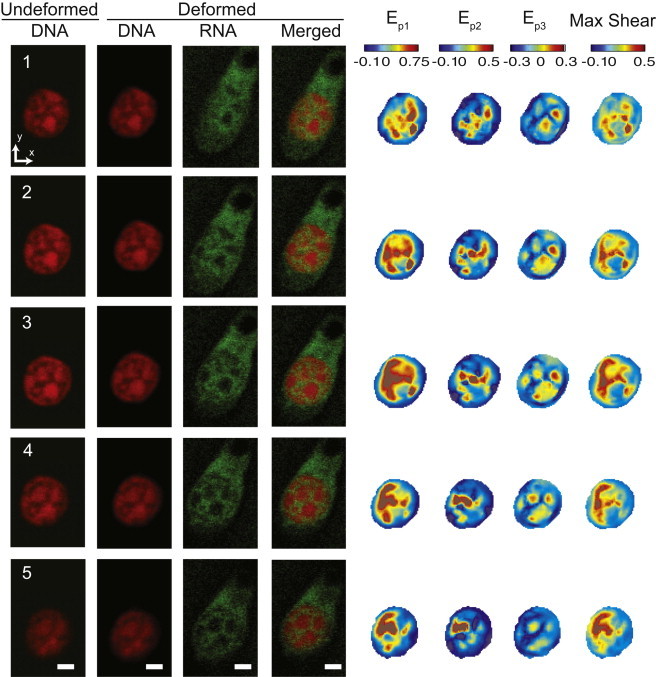
Intranuclear strain patterns in a single cell are spatially complex in three dimensions and heterogeneous even during simple shear at the tissue surface. (a) Five z-slices from an undeformed and deformed nucleus showing nascent RNA and the merged DNA-RNA image. Spatial patterns of strain, and DNA and nascent RNA images, vary by slice location. (b) However, overlaid graphs (Fig. 8) show that the relationship between strains and RNA or DNA intensity had very little variation between slices. (Scale bars = 2 μm) To see this figure in color, go online.
Validation: measurement of error in displacement fields using simulations
To validate our hybrid method, in particular the use of hyperelastic warping to quantify strain fields in the interior of small nuclear structures, and to determine the error associated with the hybrid technique overall, we used extensive forward finite element simulations. A 3D mesh was created from the z-stack images of a nucleus that was deformed in a finite element simulation with known displacement and strain magnitudes representative of those observed in the nuclei of living cells. A deformed image data set was created based upon the displacements of the forward finite element model. These images were analyzed using hyperelastic warping, and additionally, data were also directly compared to well-known (conventional) texture correlation techniques. Forward finite element stimulations were used to create a known displacement field and determine error (quantified in terms of root mean-square error (RMSE), bias, and precision) of the hyperelastic warping and texture correlation techniques. The forward simulation generated a known 3D displacement field and images of the deformed nucleus that was representative of experimental data. Gaussian noise was added to the image sets to vary the signal-to-noise (SNR) and contrast-to-noise (CNR) ratios to span the range of experimentally observed values. In-plane (x and y displacement) comparisons between the techniques were made in image slices through the center and at the edge of the image volume. Differences between calculated and known displacements were used to estimate the average RMSE, bias, and precision, over the range of simulated SNR and CNR ratios.
Nascent RNA synthesis in situ
A Click-iT RNA Alexa Fluor 488 Imaging Kit (Invitrogen) was used to tag and image newly synthesized RNA over a 60 min period of deformation (Fig. 1 b). Nascent RNA detection was performed by a click chemistry reaction between an RNA incorporated 5-ethynyl uridine tag and an azide-containing dye after cell fixation and permeabilization (21,22). Briefly, after incubation during loading, the samples were fixed with 2% formaldehyde in PBS, permeabilized with 0.1% Triton X-100 in PBS, and exposed to freshly prepared Click-iT reaction cocktail, while still in the loading apparatus.
To image nascent RNA synthesis in situ, two preliminary studies were additionally performed to successfully translate the RNA detection technology from its developed use in monolayer cells (21,22) into a 3D tissue environment. First, we minimized the background autofluorescence of collagen and the nonspecific binding of the fluorescent tag. We selected an appropriate emissions range on the confocal detector to minimize signal from collagen, and in addition we applied an image enhancing blocking reagent (Image-iT FX Signal Enhancer, Invitrogen) to help minimize nonspecific binding (Fig. 3). To remove nonspecific background staining in our studies, a 60 min incubation of the blocking reagent was used before the final image acquisition. Second, we determined the duration of 5-ethynyl uridine incubation to enhance RNA signal detection following mechanical shear loading. It should be noted that excessive incubation times resulted in an observed RNA signal from the combined effects of shear deformation and routine (e.g., housekeeping) cellular RNA synthesis (21). Using time duration studies, we determined that routine RNA synthesis was detected above background fluorescence levels in the cells of tissues that were unloaded and incubated for 60 min, indicating a time duration that could be used to best detect RNA signal enhancement due to mechanical loading. To further control for sample variation in routine RNA synthesis, one half of the tissue explant (i.e., one half of the hemicylinder) was loaded, whereas the second half was used as the unloaded control (Fig. 3).
Relationship between intranuclear strains and newly synthesized RNA
To explore links between the internal nuclear mechanics and newly synthesized RNA, the intranuclear strain fields were spatially compared qualitatively and using quantitative correlation analyses with custom MATLAB code (The MathWorks, Natick, MA). Correlation measures were made at three levels, i.e., within a given image slice through a single nucleus, within an image volume representing a whole single nucleus, or among nuclei from single cells identified in the field of view of the confocal images. The raw data consisted of strain values that were correlated to the image (DNA or RNA) intensity values at each voxel location of the nucleus. To facilitate correlations between strain and RNA or DNA image intensities, the data were binned according to the image intensity (bin size = 0.02). The data were binned for each image slice, and the binned results were additionally averaged for a single slice and averaged z-stacks. Correlation statistics (e.g., r2 values) were calculated from the binned data sets.
Results
Hybrid microscopy reveals complex intranuclear displacements and strains
Intranuclear deformation, defined by displacements and strains, were found to be heterogeneous and complex in living cells embedded within their native extracellular matrix. Intranuclear strains were both amplified and attenuated compared to tissue-scale stimuli. During a 15% simple shear strain, intranuclear displacements overall were consistently submicron in magnitude, with differences approaching only 10 s of nanometers depending on the size and location of the regions used for comparison (Fig. 5). Strong displacement gradients resulted in large intranuclear strains, typically <75% in magnitude, depending on location in the nucleus (Fig. 4). In contrast, nuclei in nonloaded control cartilage showed minimal intranuclear strain compared to those from loaded cartilage (Fig. S2).
Figure 5.
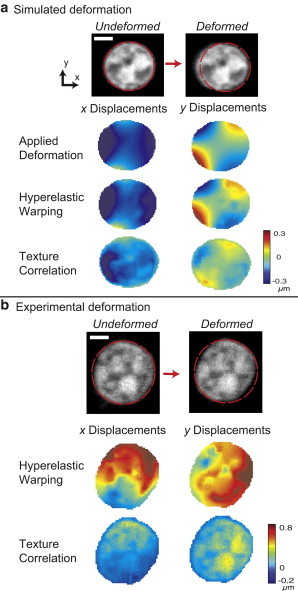
Displacement fields from hyperelastic warping, but not texture correlation, match known simulations, thus enabling the measurement of small-scale motion in individual nuclei. (a) A simulated deformation was applied to a 3D nucleus, with the middle z-slice images shown. The x and y displacement fields are shown for the known applied deformations, followed by the measured results from hyperelastic warping and texture correlation. (b) The x and y displacement fields from experimental data of a representative nucleus are shown for hyperelastic warping and texture correlation methods. Hyperelastic warping describes deformation with lower bias and increased precision compared to texture correlation (Fig. 6). (Scale bars = 2 μm) To see this figure in color, go online.
The hybrid of microscopy and hyperelastic warping quantified heterogeneous and complex intranuclear strain with minimal error. Of importance, the x and y displacement fields for the middle slice of the nucleus showed that hyperelastic warping matched more closely to the known (simulated) displacements compared to conventional (1st order) texture correlation algorithms (Fig. 5). The known displacements were related to the displacements measured by hyperelastic warping (slope = 1.11, r2 = 0.958) and texture correlation (slope = 0.97, r2 = 0.477). Hyperelastic warping and texture correlation were also compared using experimental data of a nucleus in undeformed and deformed states (Fig. 5 b). A comparison of warping and texture correlation displacement predictions with those of the forward model indicated similar qualitative displacement distributions. However, the texture correlation results were lower in magnitude than the hyperelastic warping results. We further noted that texture correlation displacement fields were biased by the bright areas in the image and around the nucleus perimeter, which was qualitatively observed in the known and experimental displacement fields (Fig. 5). Hyperelastic warping consistently resulted in displacement data with lower error (avg. RMSE = 0.017) compared to texture correlation (avg. RMSE = 0.091), and without sensitive dependences on SNR and CNR ratios (Fig. 6).
Figure 6.
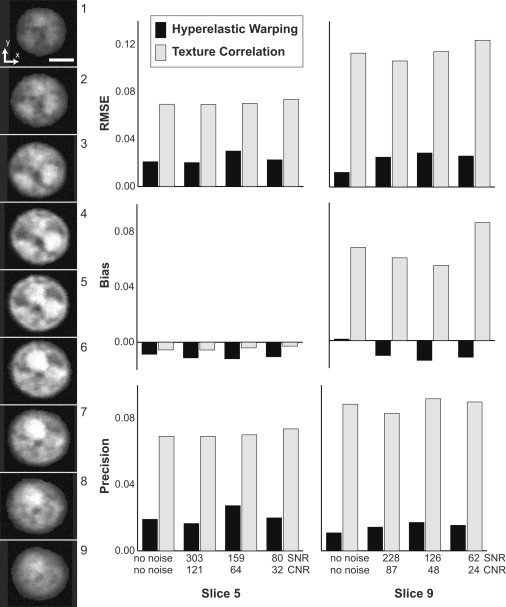
Hyperelastic warping consistently provided data with low RMSE and bias, and high precision, compared to texture correlation. SNR and CNR were calculated for each image set after the addition of Gaussian (random) image noise. Three error measurements (RMSE, bias, and precision) were used for comparison between the two techniques. The most noticeable difference between the techniques was the increase in bias seen with texture correlation between slice 5 and 9. The other measurements of error show similar performance of the techniques on each image slice. (Scale bar = 2 μm) To see this figure in color, go online.
Interchromatin regions and nascent RNA synthesis
Nascent RNA synthesis was observed in the interchromatin regions within nuclei of single cells embedded in their native extracellular matrix during shear loading at the tissue scale. For the nucleus depicted in Figs. 7 and 8, changes in chromatin position, defined in terms of strain, were also found to correspond to regions of nascent RNA synthesis. Magnitudes of principal strains and maximum shear strains approached a fivefold tensile increase over the 15% simple shear strain magnitude applied at the tissue surface in some regions of the nucleus. Different regions within the nucleus exhibited compressive or tensile strains, indicating that the magnitude of the applied shear at the tissue surface was amplified and attenuated depending on the internal region of the nucleus under investigation (Fig. 7). Principal directions for Ep2 and Ep3 were predominately in the imaging plane (i.e., in the xy plane), whereas Ep1 directions were largely through-plane. Qualitative comparisons of the strain patterns revealed a correspondence between high tensile strain regions with increased intensities of the RNA and DNA patterns for max shear (RNA) and Ep3 (DNA), respectively. Strain patterns tended to correspond visually to observed DNA and RNA patterns, suggesting that specific stimuli, e.g., localized maximum strain, may cause changes in the chromatin structure to influence newly synthesized RNA in nuclear regions with low DNA content.
Figure 7.
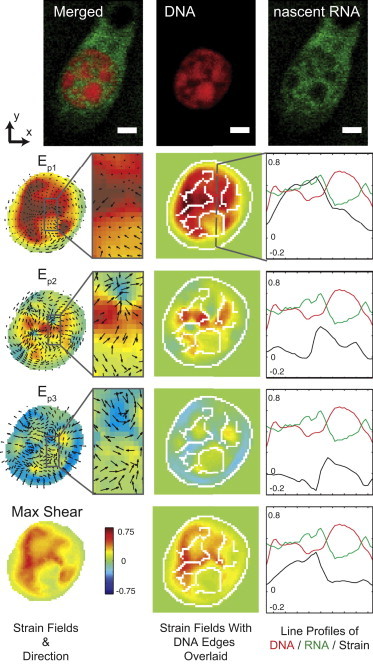
Principal strain magnitudes and directions correspond to the intensity of DNA and nascent RNA. A magnification box highlights a nuclear region with heterogeneous (amplified and attenuated) strains. The strain direction for Ep1 was predominantly through the imaging plane, whereas the Ep2,3 directions were largely in the imaging plane. Although only one representative image slice from the middle of the nucleus is shown, the results were consistent in other z-slices (Figs. 4 and 8). A single line scan at the right of the strain maps depicts the cross-sectional profiles for DNA (red), RNA (green), and strain magnitudes (black). White lines of the strain map are edges of the DNA image intensity calculated by an edge detection function in MATLAB to help spatially visualize the high and low DNA regions. (Scale bars = 2 μm) To see this figure in color, go online.
Figure 8.
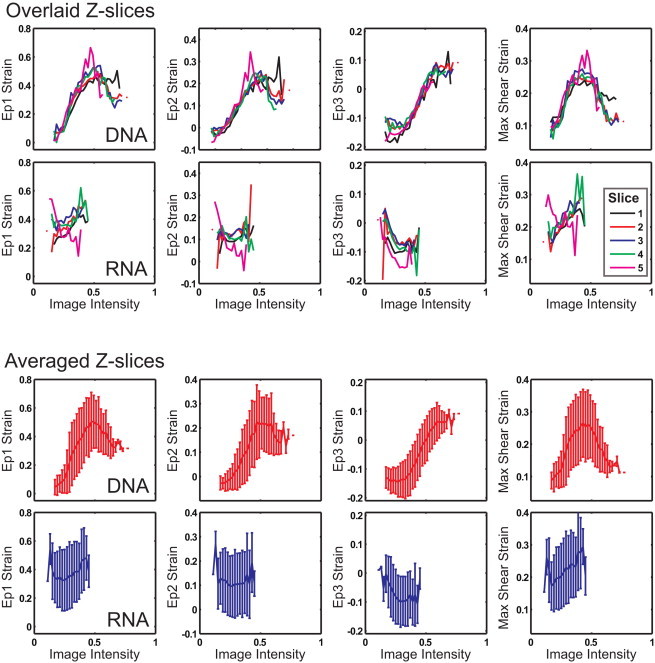
Complete 3D, DNA, and RNA image intensity to strain relationship analysis for a single nucleus from Fig. 4. The top graphs show the image intensity versus strain results for each z-slice and the bottom graphs are the averaged results. To see this figure in color, go online.
Relationships between DNA, RNA, and strain were quantified at multiple levels, i.e., within a given image slice through a single nucleus (e.g., Fig. 7), within an image volume representing a whole single nucleus (e.g., Fig. 8), or among nuclei from many single cells (e.g., Fig. 9). For the single image slice in Fig. 7, significant correlations (p < 0.007) were found between DNA intensities and Ep1 (r2 = 0.238), Ep2 (r2 = 0.506), and Ep3 (r2 = 0.833), but not max shear (r2 = 0.000; p = 0.984). Significant correlations (p < 0.001) were also found between RNA intensities and Ep1 (r2 = 0.617), Ep3 (r2 = 0.754), and max shear (r2 = 0.827), but not Ep2 (r2 = 0.214; p = 0.096). Similar correlations were observed for a whole single nucleus (Fig. 8), after pooling r2 values from each slice of the image volume, with DNA and RNA related to Ep1 (r2 = 0.543, r2 = 0.668), Ep2 (r2 = 0.677, r2 = 0.471), Ep3 (r2 = 0.889, r2 = 0.473), and max shear (r2 = 0.192, r2 = 0.694), respectively.
Figure 9.
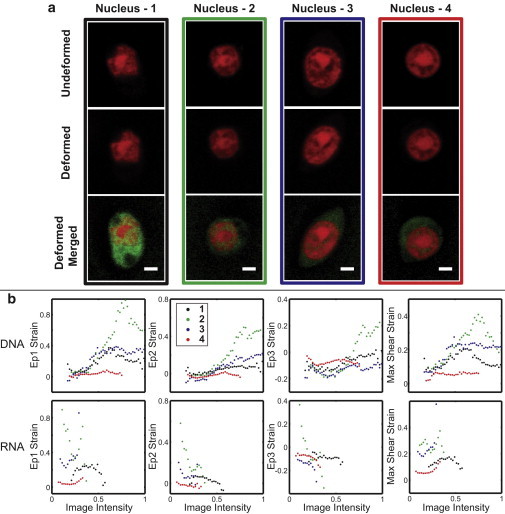
Strain magnitudes and nascent RNA expressions in nuclei vary among single cells within in a tissue volume subjected to uniform shear. (a) The undeformed DNA image, deformed DNA, and merged DNA-RNA images are shown for four cells from the same imaging field of view seen in Fig. 3. The nuclei were from the same region in the tissue, although there were several physical characteristics, such as shape, size, and long-axis orientation that may influence how the strain was transferred from the tissue to the nucleus. (b) The average image intensity versus strain for the four nuclei were graphed in matching colors as boxed in (a). Three of the four nuclei (excluding nucleus two) exhibited similar trends in DNA and RNA intensity patterns. The RNA graphs also show that there were different RNA image intensities between nuclei, which were qualitatively observed in panel (a). (Scale bars = 2 μm) To see this figure in color, go online.
Relationships between strains and DNA and RNA intensities varied among the cell nuclei studied (Fig. 9). Nascent RNA synthesis was consistently observed in the interchromatin regions, although specific intranuclear statistical correlations covered a broader range when compared cell to cell. Significant statistical correlations (p < 0.015) were found in all cells studied between DNA intensities and Ep1 (r2 = 0.575) and Ep2 (r2 = 0.735), and between RNA intensities and Ep3 (r2 = 0.641), with aforementioned coefficients of determination pooled over all nuclei shown in Fig. 9. Statistical correlations varied among cells between DNA intensities and Ep3 (r2 = 0.550, p < 0.443) and max shear (r2 = 0.575, p < 0.306), and between RNA intensities and Ep1 (r2 = 0.357, p < 0.379), Ep2 (r2 = 0.492, p < 0.295), and max shear (r2 = 0.374, p < 0.954), with 6 of 20 total possible correlations (i.e., five relationships for the four cells shown in Fig. 9) not significant (p > 0.040).
Discussion
Diffusive or biochemical processes are thought to drive the movement of genes to different regions of the nucleus, perhaps due to changes in binding affinities in addition to conformational alterations in the chromatin structure. Additional studies, involving the use of relatively insensitive methods based on texture correlation, indicate that the deformation in the nucleus is minimal compared to cell and extracellular matrix strains (23). These studies imply that the relative stiffness of the nucleus is high compared to surrounding cellular structures, resulting in minimal internal deformation for a given applied load. The concept of a relatively stiff nucleus would indirectly support the idea that specific (e.g., diffusive) mechanisms alone may drive gene expression, because the nucleus interior would be more isolated from physical deformation occurring in the extranuclear regions, and would require alternative mechanisms for transcription and other regulators to access DNA.
Here, we find that movement of the nuclear structures, quantified by strain, is highly heterogeneous and is both amplified and attenuated during even simple mechanical loading at the tissue scale. Given a reasonable compliance of chromatin, the heterogeneous strains would be expected to shift and reposition the relative internal position of genes, thereby altering the dynamics of regulation. Interestingly, the compliance of individual chromatin fibers has also been noted as a possible physical basis for DNA accessibility (24). However, we do not yet know the extent that either chromatin remodeling, or passive chromatin deformation in response to the applied load, explains the intranuclear strain patterns described. In light of this current limitation, and in contrast to single molecule studies conducted in controlled in vitro experiments, we overcame technical challenges in obtaining measurements within the nuclei of cells embedded in extracellular matrix in situ. The experimental setup and hybrid microscopy technique allowed us to propagate realistic and physiologically relevant mechanical forces through native structures to better quantify the extent that strain transfer may directly influence nuclear mechanics. The hybrid method, based on microscopy and hyperelastic warping, allowed the measurement of internal deformation (displacements and strains) in small nuclear structures at high spatial resolutions, limited most by the time constraints of image acquisition. Interestingly, the simultaneous detection of newly synthesized RNAs reveal localized expression corresponding to mechanical loading and patterns of principal and shear strains. The measurement of large (e.g., shear) strains in the interchromatin space that spatially correspond to the localization of nascent RNA expression supports the hypothesis that localized movement of chromatin in the nucleus may not be due to random or biochemical mechanisms alone, but instead can occur simply as a result of mechanical force transfer applied at a distant tissue surface. However, significant transport of RNA over the incubation time (e.g., Fig. 1) indicates that the nascent RNAs observed may not be a true response of the nucleus to deformation. Additionally, RNA expression was observed in nucleus regions that were expected to be more naturally transcriptionally active. Nevertheless, these regions corresponded with unexpectedly high levels of strain, suggesting that the nucleus structure may be routinely regulated through a variety of casual physical activities that involve tissue loading, which in turn allow for transcriptional and other regulators of biological activity and gene expression.
Our experimental analysis demonstrated the hierarchical transfer of strain over large distances and log scales from the tissue surface to the interior of individual nuclei in situ. In our study, simple shear strain applied at a distant tissue surface transfer to individual nuclei, amplifying strain up to fivefold in localized nuclear regions. Interestingly, we note reports of novel quantitative approaches to measure detailed internal biomechanics in individual cells, but these techniques largely ignore the intranuclear strain and gene expression (12). Quantification of intranuclear strains is important, because they possibly extend the concept of nuclear mechanics arising due to physical links to the cytoskeleton or extracellular matrix (6,7), to also include remote links through pericellular and extracellular molecules, e.g., type VI and type II collagens, respectively, in the hierarchical organization of complex tissues like cartilage. Using the hybrid method, we did not yet in the current configuration tease apart the relative influence of load transfer through solid or fluid phases (25), or specify candidate matrix or cytoskeletal molecules that result in strain transfer. However, candidate molecules may be identified and visualized by fluorescent tagging in subsequent studies. In this work, it is important to emphasize the nondestructive nature of the hybrid technique, coupled with the ability to reveal internal spatiotemporal dynamics in living cell nuclei in situ for the first time, which enables the study of diverse cell-laden materials, including hydrogel or polymer constructs, and diseased tissues, that contain unique or rare cell populations.
The nucleus-to-nucleus variation of intranuclear strain field patterns (Figs. 4, 8, and 9) suggested that the applied tissue load was not uniformly transferred to every nucleus, possibly due to the spatially heterogeneous mechanics of cartilage explants that arise from cell location and spatial density in the tissue (26). This observation indicates the possibility of subtle and variable underlying cell-matrix connections or other structural parameters that dictate how load is shared over hierarchical scales. Interestingly, newly synthesized RNA was also observed outside of the nuclear region of the cell, illustrating transport during the short incubation time, which has also been reported in other cell types following treatments with soluble factors (21,22). Furthermore, we observe that the time-dependent response of the individual nuclei varies among nuclei to the applied shear (Fig. S3), suggesting more complicated (e.g., viscoelastic) mechanisms may play a role in the biomechanics and RNA expression in some cells. For example, the arrangement of structural elements in the microenvironment, and the local composition of extracellular and pericellular molecules (e.g., proteoglycans), influence the rate of force transmission to the cell, and were not measured in this study. Variations in cell shape and orientation were observed, suggesting that variations in other physical structures were present as well. Moreover, the cartilage may not yet have achieved a mechanical equilibrium, with time-dependent changes in interstitial fluid pressure and flow, in combination with constituents in the local microenvironment, possibly influencing strain patterns observed. Strain maps measured from DNA images taken at 10 min and 60 min postdeformation indicated an increase, decrease, or no change in the intranuclear strain patterns and magnitudes between the two time points. In contrast, very small differences in aspect ratios of nuclei between 10 and 60 min indicate little to no bulk deformation of the nucleus between the two time points (Fig. S3).
Conclusion
These data suggest that cells in situ can sense an applied load that is transferred over relatively large distances and log scales to alter intranuclear deformation and to possibly directly influence new RNA synthesis, and may regulate other actions as well, including the transport of mRNA or other molecules through nuclear pores. The direct correspondence of strain and DNA or RNA patterns may suggest specific mechanical stimuli, e.g., shear strain, that influence spatially localized gene expression in individual cells, although it remains to be determined the extent that mechanics directly regulate nuclear mechanobiology in larger cell populations. We expect that this hybrid method, based on microscopy and hyperelastic warping, will enable a wide variety of future investigations into mechanotransduction mechanisms, including transcription of specific RNAs, and translation and control of downstream protein synthesis.
Acknowledgments
The manuscript benefitted from helpful comments from Tyler Novak, Deva Chan, Sarah Calve, Russell Main, and Eric Nauman.
The authors gratefully acknowledge funding from the Andrews Fellowship (J.T.H.).
Supporting Material
References
- 1.Misteli T. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- 2.Cremer T., Cremer M., Fakan S. Chromosome territories—a functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 4.Polach K.J., Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J. Mol. Biol. 1996;258:800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- 5.Cairns B.R. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang N., Butler J.P., Ingber D.E. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 7.Wang N., Tytell J.D., Ingber D.E. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 8.Mammoto A., Mammoto T., Ingber D.E. Mechanosensitive mechanisms in transcriptional regulation. J. Cell Sci. 2012;125:3061–3073. doi: 10.1242/jcs.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardoso M.C., Schneider K., Leonhardt H. Structure, function and dynamics of nuclear subcompartments. Curr. Opin. Cell Biol. 2012;24:79–85. doi: 10.1016/j.ceb.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Guilak F. Compression-induced changes in the shape and volume of the chondrocyte nucleus. J. Biomech. 1995;28:1529–1541. doi: 10.1016/0021-9290(95)00100-x. [DOI] [PubMed] [Google Scholar]
- 11.Knight M.M., van de Breevaart Bravenboer J., Bader D.L. Cell and nucleus deformation in compressed chondrocyte-alginate constructs: temporal changes and calculation of cell modulus. Biochim. Biophys. Acta. 2002;1570:1–8. doi: 10.1016/s0304-4165(02)00144-7. [DOI] [PubMed] [Google Scholar]
- 12.Legant W.R., Miller J.S., Chen C.S. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat. Methods. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iyer K.V., Pulford S., Shivashankar G.V. Mechanical activation of cells induces chromatin remodeling preceding MKL nuclear transport. Biophys. J. 2012;103:1416–1428. doi: 10.1016/j.bpj.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouyang, M., J. Sun, …, Y. Wang. 2008. Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proceedings of the National Academy of Sciences of the United States of America. 105:14353–14358. [DOI] [PMC free article] [PubMed]
- 15.Chan D.D., Neu C.P. Transient and microscale deformations and strains measured under exogenous loading by noninvasive magnetic resonance. PLoS ONE. 2012;7:e33463. doi: 10.1371/journal.pone.0033463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neu C.P., Reddi A.H., Di Cesare P.E. Increased friction coefficient and superficial zone protein expression in patients with advanced osteoarthritis. Arthritis Rheum. 2010;62:2680–2687. doi: 10.1002/art.27577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phatak N.S., Sun Q., Weiss J.A. Noninvasive determination of ligament strain with deformable image registration. Ann. Biomed. Eng. 2007;35:1175–1187. doi: 10.1007/s10439-007-9287-9. [DOI] [PubMed] [Google Scholar]
- 18.Veress A., Phatak N., Weiss J. Deformable image registration with hyperelastic warping. In: Suri J., Willson D., Laxminarayan S., editors. Handbook of Biomedical Image Analysis Volume III: Registration Models. Kluwer Academic/Plenum Publishers; New York, Boston, Dordrecht, London, Moscow: 2005. pp. 487–533. [Google Scholar]
- 19.Veress A.I., Weiss J.A., Rabbitt R.D. Strain measurement in coronary arteries using intravascular ultrasound and deformable images. J. Biomech. Eng. 2002;124:734–741. doi: 10.1115/1.1519279. [DOI] [PubMed] [Google Scholar]
- 20.Veress A.I., Weiss J.A., Gullberg G.T. Measuring regional changes in the diastolic deformation of the left ventricle of SHR rats using microPET technology and hyperelastic warping. Ann. Biomed. Eng. 2008;36:1104–1117. doi: 10.1007/s10439-008-9497-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jao, C. Y., and A. Salic. 2008. Exploring RNA transcription and turnover in vivo by using click chemistry. Proceedings of the National Academy of Sciences of the United States of America. 105:15779–15784. [DOI] [PMC free article] [PubMed]
- 22.Paredes E., Das S.R. Click chemistry for rapid labeling and ligation of RNA. ChemBioChem. 2011;12:125–131. doi: 10.1002/cbic.201000466. [DOI] [PubMed] [Google Scholar]
- 23.Gilchrist C.L., Witvoet-Braam S.W., Setton L.A. Measurement of intracellular strain on deformable substrates with texture correlation. J. Biomech. 2007;40:786–794. doi: 10.1016/j.jbiomech.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Kruithof M., Chien F.T., van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- 25.Kim E., Guilak F., Haider M.A. The dynamic mechanical environment of the chondrocyte: a biphasic finite element model of cell-matrix interactions under cyclic compressive loading. J. Biomech. Eng. 2008;130:061009. doi: 10.1115/1.2978991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neu C.P., Hull M.L., Walton J.H. Heterogeneous three-dimensional strain fields during unconfined cyclic compression in bovine articular cartilage explants. J. Orthop. Res. 2005;23:1390–1398. doi: 10.1016/j.orthres.2005.03.022.1100230622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


