Abstract
Objective
To aid in understanding longterm health consequences of intrauterine infections in preterm birth, we evaluated DNA methylation at nine differentially methylated regions (DMRs) that regulate imprinted genes by type of preterm birth [spontaneous preterm labor (PTL), preterm premature rupture of membranes (PPROM) or medically indicated (fetal growth restriction and pre-eclampsia)] and infection status (chorioamnionitis or funisitis).
Study Design
Data on type of preterm birth and infection status were abstracted from medical records and standardized pathology reports in 73 preterm infants enrolled in the Newborn Epigenetics STudy (NEST), a prospective cohort study of mother-infant dyads in Durham, NC. Cord blood was collected at birth, and infant DNA methylation levels at the H19, IGF2, MEG3, MEST, SGCE/PEG10, PEG3, NNAT, and PLAGL1 DMRs were measured using bisulfite pyrosequencing. One-way ANOVA and logistic regression models were used to compare DNA methylation levels by type of preterm birth and infection status.
Results
DNA methylation levels did not differ at any of the regions (p>0.20) between infants born via PTL (average n=29), PPROM (average n=17), or medically indicated preterm birth (average n=40). Levels were significantly increased at PLAGL1 in infants with chorioamnionitis (n=10, 64.4%) compared to infants without chorioamnionitis (n=63, 57.9%) p<0.01. DNA methylation levels were also increased at PLAGL1 for infants with funisitis (n=7, 63.3%) compared to infants without funisitis (n=66, 58.3%) p<0.05.
Conclusion
Dysregulation of PLAGL1 has been associated with abnormal development and cancer. Early-life exposures, including infection/inflammation, may affect epigenetic changes that increase susceptibility to later chronic disease.
Keywords: chorioamnionitis, epigenetic, preterm birth, funisitis, imprinting
Introduction
In the U.S., about 12% of all deliveries result in preterm births (PTB), defined as births occurring at less than 37 weeks gestation.1 PTB and low birth weight (LBW) are associated with not only significant perinatal and childhood morbidity but also longterm morbidity and increased risk of chronic diseases in adulthood.2–4 In addition, PTB is a heterogenous process that includes three different obstetric phenotypes: medically indicated PTB, Preterm Premature Rupture of Membranes (PPROM), and spontaneous preterm labor (PTL).5 Although there are multiple risk factors for PTB, including infections such as chorioamnionitis and funisitis, mechanisms linking infection, PTB, and perinatal and longterm morbidity, which could be used for risk stratification, are still unclear.
Epigenetics has been postulated as the mechanism linking the early developmental environment to adult disease.6, 7 Studies of survivors of the Dutch and Chinese famines have shown that those exposed to famine during the periconceptional period had increased risk of obesity and chronic diseases in adulthood as well as persistent epigenetic changes at multiple imprinted genes.8–13 In support of the role of epigenetics in preterm birth, recent studies have also found associations between DNA methylation levels of repetitive sequences or long interspersed nucleotide elements (LINE-1) and gestational age/PTB.14 In addition, increased homocysteine and global DNA methylation levels were seen in the placenta of women with pre-eclampsia compared to normotensive women.15 Moreover, a recent study found hypermethylation of multiple imprinted genes associated with spontaneous abortions and stillbirths, suggesting methylation alterations may play a role in pregnancy loss.16
The developmental origins of adult disease hypothesis postulates that epigenetic modifications from prenatal exposures may contribute to increased risk for poor birth outcomes and may potentially be carried on to affect later adult chronic disease; however, mechanisms are still unclear.17, 18 We sought to examine DNA methylation at differentially methylated regulatory regions associated with multiple imprinted genes. These genes function as critical growth effectors and regulators of development.8,13,19 Our study was designed to evaluate these differentially methylated regions (DMRs) in relation to type of preterm birth and infection status.
Materials and Methods
Study Participants
Study participants included all singleton infants born prematurely (<37 weeks) to the cohort of women enrolled in the Newborn Epigenetics STudy (NEST); a prospective study of mother/infant dyads aimed at investigating the effects of in utero exposures on epigenetic profiles and phenotypes in children. The target population and methods for participant identification and enrollment have been described in detail previously.20, 21 Briefly, between 2007 and 2011, pregnant women were recruited during their first or second trimester visits to prenatal clinics serving Duke Hospital or Durham Regional Hospital, the only two obstetrical facilities in Durham County. Eligibility criteria were age 18 years and older, intention to use Duke Hospital or Durham Regional Hospital for delivery, and English and Spanish speaking. Subjects were excluded if they planned to give up custody of their children or had HIV due to the limited research on the interaction of HIV infection with potential DNA methylation alterations in the offspring. Of the 181 singleton preterm births in this cohort, infection, complete parturition and methylation data were available in a subset of 73 mother-infant pairs. These mother-infant pairs are similar to the larger group of 181 with respect to maternal age (p=0.3284), race (p=0.638), maternal BMI (p=0.8156), type of preterm birth (p=0.106), and proportion of births with chorioamnionitis (p= 0.620) and funisitis (p=0.619). The study protocol was approved by the Duke University Institutional Review Board (IRB).
Data Collection
Demographic information was collected from mothers at enrollment. Gestational age at enrollment ranged from 5–36 weeks (mean 17.5 weeks, SD 9.0). Gestational age at enrollment was slightly higher in the group of 73 mother-infant pairs (mean 20.8 weeks, SD 9.1) as compared to the larger group of 181 (mean 17.5 weeks, SD 9.0), p=0.01. Pregnant women self-reported birthdate (maternal age was computed by subtracting delivery date from birth date), current health status, parity, marital status, and usual weight and height at last menstrual period (LMP) for BMI. Due to changes in some questions in more recent demographic surveys, this study only characterizes women as “Black” or “White”, and the remaining women (ones who chose “Asian”, “Native-American”, “Hispanic”, or “Other”) are considered “Other”. Women who chose “White” or “Caucasian” were classified as “White.” Women were classified as “Black” if they identified themselves as “Black/African-American” or if they identified as “Biracial/other” and their mother was “Black/African-American.” Smoking status was harmonized between questionnaires to three categories, none, smoking during pregnancy, and quitting during pregnancy, as previously described.22 Education level was harmonized to less than high school, high school or GED, college, and graduate school.
After delivery of the infant, trained personnel abstracted parturition data from medical records including gestational age at birth, infant sex, delivery mode, and birthweight. PTB was defined as gestational age <37 weeks, and LBW was defined as birthweight <2500grams.23 Trained abstractors examined the medical records of all preterm infants to determine the type of preterm birth. Medically indicated preterm birth was defined as emergent delivery due to a medical complication, intrauterine growth restriction (IUGR), or pre-eclampsia (defined for this study as medical order for magnesium and 24 hr urine protein level >300mg).24 PPROM was defined by a clinical diagnosis (listed on problem list) or presence of two out of three of the following positive tests (pool, fern, or nitrazine). All other preterm births were defined as spontaneous. When the medical record was unclear, an obstetrician (APM) determined the phenotype based on clinical expertise. Standard pathological reports from the placenta of preterm infants were used to assess infection status. Chorioamnionitis was defined as a pathological diagnosis of Stage II or III chorioamnionitis.25 Funisitis was also a standardized pathological diagnosis.25 All data were collected and de-identified in STATA 12.0.
DNA methylation analysis
Infant cord blood specimens were collected at birth. Samples were collected in EDTA-containing Vacutainer tubes and centrifuged using standard protocols to allow for collection of plasma and buffy coat, with buffy coat used for DNA extraction (Qiagen; Valencia, CA). Specimens were stored at −80°C until time of analysis. DNA was extracted using Puregene reagents according to the manufacturer’s protocol (Qiagen) and quantity and quality assessed using a Nanodrop 1000 Spectrophotometer (Thermo Scientific; Wilmington, DE). Infant genomic DNAs (800 ng) were modified by treatment with sodium bisulfite using the Zymo EZ DNA Methylation kit (Zymo Research; Irvine, CA). Bisulfite treatment of denatured DNA converts all unmethylated cytosines to uracils, leaving methylated cytosines unchanged, allowing for quantitative measurement of cytosine methylation status. Pyrosequencing was performed using a Pyromark Q96 MD pyrosequencer (Qiagen). The bisulfite pyrosequencing assays were utilized to quantitatively measure the level of methylation at CpG sites contained within nine imprinted DMRs. DMRs analyzed were the paternally methylated IGF2 DMR, H19 DMR, MEG3-IG DMR (located intergenic to DLK1 and MEG3) and MEG3 DMR (promoter), and the maternally methylated PEG3 DMR, MEST DMR, SGCE/PEG10 DMR, NNAT DMR and PLAGL1 DMR. For all DMRs except PLAGL1, pyrosequencing assay design, genomic coordinates, assay conditions and assay validation are described in detail elsewhere.26, 27 Assays were designed to query established imprinted gene DMRs using the Pyromark Assay Design Software (Qiagen). For amplification of PLAGL1 from bisulfite modified DNAs, PCR primers were forward, 5′-GTA GGG TAG GTG TTT GGG TGT T-3′ and reverse, 5′-[biotin]CRA CAA AAA CAC ACC CTC CTC-3′. PCR was performed as previously described26 using the following conditions: 95°C for 15m, 55 cycles of 94°C for 30s, 68°C for 30s and 72°C for 30s, and a final 10m extension at 72°C. Pyrosequencing was performed using primer 5′-TGA GGT GTT TGG GTG TT-3′ to analyze sequence at genomic coordinates 144,329,179 to 144,329,209 on chromosome 6 (UCSC Genome Browser, Feb. 2009 release, GRCh37/hg19). PCR conditions were optimized to produce a single, robust amplification product. Defined mixtures of fully methylated and unmethylated control DNAs were used to show a linear increase in detection of methylation values as the level of input DNA methylation increased (Pearson r >0.98 for all DMRs). Once optimal conditions were defined, each DMR was analyzed using the same amount of input DNA from each specimen (40 ng, assuming complete recovery following bisulfite modification). Percent methylation for each CpG cytosine was determined using Pyro Q-CpG Software (Qiagen).
Statistical Analysis
Fisher’s exact tests were used to determine associations between type of preterm birth and infection status (chorioamnionitis and funisitis). To examine the role of epigenetics, we then assessed each DMR for normality using the Kolmogorov-Smirnov test. We found that with the exception of SGCE/PEG10 (p<0.01), all other infant DMRs were normally distributed (p>0.05). Confirmatory factor analysis for individual maternal and infant CpGs revealed Cronbach’s alphas for all DMRs were >0.74, suggesting mean methylation levels for each DMR could be used in models. One-way ANOVA was used to compare infant DNA methylation differences at the nine DMRs by type of preterm birth (medically indicated, PPROM, or spontaneous PTL). T-tests were used to compare infant DNA methylation differences at DMRs by infection status (chorioamnionitis or funisitis). Wilcoxon-rank sum tests were used to confirm differences found by t-testing in all DMRs that were not normally distributed. For DMRs that differed by type of PTB or infection status, logistic regression models were fit to examine associations between PTB or infection status and DNA methylation at DMRS. Models were initially fit with all variables considered clinically relevant including maternal age at delivery, maternal race, health status, parity, marital status, maternal BMI, smoking, education level, delivery mode, infant sex, infant birthweight, and gestational age at delivery. A backward stepwise approach was used to refine the model, and log likelihood tests for individual covariates were used to create the final parsimonious model.
Results
Demographic Characteristics
Table 1 describes the characteristics of the 181 preterm mother-infant dyads. Maternal age ranged from 18–49, with mean age of 30 years and standard deviation of 6.6 years. 38% of women were married, 31% were never married, and the rest were either living with a partner, divorced/separated or chose “other.” 54% of the mothers were Black, 28% were White, and 18% were grouped as Other, which included women choosing a race/ethnicity of Hispanic, Asian, Native American, or Other. The majority of the women were multiparous (68%). BMI ranged from 16–67 with a mean BMI of 28, standard deviation of 7.7. The majority of the women endorsed good, very good, or excellent health with 17% endorsing fair or poor health. 20% of women smoked during pregnancy while 17% reported quitting. The majority (>50%) of women had some college education or more. 51% of the births were delivered vaginally, and 49% were delivered via C-section. 56% of the infants delivered were male, and 44% were female. Of the preterm births, 37% were medically indicated, 24% were a result of PPROM, and 39% were spontaneous. Birthweight ranged from 580–3765 grams with a mean birthweight of 2346 grams (SD 677 grams). Gestational age at birth ranged from 24–36 weeks with a mean of 33.9 weeks (st. dev. 2.9 weeks). 16% of the PTB infants had chorioamnionitis, and 12% had funisitis. Of the 22 cases of chorioamnionitis and 16 cases of funisitis, 13 cases overlapped and had pathological features of both.
Table 1.
Summary of Characteristics of Preterm Mother-infant Dyads in NEST
| Variable | n(%) |
|---|---|
| Age (years) | |
| <20 | 6 (3%) |
| 20–24 | 51 (28%) |
| 25–29 | 33 (18%) |
| 30–34 | 48 (27%) |
| 35–39 | 36 (20%) |
| >40 | 7 (4%) |
|
| |
| Marital Status | |
| Never Married | 54 (31%) |
| Married | 65 (38%) |
| Widowed | 0 (0%) |
| Living with Partner | 38 (22%) |
| Divorced/Separated | 13 (8%) |
| Other | 1 (1%) |
|
| |
| Race | |
| White | 47 (28%) |
| Black | 93 (54%) |
| Other | 31 (18%) |
|
| |
| Parity | |
| Nulliparous | 54 (32%) |
| Multiparous | 117 (68%) |
|
| |
| BMI | |
| <18.5 | 9 (6%) |
| 18.5–<25 | 41 (29%) |
| 25–<30 | 41 (29%) |
| 30–<35 | 30 (21%) |
| 35–<40 | 13 (10%) |
| >40 | 7 (5%) |
|
| |
| Health | |
| Excellent | 17 (10%) |
| Very Good | 49 (29%) |
| Good | 76 (44%) |
| Fair | 26 (15%) |
| Poor | 3 (2%) |
|
| |
| Smoking | |
| None | 109 (63%) |
| Smoking | 35 (20%) |
| Quit | 30 (17%) |
|
| |
| Education | |
| <High School | 14 (8%) |
| High School/GED | 58 (34%) |
| Some College | 82 (48%) |
| Graduate School | 17 (10%) |
|
| |
| Delivery Mode | |
| Vaginal | 91 (51%) |
| C-section | 90 (49%) |
|
| |
| Baby Sex | |
| Male | 102 (56%) |
| Female | 79 (44%) |
|
| |
| Type of Preterm Birth | |
| Medically Indicated | 67 (37%) |
| PPROM | 43 (24%) |
| Spontaneous | 71 (39%) |
|
| |
| Birthweight | |
| Range | 580–3765 grams |
| Mean Birthweight | 2346 grams |
| Standard Deviation | 677 grams |
|
| |
| Chorioamnionitis | |
| Chorioamnionitis | 22 (16%) |
| None | 113 (84%) |
|
| |
| Funisitis | |
| Funisitis | 16 (12%) |
| None | 119 (88%) |
|
| |
| Gestational Age at Birth | |
| Range | 24–36 weeks |
| Mean | 33.9 weeks |
| Standard Deviation | 2.9 weeks |
Types of PTB and Infection
When examining type of preterm birth by infection status, more cases of chorioamnionitis occurred in preterm births that were the result of PPROM (n=14) or spontaneous PTL (n=8) when compared to medically indicated PTB (n=0), Fisher’s exact p-value <0.001), which was expected based on previous epidemiological studies.5 A similar phenomenon was observed in those infants with funisitis with more cases in preterm births that occurred as a result of PPROM (n=8) or spontaneous PTL (n=7) when compared to medically indicated PTB (n=1), Fisher’s exact p-value =0.003. In addition, chorioamnionitis and funisitis were highly correlated (correlation coefficient 0.6449, p<0.001).
Type of PTB, Infection and DNA Methylation at Imprint Regulatory Regions
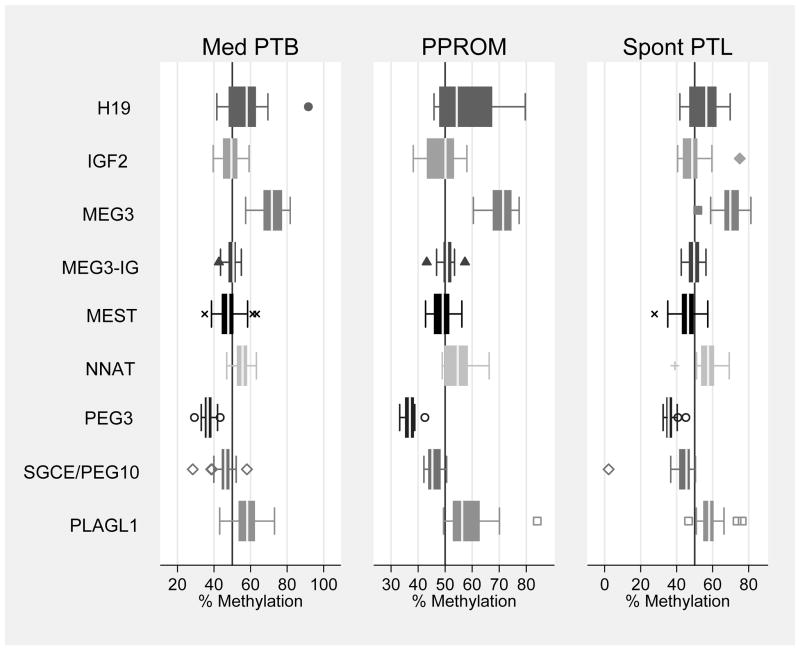
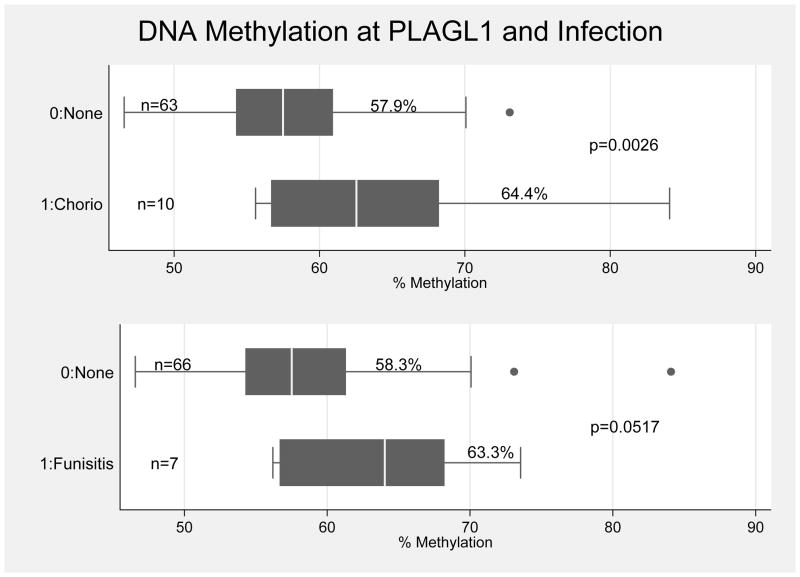
When examining the role of epigenetics in our subset of 73 preterm infants with completed methylation data, infant DNA methylation levels did not differ by type of PTB (medically indicated, PPROM, and spontaneous PTL) at any of the nine DMRs examined, p>0.20 (Figure 1). We found that infants born with pathological evidence of chorioamnionitis had higher methylation levels at the DMRs regulating the imprinted genes PLAGL1 and PEG3 (Table 2, p<0.05). We also found that infants born with pathological evidence of funisitis had higher methylation levels at the DMR regulating the imprinted gene PLAGL1 (Table 2, p<0.05). After Bonferroni correction for multiple testing, differences at PLAGL1 by chorioamnionitis persisted (p=0.0026). In logistic regression models (Table 3), this association persisted, and increasing levels of methylation at PLAGL1 was associated with increased risk for chorioamnionitis, after adjusting for the significant covariates maternal BMI and gestational age at birth (OR 1.22 95% CI 1.04–1.44, p=0.015). Decreasing gestational age at birth and increasing maternal BMI were also associated with increased risk of chorioamnionitis. Infant birthweight and gestational age at birth were highly correlated (correlation coefficient 0.7661, p<0.0001). As a result, although both were associated with chorioamnionitis, only gestational age at birth was included in overall models. After adjusting for significant covariates maternal age and infant sex, increasing levels of methylation at PLAGL1 was associated with increased risk for funisitis (OR 1.17 95% CI 1.03–1.32, p=0.016). Increasing maternal age and male gender were also associated with increased risk for funisitis. Gestational age at birth did not differ for funisitis (p=0.380), and it was not included in overall models. Figure 2 shows the distribution of DNA methylation levels at PLAGL1 by infection status (chorioamnionitis or funisitis).
Figure 1.
Distribution of DMRs by Type of PTB
Summary: Median and IQR of infant DNA methylation levels at nine DMRs by type of PTB. There are no significant differences between mean methylation levels at any of the nine DMRs by type of PTB, p>0.20. (Reference line represents 50% methylation).
Table 2.
Infant DNA Methylation by Infection Status
| Chorioamnionitis | ||||||
|---|---|---|---|---|---|---|
| None | Chorioamnionitis | |||||
| DMR | n | Mean % Methylation (SE) |
n | Mean % Methylation (SE) |
T-test P-value |
Ranksum P-value |
| H19 | 43 | 54.6 (1.25) | 6 | 54.1 (2.68) | 0.8882 | 0.9999 |
| IGF2 | 38 | 49.4 (1.07) | 5 | 48.9 (3.05) | 0.8824 | 0.8945 |
| MEG3-IG | 58 | 49.6 (0.40) | 10 | 50.6 (1.19) | 0.3829 | 0.4358 |
| MEG3 | 58 | 71.0 (0.81) | 9 | 70.1 (1.84) | 0.6858 | 0.6994 |
| MEST | 59 | 47.7 (0.68) | 9 | 44.9 (2.35) | 0.1670 | 0.4311 |
| PEG3 | 62 | 36.2 (0.27) | 10 | 38.1 (0.49) | 0.0081 | 0.0050 |
| NNAT | 58 | 55.5 (0.54) | 6 | 58.2 (2.20) | 0.1282 | 0.1971 |
| SGCE/PEG10 | 61 | 45.6 (0.54) | 10 | 42.9 (4.57) | 0.2239 | 0.4670 |
| PLAGL1* | 63 | 57.9 (0.70) | 10 | 64.4 (2.87) | 0.0026 | 0.0218 |
| Funisitis | ||||||
|---|---|---|---|---|---|---|
| None | Funisitis | |||||
| DMR | n | Mean % Methylation (SE) |
n | Mean % Methylation (SE) |
T-test P-value |
Ranksum P-value |
| H19 | 46 | 54.6 (1.19) | 3 | 53.3 (4.16) | 0.7824 | 0.8348 |
| IGF2 | 41 | 49.1 (1.02) | 2 | 53.5 (4.59) | 0.3685 | 0.2732 |
| MEG3-IG | 61 | 49.8 (0.39) | 7 | 49.4 (1.57) | 0.7398 | 0.3800 |
| MEG3 | 61 | 70.6 (0.78) | 6 | 73.0 (1.98) | 0.3606 | 0.3564 |
| MEST | 61 | 47.7 (0.66) | 7 | 43.8 (2.82) | 0.0717 | 0.2459 |
| PEG3 | 65 | 36.3 (0.27) | 7 | 37.6 (0.63) | 0.1105 | 0.0837 |
| NNAT | 60 | 55.7 (0.54) | 4 | 55.8 (2.73) | 0.9573 | 0.5982 |
| SGCE/PEG10 | 64 | 45.6 (0.52) | 7 | 41.4 (6.58) | 0.1065 | 0.4234 |
| PLAGL1 | 66 | 58.3 (0.78) | 7 | 63.3 (2.48) | 0.0517 | 0.0492 |
Bonferroni correction for multiple testing, p<0.005
Table 3.
Logistic Regression Model of Infection and PLAGL1 with Covariates
| chorioamnionitis | |||
|---|---|---|---|
| Variable | OR* | 95% CI | p-value |
| PLAGL1 | 1.22 | 1.04–1.44 | 0.015 |
| Gestational Age at Birth | 0.695 | 0.527–0.917 | 0.010 |
| Maternal BMI | 1.06 | 0.980–1.14 | 0.147 |
| Funisitis | |||
|---|---|---|---|
| Variable | OR* | 95% CI | p-value |
| PLAGL1 | 1.17 | 1.03–1.32 | 0.016 |
| Maternal Age | 1.09 | 0.97–1.23 | 0.134 |
| Gender (M->F) | 0.11 | 0.009–1.35 | 0.085 |
OR mutually adjusted for all the covariates listed in each model
Figure 2.
Infant DNA Methylation at PLAGL1 by Infection
Summary: Median and IQR of infant DNA methylation levels at PLAGL1 by chorioamnionitis and funisitis. Mean DNA methylation levels at PLAGL1 are higher in infants who experienced chorioamnionitis (difference 6.5%, p=0.0026) and funisitis (difference 5.0%, p=0.0517) compared with infants who experienced no infection at birth.
Comment
We examined the role of DNA methylation at multiple imprint regulatory regions implicated in growth and development by type of preterm birth and infection status. Both chorioamnionitis and funisitis were more common in PPROM and spontaneous PTL as compared to medically indicated PTB. We found no differences in DNA methylation at any of the nine DMRs examined by type of preterm birth. However, in preterm infants with pathologically defined chorioamnionitis or funisitis, DNA methylation may be increased at the PLAGL1 DMR, an association which persisted after adjustment for significant covariates.
PTB has been associated with significant mortality and morbidity, both at birth and later in development. Children born preterm are at increased risk for respiratory distress and apnea, hypoglycemia and infant death.4 Although longterm effects of PTB are still poorly understood, children and adolescents born preterm are at increased risk for behavioral and developmental disorders.4 In addition, epidemiological studies have seen associations between LBW and PTB and increased risk for obesity and cardiovascular disease in adulthood.6 Environmental factors such as nutrition, inflammation, and toxic exposures may increase susceptibility for PTB or infection through epigenetic perturbations and subsequent alterations in early growth and development.28 Chorioamnionitis has been associated with inflammation and changes in chemokine profiles and signaling.29 These endocrine disruptors may alter appropriate maintenance of epigenetic profiles during pregnancy, resulting in changes that adversely affect birth outcomes and may also be perpetuated through somatic cell division to increase susceptibility to disease in adulthood.30
Our study found differences in DNA methylation at the PLAGL1 DMR associated with chorioamnionitis and funisitis. PLAGL1 is located at chromosome 6q24.2 and encodes a zinc-finger transcription factor thought to be involved in tumor development and growth via IGF2 signaling.31 Aberrant epigenetic marks at this site are also associated with transient neonatal diabetes mellitus (TNDM), a disorder of growth restriction and hyperglycemia.32 Moreover, the PLAGL1 gene product is thought to function as a major regulatory hub that coordinates the expression of a network of genes, including many that are imprinted such as IGF2, H19 and MEST.33 The IGF2/H19 domain is one of the best characterized imprinted regions, located at chromosome 11p15.5, and was originally associated with Beckwith-Wiedemann syndrome, a somatic overgrowth disorder associated with increased risk of Wilms tumor and hepatoblastoma34 Elevated levels of DNA methylation at H19 have also been observed in assisted reproductive technology (ART-related) pregnancy loss as compared to spontaneously conceived pregnancy loss.35
We found changes at the PLAGL1 DMR associated with infection, and these alterations may not only affect growth/development at birth but also be maintained throughout life to increase susceptibility to adult-onset disease, as postulated by the developmental origins of adult disease hypothesis. This has potential implications for early screening of infection during pregnancy and novel epigenetic-based therapies to modulate clinical sequelae of infection related preterm birth. It is also possible that these marks were established earlier in pregnancy and may have increased risk for infection during pregnancy; however our study cannot differentiate between these two possibilities. Interestingly, we found no differences in DNA methylation at any of the nine DMRs examined by type of preterm birth, and this may reflect the heterogeneity in causes and risk factors for PTB.
Our study is one of the first to examine epigenetic regions in regards to type of preterm birth and infection. We used rigorous definitions verified by obstetrical experts for the various types of PTB and standardized pathological reports to define infection status. We also investigated methylation at multiple regions thought to be important in growth and development. Interestingly, we found that male sex was associated with increased risk of funisitis as compared to female sex, which may relate to sex-dependent epigenetic findings reported previously.13, 22 Limitations include the small sample size, cross-sectional nature of the study, which prevents assessment of causation, and relatively small regions of the imprintome examined. Although multiple testing may be a concern, the finding of differences in methylation at PLAGL1 by chorioamnionitis persisted after Bonferroni correction, and the difference by funisitis may be limited by small sample size. In addition, only cord blood methylation was examined in this study; however, prior studies have shown that DNA methylation at DMRs of these imprinted genes is consistent across tissues and cord blood fractions26 including PLAGL1, for which methylation of cord blood Polymorphonuclear Cells (PMNs) is slightly lower than that in matched Peripheral Blood Mononuclear Cells (PBMCs) by 0.68% (N=25; SKM, unpublished data). Future larger studies are required to verify these intriguing preliminary results and to further characterize these complex epigenetic networks and their interactions.
Acknowledgments
Financial Support: R01ES016772; R01DK085173; R21ES014947
We are especially grateful to the women and families involved in the Newborn Epigenetics STudy and acknowledge the expert contributions of study coordinator Stacy Murray, research nurse Tammy Bishop, and laboratory technicians Carole Grenier, Erin Erginer, Cara Davis and Allison Barratt.
Abbreviations
- LBW
Low Birthweight
- DMR
Differentially Methylated Region
- PTB
Preterm Birth
- PPROM
Preterm Premature Rupture of Membranes
- PTL
Preterm Labor
- BMI
Body Mass Index
- LMP
Last Menstrual Period
- NEST
Newborn Epigenetics STudy
- IQR
Interquartile Range
- C-section
Cesarean Section
- SE
Standard Error
Footnotes
Conflict of Interest: The authors report no conflicts of interest.
Presentations: Platform presentation, 33rd Annual Meeting, Society of Maternal-Fetal Medicine, February 11, 2013 - February 16, 2013, San Francisco, CA
References
- 1.Martin JA, Hamilton BE, Sutton PD, et al. Births: final data for 2007. National Vital Statistics Reports. 2010;58(24):1–125. [PubMed] [Google Scholar]
- 2.Iams JD, Romero R, Culhane JF, Goldenberg RL. Primary, secondary, and tertiary interventions to reduce the morbidity and mortality of preterm birth. Lancet. 2008 Jan 12;371(9607):164–175. doi: 10.1016/S0140-6736(08)60108-7. [DOI] [PubMed] [Google Scholar]
- 3.McCormick MC. The contribution of low birth weight to infant mortality and childhood morbidity. N Engl J Med. 1985 Jan 10;312(2):82–90. doi: 10.1056/NEJM198501103120204. [DOI] [PubMed] [Google Scholar]
- 4.Saigal S, Doyle LW. An overview of mortality and sequelae of preterm birth from infancy to adulthood. Lancet. 2008 Jan 19;371(9608):261–269. doi: 10.1016/S0140-6736(08)60136-1. [DOI] [PubMed] [Google Scholar]
- 5.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008 Jan 5;371(9606):75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007 May;261(5):412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 7.Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007 Apr-May;23(3):297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008 Nov 4;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, Li Z, Wang M, Martorell R. Early life exposure to the 1959–1961 Chinese famine has long-term health consequences. J Nutr. 2010 Oct;140(10):1874–1878. doi: 10.3945/jn.110.121293. [DOI] [PubMed] [Google Scholar]
- 10.Lumey LH, Stein AD, Kahn HS, Romijn JA. Lipid profiles in middle-aged men and women after famine exposure during gestation: the Dutch Hunger Winter Families Study. Am J Clin Nutr. 2009 Jun;89(6):1737–1743. doi: 10.3945/ajcn.2008.27038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravelli GP, Stein ZA, Susser MW. Obesity in young men after famine exposure in utero and early infancy. N Engl J Med. 1976 Aug 12;295(7):349–353. doi: 10.1056/NEJM197608122950701. [DOI] [PubMed] [Google Scholar]
- 12.St Clair D, Xu M, Wang P, et al. Rates of adult schizophrenia following prenatal exposure to the Chinese famine of 1959–1961. JAMA. 2005 Aug 3;294(5):557–562. doi: 10.1001/jama.294.5.557. [DOI] [PubMed] [Google Scholar]
- 13.Tobi EW, Lumey LH, Talens RP, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009 Nov 1;18(21):4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burris HH, Rifas-Shiman SL, Baccarelli A, et al. Associations of LINE-1 DNA Methylation with Preterm Birth in a Prospective Cohort Study. J Dev Orig Health Dis. 2012 Jun;3(3):173–181. doi: 10.1017/s2040174412000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulkarni A, Chavan-Gautam P, Mehendale S, Yadav H, Joshi S. Global DNA methylation patterns in placenta and its association with maternal hypertension in pre-eclampsia. DNA Cell Biol. 2011 Feb;30(2):79–84. doi: 10.1089/dna.2010.1084. [DOI] [PubMed] [Google Scholar]
- 16.Pliushch G, Schneider E, Weise D, et al. Extreme methylation values of imprinted genes in human abortions and stillbirths. Am J Pathol. 2010 Mar;176(3):1084–1090. doi: 10.2353/ajpath.2010.090764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008 Jul 3;359(1):61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menon R, Conneely KN, Smith AK. DNA methylation: an epigenetic risk factor in preterm birth. Reprod Sci. 2012 Jan;19(1):6–13. doi: 10.1177/1933719111424446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skaar DA, Li Y, Bernal AJ, Hoyo C, Murphy SK, Jirtle RL. The human imprintome: Regulatory mechanisms, methods of ascertainment, and roles in disease susceptibility. ILAR Journal. 2012;53:337–354. doi: 10.1093/ilar.53.3-4.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Murphy SK, Murtha AP, et al. Depression in pregnancy, infant birth weight and DNA methylation of imprint regulatory elements. Epigenetics. 2012 Jul;7(7):735–746. doi: 10.4161/epi.20734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoyo C, Murtha AP, Schildkraut JM, et al. Folic acid supplementation before and during pregnancy in the Newborn Epigenetics STudy (NEST) BMC Public Health. 2011;11(1):46. doi: 10.1186/1471-2458-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy SK, Adigun A, Huang Z, et al. Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene. 2012 Feb 15;494(1):36–43. doi: 10.1016/j.gene.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer MS. The epidemiology of adverse pregnancy outcomes: an overview. The Journal of nutrition. 2003;133(5):1592S–1596S. doi: 10.1093/jn/133.5.1592S. [DOI] [PubMed] [Google Scholar]
- 24.ACOG. Practice Bulletin. Diagnosis and management of preeclampsia and eclampsia. Number 33, January 2002. American College of Obstetricians and Gynecologists. Int J Gynaecol Obstet. 2002;77(1):67–75. [PubMed] [Google Scholar]
- 25.Redline RW, Faye-Petersen O, Heller D, Qureshi F, Savell V, Vogler C. Amniotic infection syndrome: nosology and reproducibility of placental reaction patterns. Pediatr Dev Pathol. 2003 Sep-Oct;6(5):435–448. doi: 10.1007/s10024-003-7070-y. [DOI] [PubMed] [Google Scholar]
- 26.Murphy SK, Huang Z, Hoyo C. Differentially methylated regions of imprinted genes in prenatal, perinatal and postnatal human tissues. PLoS One. 2012;7(7):e40924. doi: 10.1371/journal.pone.0040924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nye MD, Hoyo C, Huang Z, et al. Associations between methylation of Paternally Expressed Gene 3 (PEG3), cervical intraepithelial neoplasia and invasive cervical cancer. PLoS One. 2013 doi: 10.1371/journal.pone.0056325. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 29.Redline RW. Inflammatory response in acute chorioamnionitis. Semin Fetal Neonatal Med. 2012 Feb;17(1):20–25. doi: 10.1016/j.siny.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Robins JC, Marsit CJ, Padbury JF, Sharma SS. Endocrine disruptors, environmental oxygen, epigenetics and pregnancy. Front Biosci (Elite Ed) 2011;3:690–700. doi: 10.2741/e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Dyck F, Declercq J, Braem CV, Van de Ven WJ. PLAG1, the prototype of the PLAG gene family: versatility in tumour development (review) Int J Oncol. 2007 Apr;30(4):765–774. [PubMed] [Google Scholar]
- 32.Temple IK, Mackay DJG, Docherty LE. Diabetes Mellitus, 6q24-Related Transient Neonatal. 1993 [Google Scholar]
- 33.Varrault A, Gueydan C, Delalbre A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006 Nov;11(5):711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Engel JR, Smallwood A, Harper A, et al. Epigenotype-phenotype correlations in Beckwith-Wiedemann syndrome. J Med Genet. 2000 Dec;37(12):921–926. doi: 10.1136/jmg.37.12.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zechner U, Pliushch G, Schneider E, et al. Quantitative methylation analysis of developmentally important genes in human pregnancy losses after ART and spontaneous conception. Mol Hum Reprod. 2010 Sep;16(9):704–713. doi: 10.1093/molehr/gap107. [DOI] [PubMed] [Google Scholar]