Abstract
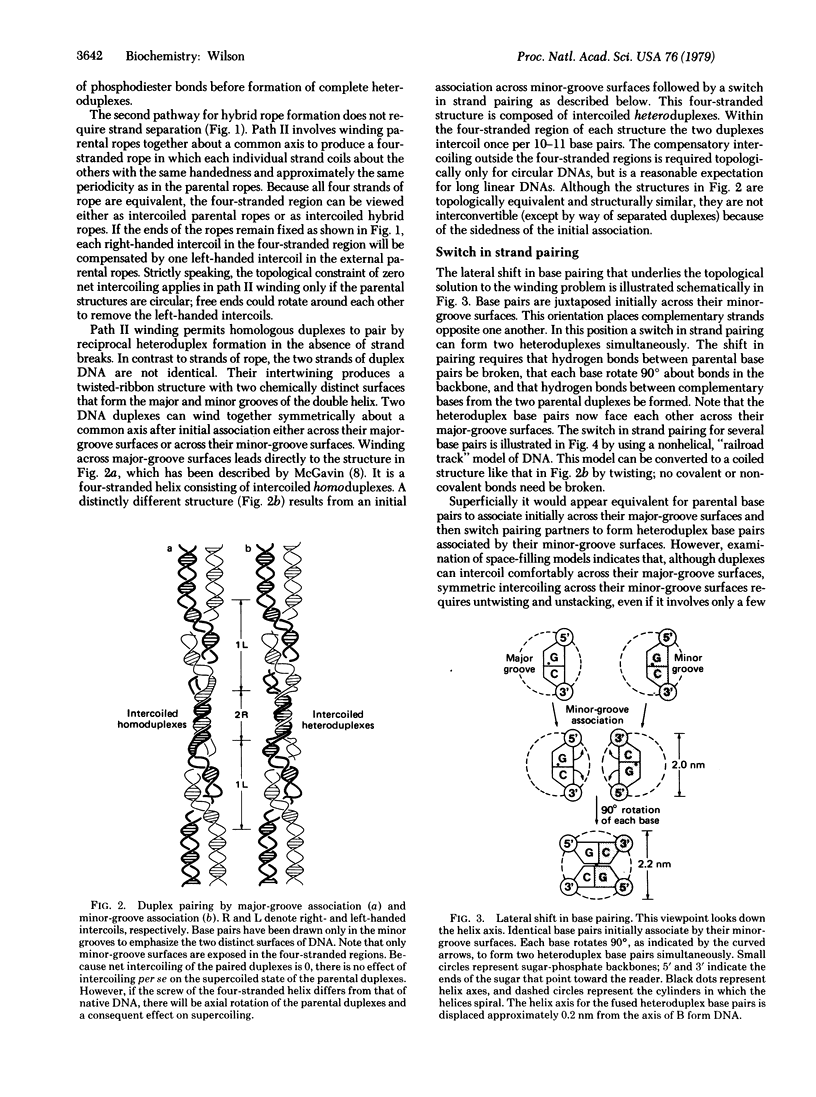
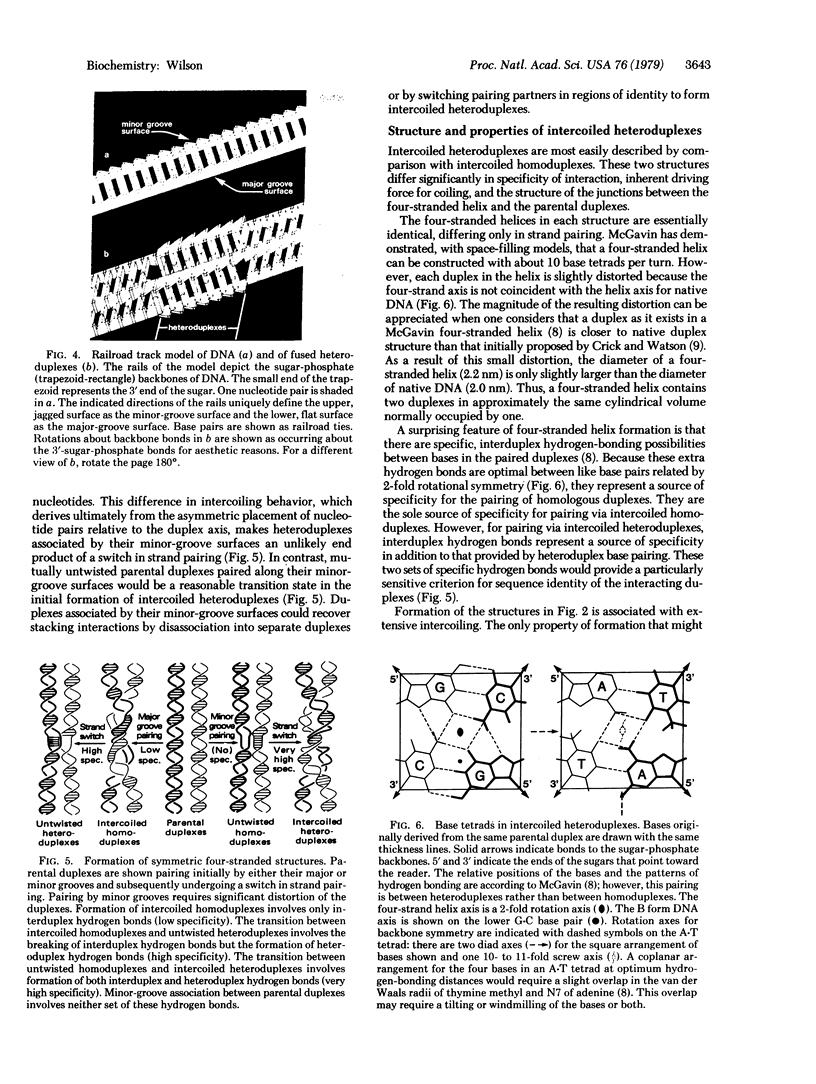
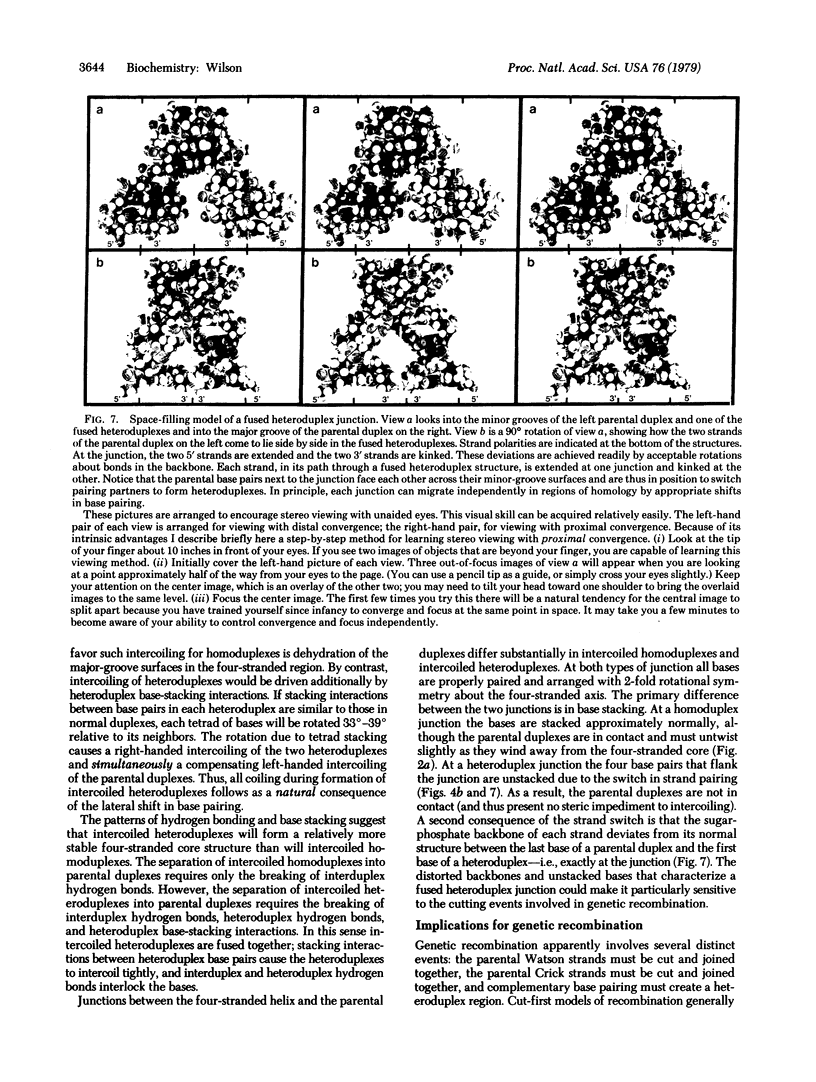
Because the individual strands of DNA are intertwined, formation of heteroduplex structures between duplexes--as in presumed recombination intermediates--presents a topological puzzle, known as the winding problem. Previous approaches to this problem have assumed that single-strand breaks are required to permit formation of fully coiled heteroduplexes. This paper describes a simple, nick-free solution to the winding problem that satisfies all topological constraints. Homologous duplexes associated by their minor-groove surfaces can switch strand pairing to form reciprocal heteroduplexes that coil together into a compact, four-stranded helix throughout the region of pairing. Model building shows that this fused heteroduplex structure is plausible, being composed entirely of right-handed primary helices with Watson-Crick base pairing throughout. Its simplicity of formation, structural symmetry, and high degree of specificity are suggestive of a natural mechanism for alignment by base pairing between intact homologous duplexes. Implications for genetic recombination are discussed.
Full text
PDF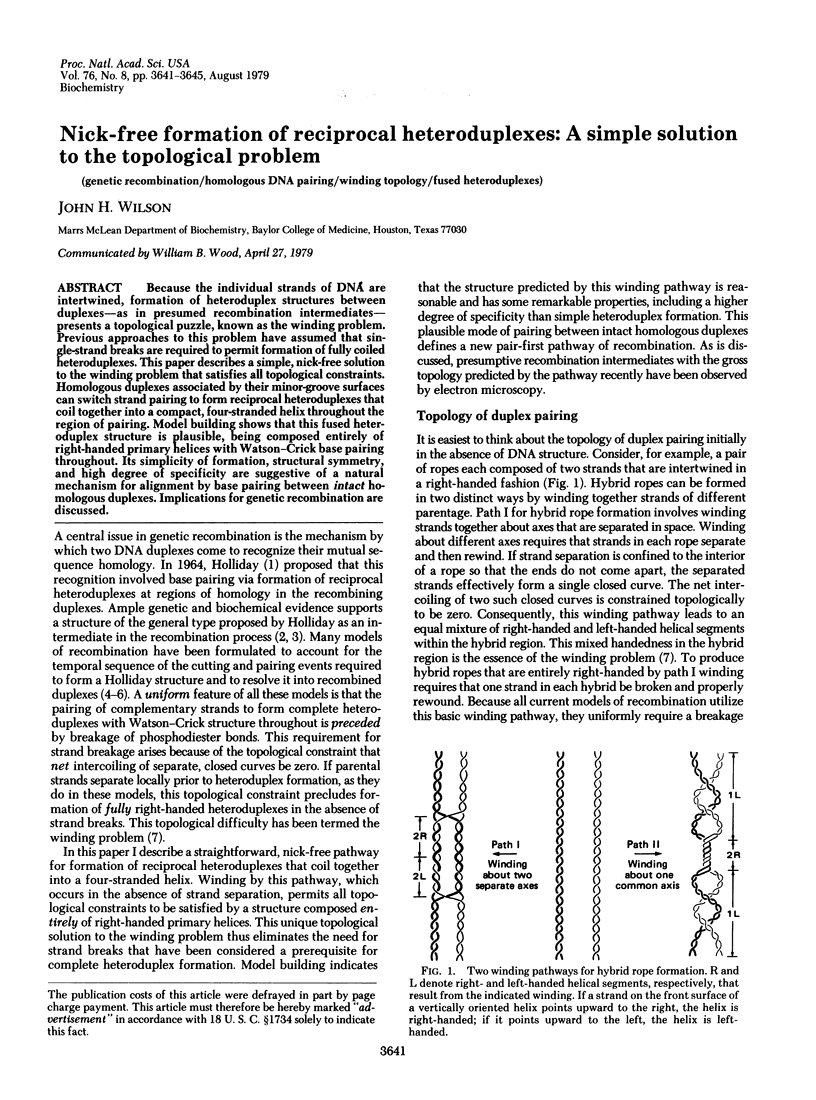

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benbow R. M., Krauss M. R. Recombinant DNA formation in a cell-free system from Xenopus laevis eggs. Cell. 1977 Sep;12(1):191–204. doi: 10.1016/0092-8674(77)90197-0. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. Renaturation of complementary single-stranded DNA circles: complete rewinding facilitated by the DNA untwisting enzyme. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5328–5332. doi: 10.1073/pnas.74.12.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings P. J. Some aspects of recombination in eukaryotic organisms. Annu Rev Genet. 1975;9:129–144. doi: 10.1146/annurev.ge.09.120175.001021. [DOI] [PubMed] [Google Scholar]
- Hotchkiss R. D. Models of genetic recombination. Annu Rev Microbiol. 1974;28(0):445–468. doi: 10.1146/annurev.mi.28.100174.002305. [DOI] [PubMed] [Google Scholar]
- McGavin S. Models of specifically paired like (homologous) nucleic acid structures. J Mol Biol. 1971 Jan 28;55(2):293–298. doi: 10.1016/0022-2836(71)90201-4. [DOI] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter H., Dressler D. In vitro system from Escherichia coli that catalyzes generalized genetic recombination. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3698–3702. doi: 10.1073/pnas.75.8.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C. M. Genetic recombination: strand transfer and mismatch repair. Annu Rev Biochem. 1978;47:847–880. doi: 10.1146/annurev.bi.47.070178.004215. [DOI] [PubMed] [Google Scholar]
- Sobell H. M. Molecular mechanism for genetic recombination. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2483–2487. doi: 10.1073/pnas.69.9.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]