Abstract
Polymeric nanoparticles are promising for gene therapy and stem cell reprogramming using non-viral vectors. A novel assay utilizing nanoparticle tracking analysis (left) is developed here to easily quantify the number of plasmids within polymeric nanoparticles while in aqueous solution. Particles effective at co-transfecting primary human fibroblasts (right) are ~100 nm in diameter and contain ~100 plasmids per particle.
Keywords: polymers, nanoparticles, nanoparticle tracking, gene expression, regenerative medicine
Gene delivery is attractive both to treat genetic diseases[1] and as a technology for regenerative medicine.[2] For successful clinical translation of a cell-based gene therapy approach[3], it is important to address two key challenges: 1) to develop safe and effective gene delivery vectors[4] and 2) to find a replenishable and genetically matched source of cells.[5, 6] Non-viral gene delivery is a safer alternative to viral vectors.[7–10] Cationic polymers condense DNA via electrostatic interaction with the negatively charged DNA backbone to form self-assembled complexes on the order of ~100 nm and are commonly termed polyplexes or polymeric nanoparticles. Polyethylenimine (PEI) is a widely-used off-the-shelf cationic polymer that forms polymeric nanoparticles with DNA and is non-degradable.[11] Poly(beta-amino ester)s (PBAEs) are a newer class of cationic polymers that similarly form polymeric nanoparticles, but are hydrolytically degradable.[12] End-modification of PBAEs has shown improved gene delivery efficacy in certain cell types.[13–15]
Recently, human fibroblasts have been successfully reprogrammed to induced pluripotent stem cells (iPSCs)[16] that offer potential applications in regenerative medicine as a replenishable source of autologous stem cells.[16–18] Many reprogramming studies utilize viral vectors that have higher efficiency but are unsafe for clinical applications, while non-viral methods, such as nanoparticle-based methods, have reported low reprogramming efficiency.[6] Reprogramming requires transfection of a combination of plasmids that encode reprogramming factors.[6, 19] Since the number of plasmids complexed per particle affects transfection efficiency, a quantitative determination of this number would benefit the development of nanoparticle-based gene delivery strategies, especially for co-delivery of multiple plasmids to the same target as required in factor-based reprogramming. Few studies have investigated the composition of gene delivery nanoparticles in terms of the number of plasmids associated with a single nanoparticle, and the assays used have had significant drawbacks that affect the estimation.[20–23] In the present study, we developed a novel assay to easily quantify the number of plasmids within polymeric nanoparticles in physiologically relevant aqueous conditions using a nanoparticle tracking analysis technique. We investigated PEI and six formulations of PBAEs optimized for gene delivery to human primary fibroblasts (IMR90). We studied how small modifications, including to the polymer structure and polymer to DNA weight ratio, affect the composition of the nanoparticles in terms of plasmids associated per particle and its implications in co-transfection of multiple plasmids to the same cells. Particles effective at transfecting primary human fibroblasts had 30–120 plasmids per particle, depending on polymer structure. Nanoparticles that contain a high level of plasmids per particle (120 vs. 30) proved promising for coexpression. This quantitative understanding has implications for co-delivery of therapeutic genes and for stem cell reprogramming using nanoparticles.
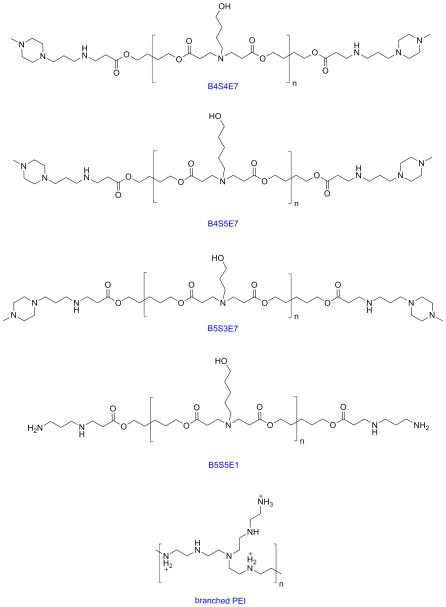
The structures of the polymers used in this study are shown in Figure 1. We focused on six end-capped PBAE formulations: B4S4E7, with base polymer synthesized at a 1.2:1 monomer ratio and formulated at 40 weight polymer to weight DNA (wt/wt), B4S4E7 (1.2:1, 60 wt/wt), B5S3E7 (1.05:1, 60 wt/wt), B5S3E7 (1.05:1, 100 wt/wt), B4S5E7 (1.2:1, 100 wt/wt), and B5S5E1 (1.2:1, 100 wt/wt). The nanoparticles were formed by complexing polymers with enhanced green fluorescent (EGFP-N1) plasmid DNA in 25 mM, pH 5 sodium acetate (NaAc) solution. The 25 kDa branched PEI-based control nanoparticles were prepared in 150 mM, pH 5.5 sodium chloride (NaCl) solution at 2 wt/wt ratio (N/P ratio of 16).
Figure 1.
The chemical structures of the PBAEs and PEI used in this study.
The nanoparticles were used for transfecting human fetal fibroblasts (IMR90) with EGFP and DsRed plasmids. The cells were transfected in three different conditions, a) transfection with complexes containing both EGFP and DsRed plasmids for 4 hours, b) transfection with exclusively EGFP containing complexes for 2 hours, followed by transfection with the same dose of exclusively DsRed containing complexes for 2 hours after media change and c) transfection with exclusively EGFP containing complexes for 4 hours, followed by transfection with the same dose of exclusively DsRed containing complexes for 4 hours after a gap of 24 hours.
The nanoparticles were analyzed by nanoparticle tracking analysis (NTA) using a Nanosight LM10 that tracks the random Brownian motion of individual nanoparticles suspended in an aqueous solution. The particle characterization performed using NTA was confirmed by dynamic light scattering (DLS) using Malvern Zetasizer NanoZS. Unlike the intensity-based distribution of particle size given by DLS, NTA gives a direct number-averaged distribution of the particle hydrodynamic diameter by tracking individual particles in real-time as light scattering centroids using a sensitive CCD camera. The number mean particle size (diameter in nanometers) and absolute particle concentration (number of particles/mL) were determined using NTA.
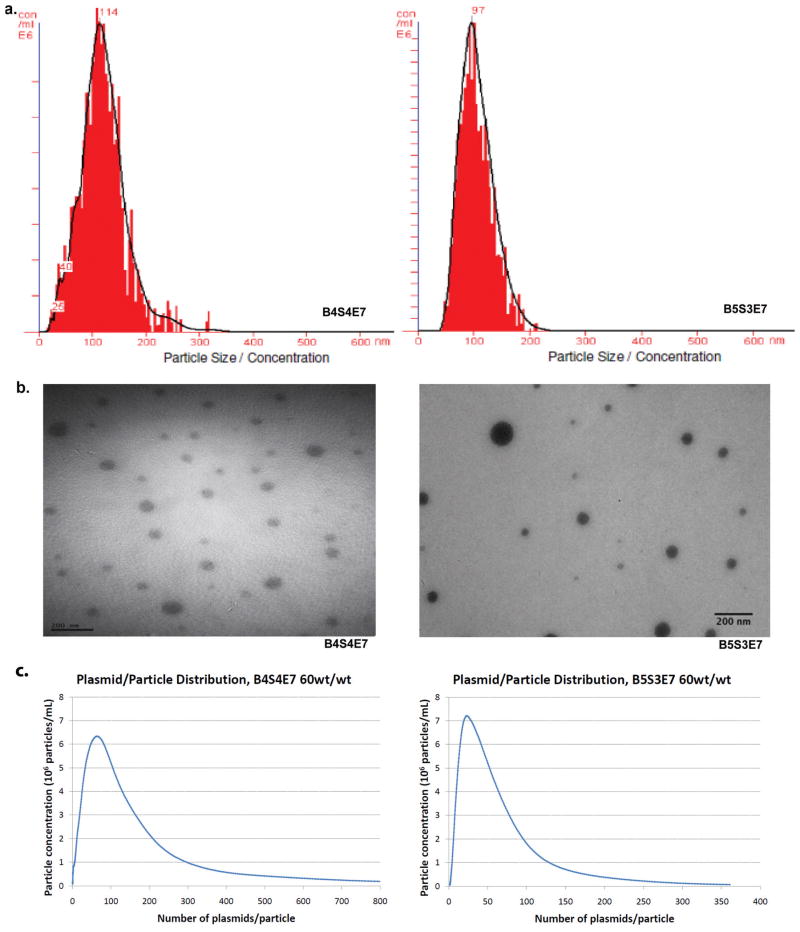
All polymers used in this study formed monodisperse particle size distributions as verified by a unimodal distribution by NTA (Figure 2a). Transmission Electron Microscopy (TEM) was also performed with dried nanoparticles and gave comparable sizes to the hydrated nanoparticles (Figure 2b). The samples were analyzed by gel electrophoresis to evaluate whether 100% of plasmid was complexed into particles at the wt/wt ratios used for NTA measurements. Each lane in the gel contained nanoparticles formulated at a particular wt/wt and the first lane contained a control sample of free DNA by itself (Figure S1). The gel electrophoresis results validate that free DNA is not present in these samples and all plasmid for these formulations is complexed within nanoparticles. The known mass (calculated from concentration and total volume) and molecular weight of the EGFP-N1 plasmid (4.7 kilobase pairs, kbp) used for particle formulation was used to calculate the number of plasmids per mL. As all of the DNA was complexed within nanoparticles and all the nanoparticles were monodisperse, the ratio of plasmids per mL to particles per mL (concentration measured using NTA) determines the average number of plasmids per particle. The analysis was performed for PEI and the six different PBAE formulations to study the effect of varying polymer structure and wt/wt ratio on the number of plasmids associated per nanoparticle.
Figure 2.
Size distribution and plasmid per particle distribution data of PBAE (B4S4E7 and B5S3E7, both 60wt/wt) based nanoparticles. (a) particle size from nanoparticle tracking analysis (b) particle size from transmission electron microscopy (c) plasmid per particle distribution from nanoparticle tracking analysis.
NTA tracks individual nanoparticles so that number fractions can be directly calculated. The nanoparticle sizes were binned rounding to the nearest nanometer, and the following equations were used to determine average nanoparticle size (Table 1). The number mean diameter, d̄, reported by NTA is determined using eq.1. The mean volume based-diameter, , was calculated using eq.2 and used to estimate the average theoretical maximum number of plasmids per particle. This average quantity gives the average volume of a particle randomly selected (or taken up by the cell) from the population of particles.
Table 1.
Nanoparticle size averages of the seven formulations. Z-average determined by dynamic light scattering (DLS), with all other sizing determined by nanoparticle tracking analysis (NTA). Error reported is standard error of the mean.
| Polymer | Molecular Weight (Da) | wt/wt | Number-weighted mean particle diameter (nm) | Mean volume based-diameter (nm) | Z-average particle diameter (nm) |
|---|---|---|---|---|---|
| B4S4E7 | 28590 | 40 | 120±10 | 130±10 | 291±9 |
| B4S4E7 | 28590 | 60 | 130±10 | 150±10 | 379±9 |
| B4S5E7 | 11072 | 60 | 110±10 | 130±10 | 106±9 |
| B5S3E7 | 4632 | 60 | 114±3 | 124±4 | 110±10 |
| B5S3E7 | 4632 | 100 | 101±2 | 112±4 | 120±10 |
| B5S5E1 | 2873 | 100 | 106±1 | 115±1 | 113±5 |
| PEI 25kDa | 25000 | 2 | 150±10 | 170±10 | 317±2 |
| (1) |
| (2) |
d̄ = Number - weighted Mean Diameter
Single condensed plasmid size was estimated from literature data to determine the average plasmid volume. Dividing the average particle volume by the volume of a condensed plasmid determines the average theoretical maximum number of plasmids per particle. Based on branched 25 kDa PEI saturated DNA condensate sizes and cationic thiol-detergent compacted single plasmid DNA dimensions reported in previous publications, the diameter used for a single condensed 5 kbp plasmid DNA can range from 20–40 nm.[24, 25] The choice of a plasmid size from within this range can dramatically affect the theoretical maximum calculation. Therefore, a likely minimum condensed plasmid size was calculated using the result that the distance between DNA helices in DNA condensates is 2.7 nm.[26] This value has been reported to range from 2.4–2.8 nm,[27, 28] but this variation has a smaller effect on the calculations than varying the plasmid diameter. Assuming a cylindrical shape, the volume of a minimum condensed plasmid can then be calculated to be 9140 nm3 ( = π*(2.7/2 nm)2*(0.34 nm per base pair)*(4.7 kbp), where 0.34 nm equals the length of a DNA rod per base pair[29] and each plasmid contains 4.7 kbp). A volume of 9140 nm3 corresponds to an equivalent spherical diameter of 25.9 nm. In general, the actual number of plasmids per particle for any sized particle is less than the theoretical maximum. This may be due to nanoparticle volume taken up by extra polymer that is not tightly associated to condensed DNA as well as a possible range in the degree of plasmid condensation.
Another important point is that there is a distribution of plasmid/particle numbers, reflecting the distribution of particle sizes observed within a population. Therefore, there will be particles with plasmid/particle numbers both greater than and less than the average numbers. An example of the profiles can be seen in Figure 2C, as well as in supplemental Figure S2, which together show the plasmid per particle distribution data for all formulations as determined by calculating the number of plasmids per particle for each 1 nm increment (see methods).
Table 2 shows the particle per plasmid quantification. Each nanoparticle condition was fabricated and analyzed in at least duplicate independent batches and each of these replicates included a minimum of 500 completed tracks by NTA to ensure good statistics for the number of particles and their size. The 25 kDa branched PEI-based nanoparticles had 90±10 plasmids per particle on average as calculated by NTA with an average theoretical maximum estimate of 267 plasmids per particle. The average plasmids per particle counts, calculated by NTA, for the six PBAE formulations ranged from 30±2 to 120±20 and the average theoretical maximum average estimates ranged from 80 to 195. Similar to PEI-based nanoparticles, B4S4E7 nanoparticles at 60 wt/wt had 110±10 plasmids per particle and at 40 wt/wt, 120±20 plasmids per particle as calculated by NTA. Changing the polymer to DNA weight ratio, from 40 to 60 wt/wt for B4S4E7-based nanoparticles and 60 to 100 wt/wt for B5S3E7-based nanoparticles did not significantly change the number of plasmids associated per particle. Interestingly, B4S4E7 nanoparticles, which are highly effective at transfecting IMR90 cells, have the highest plasmid per particle counts in the PBAE-DNA nanoparticles tested. This indicates that, in addition to polymer structure, plasmid per particle count maybe an important parameter to determine the transfection efficacy of PBAE-based nanoparticles for a specific cell type. A study conducted by Ogris et al. correlated DNA/PEI (with or without transferrin ligand) complex size with transfection efficiency.[30] Greater average intensity of green fluorescent protein (GFP) expression per cell was found following transfection with larger sized particles compared to smaller sized particles. This may be due in part to the larger particles containing more plasmids per particle than the smaller particles.
Table 2.
The average number of plasmids per nanoparticle for the seven nanoparticle formulations as determined by nanoparticle tracking analysis (NTA).
| Polymer | wt/wt | Mode plasmid/particle | Average plasmid/particle | Standard Error | Number of Tracks | Average theoretical max plasmid/particle | Average kbp/particle | Average theoretical max kbp/particle |
|---|---|---|---|---|---|---|---|---|
| B4S4E7 | 40 | 57 | 120 | 20 | >500 | 127 | 564 | 597 |
| B4S4E7 | 60 | 63 | 110 | 10 | >500 | 195 | 517 | 917 |
| B4S5E7 | 60 | 29 | 90 | 30 | >500 | 119 | 423 | 559 |
| B5S3E7 | 60 | 23 | 45 | 9 | >500 | 110 | 212 | 517 |
| B5S3E7 | 100 | 17 | 30 | 2 | >500 | 80 | 141 | 376 |
| B5S5E1 | 100 | 23 | 35 | 2 | >500 | 86 | 165 | 404 |
| PEI 25kDa | 2 | 49 | 90 | 10 | >500 | 267 | 423 | 1255 |
The three B4-based nanoparticles have similar plasmids per particle counts (90–120) and the three B5-based nanoparticles have similar plasmids per particle counts (30–45). Intriguingly, the B4-based nanoparticles have approximately 3-fold the DNA carrying capacity per particle even though the structure of B4 and B5 differ by only a single carbon to the repeating backbone and both B4-based and B5-based nanoparticles have similar ~100 nm particle size as measured by NTA analysis. This finding highlights how the number of plasmids/particle is an important parameter for consideration in nanoparticle design and characterization and it can be tuned through changes to biomaterial structure.
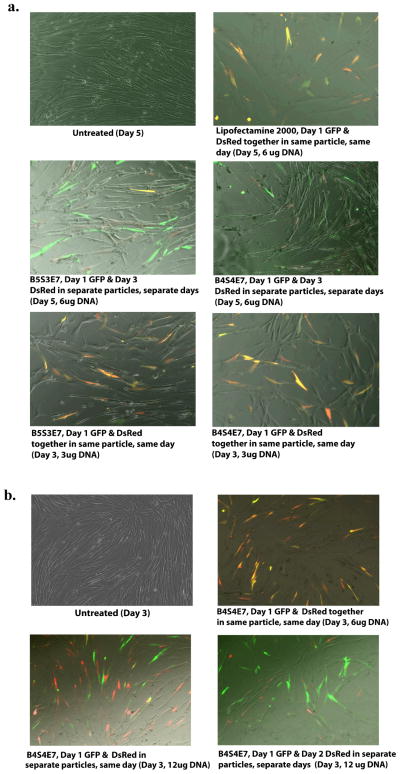
A critical barrier to the generation of induced pluripotent stem cells and to achieving high reprogramming efficiency with non-viral vectors is the low probability of uptake and expression of multiple plasmids (carrying multiple reprogramming factors) by the same cell. Accordingly, co-transfection experiments using EGFP and DsRed plasmids were conducted in IMR90s to gain insight into the process of delivering multiple plasmid types intracellularly using nanoparticles. Similar co-delivery is seen with PBAE-based nanoparticles as with Lipofectamine 2000, a leading commercially available transfection agent, but with higher cell viability for the PBAE-based nanoparticles. Gene expression following co-delivery with nanoparticles containing multiple plasmids differs from delivery of the same genes in the same type of nanoparticles when formulated and administered separately. Figure 3 shows that if the two plasmids are mixed together before nanoparticle self-assembly to form co-delivery particles, more cells coexpress the two plasmids together (orange-yellow cells) than when the two plasmids are formulated and delivered separately with the same type of nanoparticles on two different days (green and red cells, Figure 3a). When the nanoparticles are formulated separately and administered together on the same day, the trend of reduced co-delivery also holds (Figure 3b). In the case of nanoparticles that contain both GFP and DsRed plasmids, most of the cells exhibit coexpression (yellow/orange cells). When the same type of nanoparticles are used on the same day at the same dose, but containing exclusively GFP and then exclusively DsRed, very few cells exhibit coexpression (yellow/orange cells) and instead individual plasmid expression is strongly seen as either EGFP (green cells) or DsRed (red cells). Cells also do not exhibit coexpression (yellow/orange cells) of the GFP and DsRed plasmids in the separate particles when they are administered 24 hours apart. Thus, for certain applications, co-delivery within the same nanoparticles is critical to ensure coexpression. Schwake et al. presented a stochastic model of transfection to predict co-transfection ratio and plasmid expression level distribution in Lipofectamine and PEI complexes.[31] They studied co-transfection of pre-mixed plasmids encoding yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) and post-mixed plasmids (complexes contains either CFP or YFP plasmid) and observed that the co-transfection ratio was higher (21.9%) in pre-mixed than in post-mixed complexes (12.9%), which could also be predicted by their mathematical model.[31] This observation is consistent with our observations in the co-transfection experiments in IMR90 cells. Choosing nanoparticles that contain a high level of plasmids per particle (120 vs. 30) may be a promising strategy to maximize coexpression. Flow cytometry analysis of the co-transfected cells (same day, same particle) showed that B4S4E7-based nanoparticles achieved coexpression in 15.8±0.5% of live cells, whereas B5S3E7-based nanoparticles achieved coexpression in 3.3±1% of live cells (Figure S3).
Figure 3.
Co-transfection data in IMR90 human fibroblasts. B4S4E7 and B5S3E7-based nanoparticles were used to deliver EGFP and DsRed plasmids within the same particles on the same day as well as a) within separate particles on separate days and b) within separate particles on the same day. Lipofectamine 2000 was used as a positive control.
Previous studies on the number of plasmids contained within gene delivery nanoparticles have been limited.[20–23] In studies using cryo-TEM, X-ray scattering, and DLS to evaluate lipopolyamine-DNA complexes, up to 7 plasmids (6.4 kbp) per ~50 nm lipoplex particle and 13 plasmids (3.7 kbp) per 50–100 nm lipoplex particle were calculated.[21, 23] In making these calculations, the particles were assumed as monodisperse spheres although size showed irregularities. Based on the periodicity of ordered microdomains of the particles, half the TEM volume of the ~50 nm spheres was assumed to be occupied by DNA and the number of plasmids was calculated by dividing this volume by the density of DNA as 1.66 g cm−3. In this manner, as the length of DNA was shortened, the number of plasmids per particle was calculated to increase linearly up to 55 plasmids per lipoplex particle when the DNA was 0.9 kbp in size.24 This study, which analyzed lipoplexes and did not include polymeric nanoparticles, is dependent on the observation of ordered microdomain structure and did not look at the effect of varying biomaterial structure on nanoparticle composition and properties.
In an alternative strategy, Ho et al. reported the use of quantum dot (QD) labeling combined with regular TEM for estimation of plasmids per particle in chitosan-based nanoparticles.[20] The number of quantum dots per polyplex nanoparticle can be determined by the ratio of fluorescence signal from a single polyplex compared to a single QD and by counting the electron-dense regions of QDs within a polyplex in a TEM image. Estimating 0–3 QD per plasmid, ~30 plasmids per particle were calculated for a ~60 nm particle.[20] While this QD technique works very well as a novel tool to evaluate intracellular stability and DNA unpacking, it has limitations in easily estimating plasmids per particle as the DNA must be first labeled and this may change its self-assembly properties with certain materials, unlabeled DNA that may be encapsulated is not directly detected, counting QDs per particle by TEM becomes more difficult as the number of plasmids increases due to potentially overlapping plasmids and QDs in the 2D image, and other simplifying assumptions.
A third approach at measuring plasmids per particle was developed by Collins et al. who studied (Lys)16 containing peptide/DNA complexes and quantified the number of plasmids per particle in physiologically relevant aqueous solution.[22] However, the technique used, Flow Particle Image Analysis, is only amenable to sizing larger, micron-sized particles. A polylysine-based Polymol peptide was found to form ~1 μm particles that contained an estimated 70,000 (6.0 kbp) plasmids per particle. These microparticles are approximately 10-fold larger than the nanoparticles used in the current study, and hence, their volume is ~103-fold greater.
For all formulations described in this manuscript, calculated plasmid per particle counts were lower than the estimated theoretical maximum value, but of the same order of magnitude. The calculated values determined from this novel NTA method are similar in some ways to the numbers previously reported in literature including an average of 30 plasmids per particle for chitosan-DNA nanoparticles[20] and 13 plasmids per particle for lipopolyamine-DNA lipoplex nanoparticles.[21] These are both close to the low range of the PBAE nanoparticles described. However, we also show that other PBAE nanoparticles as well as PEI nanoparticles can have a much higher DNA carrying capacity of ~100 plasmids per particle. This higher DNA loading may be especially important for the co-delivery of genetic therapeutics for the treatment of diseases such as cancer or as an enabling tool for stem cell reprogramming.
Although TEM is routinely used for nanoparticle characterization, and can be used to estimate particle concentration, there are drawbacks of using this method. They include errors introduced by processing effects such as dehydration and aggregation of “soft” self-assembled particles that are typically hydrated in their native state, limited sample size per image for sufficient statistics, consumption of the sample by the assay, and relatively high cost and time consumed for sample collection and analysis. DLS is unable to measure particle concentration and reports size that is biased towards larger particles. Thus, a new technique is required. Here, we show that NTA is a technique that is able to meet this need. In addition to its utility at measuring number-averaged size and concentration of nanoparticles, the assay is also quick and non-destructive of the sample.
In comparison to viruses, which are natural nanoparticles, the polymeric nanoparticles are found to contain up to 100-fold more copies of their delivery genome than viruses contain. For example, while adenovirus particles and the polymer-based nanoparticles evaluated here both contain double stranded DNA, are both ~100 nm in size, and have been shown to have comparable efficacy in vitro to certain human cells,[32] this work shows that each synthetic nanoparticle is dramatically less efficient in delivery. Future work is needed to improve the efficiency of synthetic nanoparticles for non-viral gene delivery.
In conclusion, a quick, easy and robust assay for quantifying the number of plasmids per nanoparticle in physiologically relevant aqueous solution was developed. PBAE-based and PEI-based polymeric nanoparticles were analyzed by varying their polymer structure and polymer to DNA weight ratio. The plasmids per particle values ranged from 30 to 120 plasmids per particle and depended on polymer structure. More plasmids per particle led to higher co-expression following gene delivery. To our knowledge, this is the first study to report characterization of polymeric nanoparticles in terms of plasmids per particle in aqueous conditions. Characterization of polymeric nanoparticles in terms of the number of plasmids per particle can allow for improved design to non-virally co-deliver multiple plasmids to the same target, a requirement for certain gene therapies and for factor-based stem cell reprogramming.
Experimental Section
Polymer synthesis and nanoparticle formation
Details of the polymer synthesis and nanoparticle formation method have been previously described.[13] Briefly, diacrylate terminated base polymers (BxSx) were first synthesized at 90 C by conjugate addition of acrylate and amine monomers for 24 hours. As a second step, the base polymers were end-capped with amine containing small molecules at room temperature in DMSO to form amine terminated end-capped PBAE versions (BxSxEx). To form nanoparticles, PBAE stock solutions (100 mg mL−1) in DMSO and EGFP-N1 DNA stock (1 mg mL−1) in deionized (DI) water were both diluted separately in sodium acetate (NaAc) solution (25 mM, pH 5) at concentrations required to achieve polymer to DNA weight ratios (wt/wt) of 40, 60 and 100 and mixed vigorously at a 1:1 ratio, with final concentrations at 0.006 or 0.06 mg DNA per mL. The nanoparticles were formed through self-assembly during a 10-minute incubation. PEI-based nanoparticles at a 2 wt/wt ratio were prepared similarly, but using a sodium chloride solution (150 mM, pH 5.5).
Cell Transfection
IMR90s maintained in Eagle’s Minimum Essential Medium supplemented with 10% FBS were plated at a density of 1.8x105 cells per well and transfected at 6 μg DNA per 100 μl particles per well in a 6-well plate. After incubating the cells with nanoparticles for 4 hours, the media was aspirated and replaced with fresh media and the fluorescence expression was analyzed using microscopy after 48 hours.
Nanoparticle characterization by NTA and TEM
A Nanosight LM10 equipped with a sensitive CCD camera was used for characterizing the nanoparticles by NTA. Two hundred microliters of diluted nanoparticle solution was loaded into the sample chamber using a 1 mL syringe, the chamber connected to a 405 nm laser source was placed on the Nanosight microscope stage, and a 60 s movie containing the Brownian motion tracking of the scattering centroids (particles) was recorded using the NTA software (Version 2.0). The movie was processed using the manufacturer recommended auto settings with manual adjustment of the gain, blur and brightness as recommended. The particle solution was diluted in DI water to adjust the sample concentration to 107–109 particles per mL. For TEM studies, nanoparticles were formed by complexing with EGFP-DNA as described earlier and 10 μL of these particles was placed on a formvar/carbon coated copper grid (FCF400-Cu, Electron Microscopy Sciences). The nanoparticles were dried for 2 hours and then imaged in a Philips/FEI BioTwin CM120 Transmission Electron Microscope at the JHU-SOM Microscope Facility. The images were processed using ImageJ software.
Plasmid per particle distribution calculations
The known total plasmid concentration was multiplied by the volume fraction of each 1 nm-spaced particle-size class to get the number of plasmids in each 1 nm-spaced particle-size class. This plasmid number was divided by the number of particles in that particle-size class to get the number of plasmids per particle for each particle-size class. The particle per plasmid distribution was determined for all replicates, the replicates were averaged and the average was rounded off to the nearest whole number to plot a graph of number of plasmids per particle (at each 1-nm spaced particle class) against the concentration of particles that contain that given number of plasmids. Both the mode and mean of the distribution were calculated, where the peak of the distribution represents the mode plasmid per particle number for each formulation. The mean of 1 nm distribution matched the mean obtained from overall average calculations.
Supplementary Material
Footnotes
The authors thank the TEDCO MSCRF (2009-MSCRFE-0098-00) and NIH R21CA152473 for support. Corey Bishop and Stephany Tzeng are thanked for technical assistance in acquiring the TEM images.
Supporting Information is available on the WWW under http://www.small-journal.com or from the author.
Contributor Information
Nupura S. Bhise, Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD 21231 (USA)
Ron B. Shmueli, Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD 21231 (USA)
Jose Gonzalez, Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD 21231 (USA).
Prof. Jordan J. Green, Email: green@jhu.edu, 400 North Broadway, Smith Building Room 5017, Baltimore, MD 21231 (USA)
References
- 1.Putnam D. Nat Mater. 2006;5:439. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 2.Sheyn D, Mizrahi O, Benjamin S, Gazit Z, Pelled G, Gazit D. Adv Drug Deliv Rev. 2010;62:683. doi: 10.1016/j.addr.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Angoulvant D, Fazel S, Weisel RD, Lai TY, Fedak PW, Chen L, Rafati S, Seneviratne CK, Degousee N, Li RK. J Thorac Cardiovasc Surg. 2009;137:471. doi: 10.1016/j.jtcvs.2008.08.031. [DOI] [PubMed] [Google Scholar]
- 4.Sunshine JC, Bishop CJ, Green JJ. Therapeutic Delivery. 2011;2:493. doi: 10.4155/tde.11.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercola M, Ruiz-Lozano P, Schneider MD. Gene Dev. 2011;25:299. doi: 10.1101/gad.2018411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu J. Science. 2009;324:1266. [Google Scholar]
- 7.Verma IM, Somia N. Nature. 1997;389:239. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 8.Hollon T. Am J Ophthalmol. 2000;129:701. doi: 10.1016/s0002-9394(00)00442-6. [DOI] [PubMed] [Google Scholar]
- 9.Check E. Nature. 2005;433:561. doi: 10.1038/433561a. [DOI] [PubMed] [Google Scholar]
- 10.Cavazzana-Calvo M, Thrasher A, Mavilio F. Nature. 2004;427:779. doi: 10.1038/427779a. [DOI] [PubMed] [Google Scholar]
- 11.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc Natl Acad Sci. 1995;92:7297. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynn DM, Langer R. J Am Chem Soc. 2000;122:10761. [Google Scholar]
- 13.Bhise NS, Gray RS, Sunshine JC, Htet S, Ewald AJ, Green JJ. Biomaterials. 2010;31:8088. doi: 10.1016/j.biomaterials.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tzeng SY, Guerrero-Cazares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ. Biomaterials. 2011 doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sunshine J, Green JJ, Mahon K, Yang F, Eltoukhy A, Nguyen DN, Langer R, Anderson DG. Adv Mater. 2009;21(48):4947. doi: 10.1002/adma.200901718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu JY, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Science. 2007;318:1917. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Cell. 2007;131:861. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Nature. 2008;451:141. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 19.Yusa K, Rad R, Takeda J, Bradley A. Nat Methods. 2009;6:363. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho YP, Chen HH, Leong KW, Wang TH. J Control Release. 2006;116:83. doi: 10.1016/j.jconrel.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreiss P, Cameron B, Rangara R, Mailhe P, Aguerre-Charriol O, Airiau M, Scherman D, Crouzet J, Pitard B. Nucleic Acids Res. 1999;27:3792. doi: 10.1093/nar/27.19.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins L, Kaszuba M, Fabre JW. Biochim Biophys Acta. 2004;1672:12. doi: 10.1016/j.bbagen.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Pitard B, Aguerre O, Airiau M, Lachages AM, Boukhnikachvili T, Byk G, Dubertret C, Herviou C, Scherman D, Mayaux JF, Crouzet J. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:14412. doi: 10.1073/pnas.94.26.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunlap DD, Maggi A, Soria MR, Monaco L. Nucleic Acids Res. 1997;25:3095. doi: 10.1093/nar/25.15.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dauty E, Behr JP, Remy JS. Gene Ther. 2002;9:743. doi: 10.1038/sj.gt.3301759. [DOI] [PubMed] [Google Scholar]
- 26.Arscott PG, Li AZ, Bloomfield VA. Biopolymers. 1990;30:619. doi: 10.1002/bip.360300514. [DOI] [PubMed] [Google Scholar]
- 27.Blessing T, Remy JS, Behr JP. J Am Chem Soc. 1998;120:8519. doi: 10.1021/ja015867r. [DOI] [PubMed] [Google Scholar]
- 28.DeRouchey J, Netz RR, Radler JO. European Physical Journal E. 2005;16:17. doi: 10.1140/epje/e2005-00003-4. [DOI] [PubMed] [Google Scholar]
- 29.Watson JD, Crick FH. Nature. 1953;171:737. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 30.Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R, Wagner E. Gene Ther. 1998;5:1425. doi: 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- 31.Schwake G, Youssef S, Kuhr JT, Gude S, David MP, Mendoza E, Frey E, Radler JO. Biotechnol Bioeng. 105:805. doi: 10.1002/bit.22604. [DOI] [PubMed] [Google Scholar]
- 32.Green JJ, Zugates GT, Tedford NC, Huang Y, Griffith LG, Lauffenburger DA, Sawicki JA, Langer R, Anderson DG. Adv Mater. 2007;19:2836. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.