Abstract
Biodegradable polyelectrolyte surfaces for gene delivery were created through electrospinning of biodegradable polycations combined with iterative solution-based multilayer coating. Poly(β-amino ester) (PBAE) poly(1,4-butanediol diacrylate-co-4-amino-1-butanol) end-capped with 1-(3-aminopropyl)-4-methylpiperazine was utilized due to its ability to electrostatically interact with anionic molecules like DNA, its biodegradability, and its low cytotoxicity. A new DNA release system was developed for sustained release of DNA over 24 hours, accompanied by high exogenous gene expression in primary human glioblastoma (GB) cells. Electrospinning a different PBAE, poly(1,4-butanediol diacrylate-co-4,4′-trimethylenedipiperidine), and its combination with polyelectrolyte 1-(3-aminopropyl)-4-methylpiperazine end-capped poly(1,4-butanediol diacrylate-co-4-amino-1-butanol)-based multilayers are promising for DNA release and intracellular delivery from a surface.
Keywords: electrospinning, layer-by-layer, poly(β-amino ester), biodegradable, gene delivery
Introduction
Cationic poly(β-amino ester)s (PBAEs) have been shown to have superior transfection efficiency and less toxicity compared to leading commercial reagents such as Lipofectamine™ 2000 in a variety of cell systems.1–4 These polymers can electrostatically condense DNA into nanoparticles between 80–180 nm in diameter and neutral to positive surface charge. These properties are conducive to intracellular delivery.5 PBAEs can also be synthesized under highly controlled conditions, allowing the polymer properties to be tailored according to the needs of a particular application.6 Even though these nanoparticles are hydrolytically degradable, we have been able to store polymer/DNA nanoparticles in a dry, ready-to-use form for several months, increasing their ability to be used in a clinical setting.3 Unlike some conventional chemotherapeutics used for the treatment of cancer, a DNA therapeutic requires an intracellular delivery system to be successful. An implantable device for gene delivery that could enable both controlled release and intracellular delivery of DNA would open new, complementary avenues for treatment of diseases like brain cancer, the disease model used in this study.
In this manuscript, we use a layer-by-layer (LBL) assembly technique to construct multilayer films. Thin films can allow nanometer-scale composition and a high degree of spatial control.7, 8 Such films have a wide range of applications with the ability to load different cargos, including proteins, peptides, nucleic acids, and a broad range of hydrophobic drugs.9–12 As another method of preparing polymer-based films, electrospinning has gained widespread application in tissue engineering and drug delivery.13 Electrospun fibers are structures that can mimic the natural extracellular matrix, and, due in part to their high surface area, they can be a useful tool for fabricating surfaces that interact with cells and can function as drug delivery carriers.14 While the 1-(3-aminopropyl)-4-methylpiperazine end-capped poly(1,4-butanediol diacrylate-co-4-amino-1-butanol) PBAE (called 447 below) is effective in nucleic acid delivery in several different systems,3, 4, 15–17 it has not been used previously in DNA delivery from a substrate, such as the LBL system presented here. Similarly, while PBAEs like poly(1,4-butanediol diacrylate-co-4,4′-trimethylenedipiperidine) (called 4P below) have been used for LBL film construction,18 previous work with such materials has necessitated the addition of a separate commercial reagent to released DNA in order to facilitate transfection. In this work, we combined traditional LBL approaches with electrospinning, combining the physical advantages of 4P--which can form solid structures at room temperature--and the transfection ability of 447--able to delivery DNA into cells with better safety and efficacy than leading commercial reagents19, 20--to prepare thin films as DNA delivery systems that can enable both controlled release and intracellular delivery. We examined the release kinetics of these films, their DNA loading efficiency, and their in vitro efficacy at transfecting human glioblastoma cells. We evaluated PBAE/DNA films prepared through an LBL iterative coating method, an electrospinning method, and a combination of the two (Figure 1).
Figure 1.
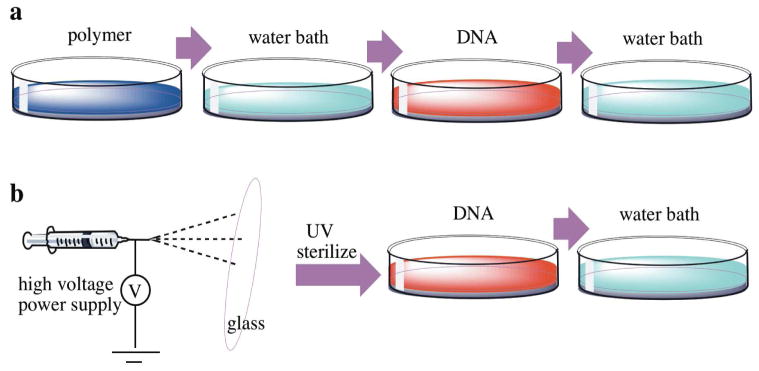
Thin layer fabrication. (A): Scheme of LBL iterative dipping process. (B): electrospinning process.
2. Materials
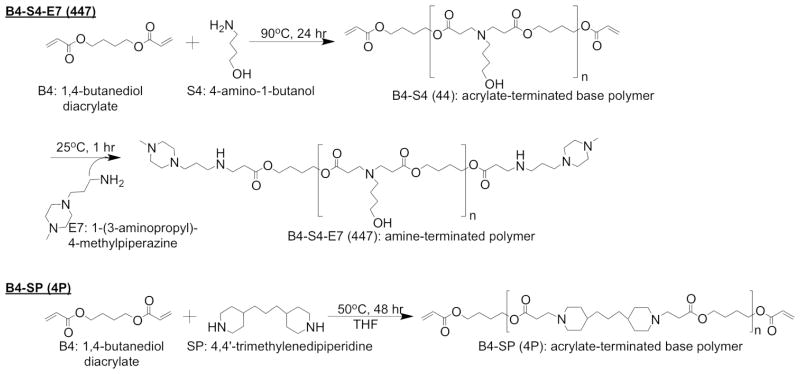
1,4-butanediol diacrylate (B4), 4-amino-1-butanol (S4), and 1-(3-aminopropyl)-4-methylpiperazine (E7) were purchased from Alfa Aesar (Ward Hill, MA). 4,4′-Trimethylenedipiperidine (SP) and poly(ε-caprolactone), 70–90 kDa (PCL) were purchased from Sigma Aldrich (St Louis, MO). pEGFP-N1 DNA was amplified by Aldevron (Fargo, ND). All cell culture media components and other reagents were used as received.
2.1 Polymer synthesis
PBAEs were synthesized by modifying previously described procedures,3 summarized in Figure 2. The naming convention for 1-(3-aminopropyl)-4-methylpiperazine end-capped poly(1,4-butanediol diacrylate-co-4-amino-1-butanol) as “B4-S4-E7” or “447” comes from our recent work on a library of PBAEs used to fabricate nanoparticles3, 6 and refers to the four carbons between acrylate groups in “B4” and the four carbons between the amine and alcohol groups in “S4.” The name “B4-SP” or “4P” for poly(1,4-butanediol diacrylate-co-4,4′-trimethylenedipiperidine) extends this naming convention (side chain “P” for piperidine) and indicates directly the structural similarity between these two polymers, with B4-S4 being more hydrophilic and B4-SP more hydrophobic. For B4-S4-E7 (447), the diacrylate backbone monomer (B4) was mixed with the amine-containing sidechain (S4) at 1.05:1 molar ratio and stirred overnight at 90°C to form the B4-S4 base polymer. Five grams of the base polymer were dissolved in anhydrous tetrahydrofuran (THF) at 167 mg/ml. 10 mL of a 1-M solution of the end-cap (E7) in THF was added, and the mixture was shaken at 5000 rpm for 1 hr at room temperature. These end-capped polymers were purified by precipitation into anhydrous diethyl ether. Excess precipitant was decanted, and the polymers were washed with ether again and the ether decanted. Traces of solvent were removed by vacuum for 48 hr. End-capped polymers were stored at −20°C either neat (for electrospinning) or as 100-mg/ml solutions in anhydrous DMSO (for dipping and conventional nanoparticle formation). For B4-SP (4P) synthesis, SP was dissolved in THF at 500 mg/ml and added to neat B4 at a 1.05:1 molar ratio (B4:SP). The mixture was stirred 2 days at 50°C. Synthesized polymer was stored as a solution in THF at −20°C.
Figure 2.
Polymer B4-S4-E7 (447) and B4-SP (4P) synthesis.
2.2. Chemical analysis of polymers
For gel permeation chromatography (GPC) of 447, polymer in DMSO was diluted in BHT-stabilized THF with piperidine (final composition of complete GPC solvent: 94% THF, 5% DMSO, 1% piperidine) at a final 5.5 mg/mL concentration of 447. The polymer solution was filtered with a 0.2-μm PTFE syringe filter to remove any particulates before flowing through a Waters (Milford, MA) GPC system and analyzed with Breeze 2 software. For nuclear magnetic resonance (NMR) analysis, 447 in DMSO was frozen and lyophilized overnight to remove most of the DMSO solvent, then redissolved in CDCl3 with 0.03% tetramethylsilane (TMS) internal standard. The spectrum was acquired with a Bruker 400 with TopSpin 2.0 software. Analysis was done using NMR Processor v.12 (ACD Labs, Toronto, Canada).
For analysis of 4P, polymer in THF was precipitated as described above, using hexane as the precipitant in place of diethyl ether. After being dried, neat polymer was dissolved in complete GPC solvent for molecular weight analysis or in CDCl3 for NMR analysis. Polymer analysis results are presented in Supporting Figure S1.
2.3 Layer-by-layer (LBL) film assembly
The LBL films were fabricated by depositing materials with complementary charge onto a glass substrate (diameter 15 mm). Briefly, the glass substrates were sterilized by incubation in 70% ethanol and exposure to UV light for 30 min. The glasses were then placed into 24-well plates and coated with a 1-M NaOH solution for 5 min. The base was removed and the glass substrates neutralized by incubating in 1-M HCl for 1 min. The glass substrates were rinsed twice with sterile UltraPure™ distilled water, dried completely, and stored under vacuum until use. For the bottom layer, substrates were coated for 5 min using either 20 or 5 mg/mL polymer 447 in sterile DMSO solution, or 20 or 5 mg/mL poly(D-lysine) (PDL) in 100-mM sodium acetate buffer (pH 5). Substrates were then washed twice in water for 1 min each. Next, the substrates were coated with 0.75 mg/ml DNA in 100-mM sodium acetate buffer for 5 min and again washed twice with water, then treated with 5 mg/ml 447 in 100-mM sodium acetate for 5 min and washed twice with water. Each bilayer contained one layer of DNA and one layer of 447. Films with 3, 4, 6, 9, and 12 bilayers were constructed in this study to determine the optimal layering protocol.
2.4 Quantitative assessment of film fabrication
Plastic 96-well plates with UV-transparent bottoms were used to measure DNA adsorption in the films. LBL films were fabricated in the 96-well plates as above. After each layer, the DNA content was measured by absorbance at 260 nm using a Synergy 2 Multiplate Reader with Gen5 software (Biotek).
2.5 Electrospinning of nanofibrous scaffolds
The basic experimental setup used for electrospinning is schematically shown in Figure 1B. Polymer 447 was dissolved in a 9:1 (v/v) mixture of DCM/DMF to obtain a final concentration of 70% (w/v). 37.5% (w/v) 4P in THF was then added to equal weight of 11% (w/v) PCL in DCM/DMF. All resultant mixtures were vortexed to form a clear and homogeneous organic phase, then loaded into a 1-ml syringe fitted with 27G blunt needle that was clamped directly to the positive electrode of the high voltage power supply. The flow rate of the solution was governed by controllable syringe pump. 447 solution was electrospun at 7 kV with solution flow rate of 0.35 mL/hr. The spinneret was fixed 8 cm away from the aluminum rotating collection plate. Glass substrates were fixed to the aluminum plate rotating at 260 rpm and were coated with nanofibers for 3 or 5 min, then transferred to 24-well plates for further layering or other studies. The SP/PCL blend was electrospun at 7 kV over 8 cm and for 10 min on a rotating mandrel. During SP/PCL electrospinning, the needle moved toward the rotating plate at a rate of 0.1 mm/sec, the plate rotated at 260 rpm, and the solution feeding rate was 0.25 mL/hr.
2.6 DNA release from films
For release studies, the films on glass substrates were placed in 24-well plates and immersed in 300 μL of 1× PBS. At predetermined time points, the solution was removed and replaced with fresh PBS. Samples of DNA and PBAE in PBS were kept frozen at −20°C until use. The concentration of released DNA was measured by NanoDrop 2000 and associated software (v.1.4.1) (Thermo Fisher, Waltham, MA).
2.7 Cell culture and DNA transfection
Primary glioblastoma (GB) cell line GB 319 was derived from intraoperative samples from a patient as previously described3 and was cultured as adherent cells in complete medium (1:1 DMEM/F-12, 10% fetal bovine serum, 1% antibiotic-antimycotic). In order to verify that DNA released from films remained active, GB cells were seeded onto complete LBL films using two different methods to measure direct transfection from the film surface. In the first method, the films were coated with an additional layer of 447 at 20 mg/mL in DMSO, which was removed from the films by aspiration after 5 min incubation. The cover glasses were then placed into 24-well plates and GB cells seeded on the films at 1.5*105 cells/mL (7.5*104 cells/LBL film). Wells were imaged by fluorescence microscopy 48 hr later. In the second method, no additional polymer was added after the standard layering protocol. Instead, GB cells were seeded on the film as above and allowed to adhere overnight. The next day, 447 was dissolved in 25 mM sodium acetate buffer at 2 mg/mL, and 100 μL (200 μg polymer) was added to the culture medium (0.33 mg/mL final polymer concentration in each well). After 2 hr incubation at 37°C, the media were aspirated and replaced with fresh culture medium. Wells were imaged 48 hr after addition of polymer.
GB cells were also transfected separately using DNA pre-released from the LBL films. Cells were seeded at 1.5*104 cells/well in 96-well plates. The DNA samples collected in PBS from release studies were isolated by precipitating in 70% ethanol with 90 mM sodium acetate (pH 5 buffer). Samples were kept at −20°C for 30 min for complete DNA precipitation, then centrifuged at 12,000 g for 15 min at 4°C. The supernatants were aspirated, the pellet washed with 70% ethanol, and the samples centrifuged again as above. The supernatants were aspirated, and the pellet was air-dried for 5 min and resuspended in 50 μL 25 mM sodium acetate buffer. The concentration of all DNA samples was quantified using NanoDrop. Polymer 447 was diluted in 25 mM sodium acetate buffer at 60 polymer-to-DNA weight ratio (w/w) according to the DNA concentration in each sample. 45 μL of polymer solution was mixed with 45 μL of DNA solution, and particles were allowed to form over 10 min. 20 μL particles were added to each well (final volume 120 μL in each well) with n=4 for all treatment groups. Cells were incubated with particles for 2 hr at 37°C before the media were changed. Gene expression was quantified 48 hr later by flow cytometry as a percentage of GFP+ cells (emission detected at 530/30 nm). FlowJo 7 was used for analysis. As a control, fresh DNA (not previously used for coating) was diluted to the same concentration as each of the release study samples and also made into particles with 447 at 60 w/w. Efficacy of released DNA was evaluated by comparing its transfection efficacy to that of fresh DNA.
Cytotoxicity of the polymer 447 alone or in complex with GFP DNA was assessed by seeding GB 319 cells in 96-well plates and transfecting them as described with 447/DNA nanoparticles at dosages ranging from 20.3 ng/mL to 20.8 μg/mL final DNA concentration per well after adding to media. Polymer:DNA ratio was held constant at 60 w/w for final polymer concentration of 1.22 μg/mL to 1.25 mg/mL. Free 447 polymer in solution was added to other cells in the absence of DNA to assess cytotoxicity of the polymer when not in nanoparticle form. Twenty-four hr after transfection, MTS assay (CellTiter 96® AQueous One Solution Proliferation Assay, Promega, Madison, WI) was used to assess viability. For other transfections, viability was similarly measured by MTS assay 24 hr after polymer or polymer/DNA nanoparticles were added to the cells or after cells were seeded onto DNA/polymer LBL films.
2.7 Gel electrophoresis
Precipitated DNA from each sample was also analyzed by gel electrophoresis. Samples in sodium acetate buffer (25 mM, pH 5) were further diluted in sodium acetate to 10 ng/μL concentration based on NanoDrop measurements. 10 μL (100 ng) were loaded into each well of a 1% agarose gel with 1 μg/mL ethidium bromide with 5:1 ratio of sample to loading buffer (30% glycerol). Samples were run for 1 hr at 100 V before visualization under UV light.
Gel retardation assay was also done to test the ability of 447 to form complexes both with fresh DNA and with plasmid layered and then released. DNA collected at the 6 hr time point from 4-bilayer films with 50 mg/mL 447 as the bottom layer was used after precipitation for the “released DNA” sample. Fresh and released DNA were diluted separately to 10 ng/μL in 25 mM sodium acetate. 447 was diluted in 25 mM sodium acetate to a range of concentrations (0–200 w/w or 447:DNA mass ratio) and then added to diluted DNA at a 1:1 v/v ratio. Samples were mixed by pipetting and allowed to self-assemble over 10 min. Loading buffer was added, and each sample was loaded into each well of the gel (with 1 μg/mL ethidium bromide). Samples were run for 1 hr at 100 V before visualization under UV light.
3. Results and discussion
3.1 Fabrication and release properties of LBL films
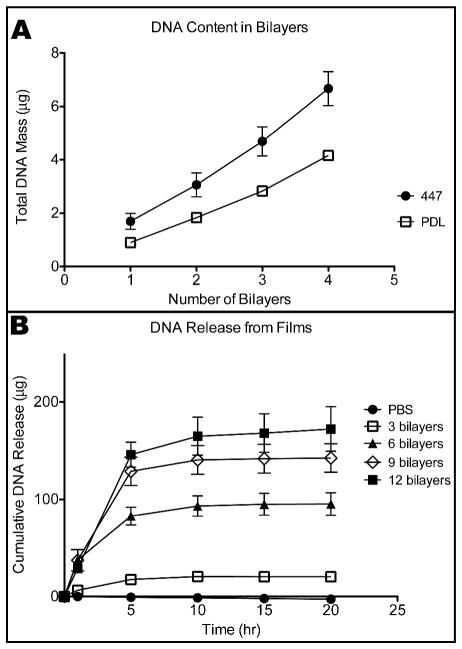
An increase in the amount of DNA incorporated into the film with each bilayer was measured by absorbance at 260 nm using a Synergy 2 Multiplate Reader. The DNA mass contained in the LBL films after each of 4 bilayers is constructed is shown in Figure 3A. The bottom layer of the films was made with either PDL or 447, both cationic polymers; PDL is commonly used to promote effective attachment to growth surfaces, while 447 has been used for DNA complexation and delivery. Films with 447 as the bottom layer had higher DNA loading compared to films made with PDL (Fig 3A). The linear increase in DNA mass with the number of bilayers demonstrates that sequential layering with polyelectrolytes of alternating charge is a viable method of loading nucleic acid onto a surface.
Figure 3.
(A) DNA loaded mass versus number of 447/DNA bilayers with 447 or PDL as the first layer. (B) Comparison of DNA release over time from films with the same structure but a different number of layers.
The amount of released DNA in the incubation solution was measured by absorbance at 260 nm using a NanoDrop 2000. Figure 3B shows the release of DNA from LBL films, again showing higher DNA loading and release with an increasing number of bilayers. These data also show DNA release over 20 hours, correlating with the dissolution or degradation of the PBAE/DNA film in PBS over this time period.
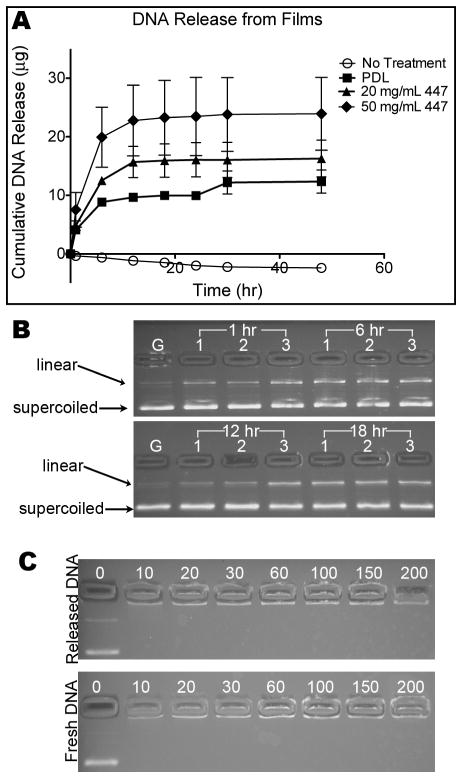
The observation in Figure 3A is confirmed and expanded in Figure 4 to show the effect of the polymer concentration in the bottom layer of the film. The in vitro DNA release profiles show highest loading and release from films using 447 as the bottom layer; increasing the 447 concentration also increases the amount of DNA released.
Figure 4.
(A) Cumulatively released DNA mass vs. incubation time for three 4-bilayer films deposited on glass substrates with the bottom layer being composed of PDL, 20 mg/mL 447, and 50 mg/mL 447. Substrates were incubated in PBS buffer at 37°C, and supernatant was collected over time. (B) Agarose gel electrophoresis corresponding to Figure 4A. Lanes are labeled with the time that supernatant samples were collected. All DNA samples were diluted to 10 μg/mL before loading into the gel. Lanes 1, 2, and 3 correspond to PDL, 20 mg/mL 447, and 50 mg/mL 447, respectively. The lanes labeled “G” used the fresh GFP as a ladder marker. (C) Agarose gel electrophoresis of DNA after mixing with 447 at the 447:DNA w/w ratios written above the lanes, showing tight complexation of DNA by 447 even at low w/w with both the fresh DNA and DNA released from 50 mg/mL 447 at 6 hr.
3.2 Preservation of function of layered and released DNA
We performed a gel electrophoresis study to compare plasmid integrity before being used in our multilayer films as well as after its release into PBS. The gel electrophoresis data are shown in Figure 4B–C. Fresh DNA (i.e. not used in layering) and released DNA both are fully complexed by 447 at 10 w/w or above, important because transfection experiments were typically done in this study at 60 w/w. As seen in 4B, fresh DNA has a primarily supercoiled structure. After deposition on and release from a film, an additional band of nicked or linear plasmid appears strongly.
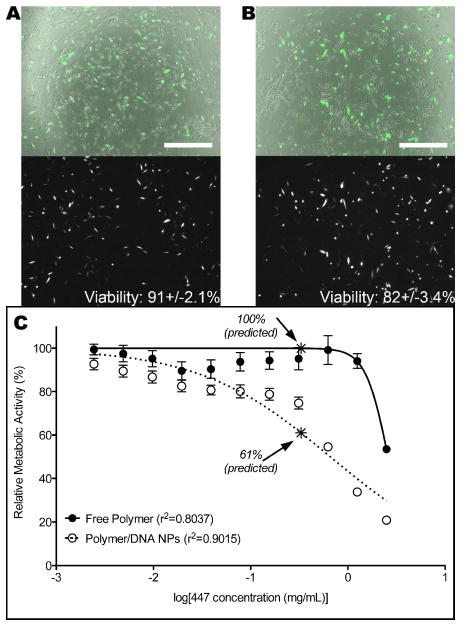
Although this nicking seems to have occurred in all groups and even at early time points, it does not seem to have a significant negative impact on the function of the plasmid, as assessed by transfection experiments. Cells seeded onto the LBL films were successfully transfected with GFP (Figure 5). Although additional polymer was required for successful transfection, adding polymer to the top layer of the films before seeding cells or adding polymer to the medium after seeding cells were both effective methods to enable high gene delivery with low cytotoxicity (91±2.1% and 82±3.4% viability, respectively). Interestingly, this shows that the DNA released from the films and additional polymer in the well were able to form complexes and transfect cells even when delivered separately. Because the amount of 447 polymer released into the culture system would vary with time in a LBL system, we tested the cytotoxicity of 447 by treating GB 319 cells with varying doses of 447, either free in solution or complexed with eGFP DNA at a 60:1 polymer:DNA weight ratio (w/w). Both showed expected dose-response curves in terms of viability, and the viability of cells transfected directed from LBL films was found to be between the two predicted values (Figure 5).
Figure 5.
Six bilayers of 447/DNA were seeded with cells for transfection directly from the film surface. (A) Additional 447 polymer was added to the medium after seeding cells. (B) Additional 447 was added as the top layer of the film before seeding cells. For both, top image: Brightfield+GFP; bottom image: GFP only; scale bar: 500 μm. (C) GB 319 cells treated with varying concentrations of 447 free polymer or 447/DNA nanoparticles at varying doses (60 w/w used for all). Cell viability of cells transfected directly on LBL films were between the predicted values from free polymer and polymer/DNA treatments.
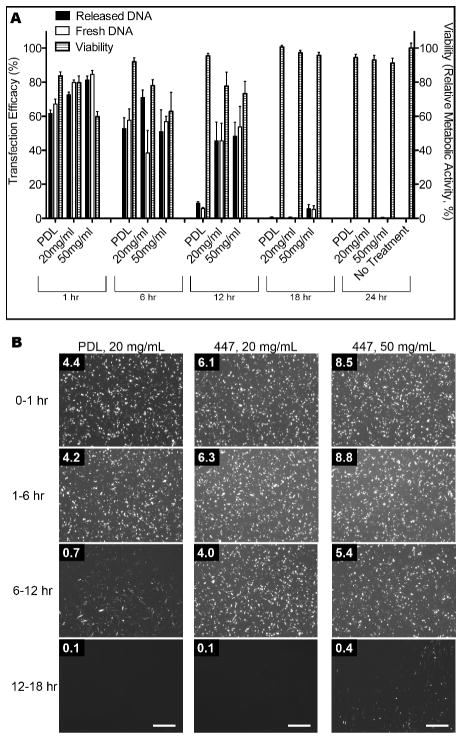
For quantitative assessment of the DNA quality after layering, transfections were also carried out with DNA plasmid released from multilayer film supernatants at various time points and isolated via ethanol precipitation. Fresh 447 polymer at 60 w/w was used to ensure successful transfection of the released DNA. Forty-eight hrs after transfection, flow cytometry analysis showed comparable or higher transfection efficiency than the positive control (fresh DNA), as shown in Figure 6. It should be noted that the DNA release and expression in each of the images shown is not cumulative but rather indicative of the amount of DNA released within the particular window of time after the previous time point and up to the time listed above the image. Thus, the amount of incremental DNA that was released even 12–18 hr after beginning the release study was still enough to cause positive transfection.
Figure 6.
Transfection efficiency and viability (relative metabolic activity) in GB 319 cells assessed by (A) flow cytometry and MTS assay and (B) fluorescence microscopy (GFP channel alone is shown). Transfection was carried out with the plasmid released from multilayer films and collected over each time period: 0–1 hr, 1–6 hrs, 6–12 hrs, and 12–18 hrs and combined with polymer 447. Transfection was measured 48 hrs post-transfection. For all, the number in the upper right-hand corner is the final concentration of DNA added to each well in that condition. For all, scale bar: 500 μm.
Using PBAE 447 as the degradable polycation enables DNA loading into LBL films and DNA release that can enable high transfection efficacy in human cells. Polymer 447 used here interacts electrostatically with DNA to form not only LBL structures but also complexes/nanoparticles with DNA isolated from release study aliquots, in contrast to previously reported PBAE LBL systems.18 This showed DNA release sustained over 18 hr and high gene expression of nearly 90% transfection using DNA that was released from LBL films, isolated from ethanol precipitation, and complexed with fresh 447. Although additional 447 polymer is needed to transfect cells seeded directly onto the surface, simply adding 447 separately into the medium or to the LBL film is sufficient for high transfection. The 447/DNA nanoparticles are hydrolytically degradable through cleavage of the PBAE ester group, and 447 facilitates DNA escape from the endosome to the cytoplasm via the proton sponge mechanism.6, 21, 22
Work previously conducted on LBL DNA delivery systems using a non-447 PBAE (polymer 4P) as the polycationic layer also showed DNA release over time but required the use of Lipofectamine™ 2000, a commercially available reagent, to achieve transfection in an easy-to-transfect cell type (COS-7),18 as PBAE 4P does not itself efficiently facilitate high transfection. The authors of this previous study suggested that the nature of the LBL system might allow the direct incorporation of next-generation materials that can effectively complex with and deliver DNA intracellularly. The PBAE used in the current study, 447, has been shown in other systems to be very effective at nucleic acid delivery to hard-to-transfect cells like the glioblastoma cells studied here,3, 19 as well as fibroblasts,4 endothelial cells,17 hepatocytes and livery cancer cells,20 and stem cells.16 Direct addition of 447 during transfection or during the layering process allows high transfection of GB cells seeded onto these films.
LBL design allows additional spatial control over a drug delivery system compared to therapeutics delivered in solution or in fluid suspension, which can diffuse or flow quickly away from the site of interest, and has been investigated for delivery of various therapeutics from the surface of implanted devices.23–25 For applications like brain cancer therapy, in which direct local access to residual tumor is often available following surgical resection, local delivery from an implantable device could be particularly useful. For example, the Gliadel® wafer,26 which releases a conventional chemotherapeutic at the tumor site, is in use in the clinic for treatment of glioma but, even in combination with other available therapies, provides only a modest improvement in prognosis; DNA-based therapeutics delivered locally from LBL films may further improve the current treatment regimens.
3.3 Electrospun films: fiber morphology and DNA release
While the LBL process with polymer 447 enabled successful gene delivery, we were also interested in varying the underlying substrate material. The extracellular matrix contributes to the mechanical properties of tissue.27 This matrix has a largely porous structure, with a wide pore size distribution. In order to mimic this natural substrate, we explored biochemical composites for the incorporation of plasmid DNA into delivery vehicles and tissue engineering scaffolds. Electrospinning is a simple, straightforward, and cost-effective method to make various types of scaffolds. Moreover, it can provide highly porous microstructure with interconnected pores and extremely large surface-area-to-volume ratio, which is conductive to tissue growth.28
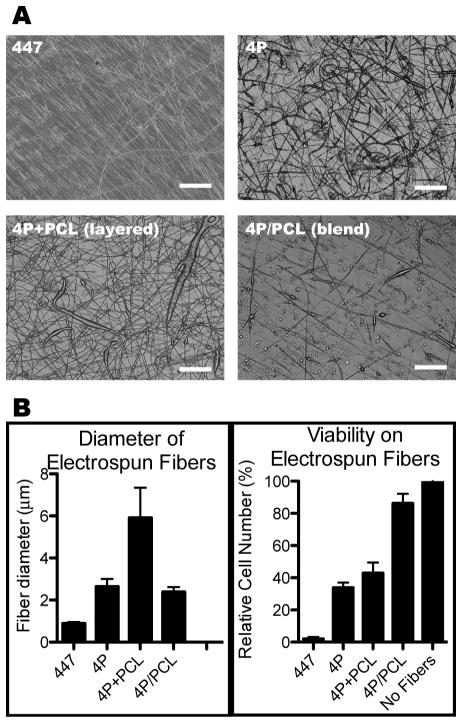
To evaluate if the cationic polymer used in this study, 447, could be used in other ways to fabricate a polymer-based surface for DNA delivery, 447 was electrospun into nanofibers on glass substrates. Although fibers could be formed, 447 substrates were viscous liquids at room temperature and tended to be easily deformed upon addition of aqueous media. Another related PBAE, 4P, which is solid at room temperature and more hydrophobic than 447, was therefore used as the primary PBAE for electrospinning in order to create a more durable substrate. PCL was also spun in some cases as an added component under the hypothesis that the more hydrophobic and slowly degrading PCL would slow the degradation of the PBAE substrate upon addition of aqueous media. Electrospinning 4P on its own, in alternating layers with PCL, or as a single layer of a 4P/PCL blend gave us not only different fiber morphologies but also variation in the ability of cells to attach to the surface. The 4P/PCL blend was the most conducive to cell attachment and viability, as shown in Figure 7. Higher cell viability was also observed on electrospun 4P than on 447.
Fig. 7.
Electrospun fiber morphology (A), average fiber diameter, and cell viability after being seeded onto fibers (B). For all, scale bar: 100 μm.
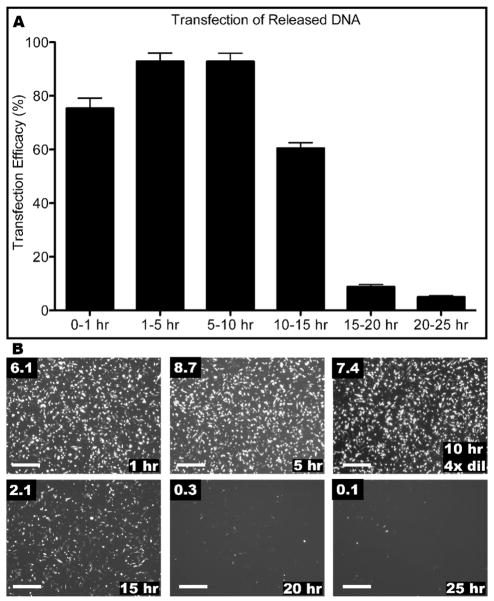
Although 4P, being of high molecular weight and hydrophobicity, was insoluble in aqueous solvents and therefore could not on its own be used in nanoparticle formation or in transfection, it served well as a base on which to build an LBL film for DNA delivery using 447. Electrospun PBAE fibers could then be combined with PBAE polyelectrolyte multilayers to create surfaces that were especially effective for non-viral gene delivery (Figure 8). During the transfection, the sample containing DNA released between 5–10 hr was so effective that, even after diluting the DNA/particles by a factor of four, intracellular DNA delivery occurred in greater than 90% of the primary human glioblastoma cells in the presence of serum. This demonstrates both the high release dosage and also the delayed release kinetics as opposed to only a large burst in the first few hours.
Figure 8.
Green fluorescent protein expression of GB 319 cells. Cells were transfected with released DNA from the films with the structure of electrospun fiber (4P/PCL blend), followed by 4 bilayers of 447 and DNA. Transfection efficiency measured by (A) flow cytometry and (B) fluorescent microscopy with the plasmid released from electrospun fibers and collected over each time period. 1 hr: 0–1 hrs, 5 hr: 1–5 hrs, 10 hr 4× dil: 5–10 hrs diluted 4-fold, etc. For all, numbers in the upper left corner refer to the final concentration of DNA added to each well in the transfection. For all, scale bar: 500 μm.
4. Conclusions
This study demonstrated that the cationic poly(β-amino ester) 1-(3-aminopropyl)-4-methylpiperazine end-capped poly(1,4-butanediol diacrylate-co-4-amino-1-butanol) (447) is a promising material for the construction of polyelectrolyte multilayers for gene delivery. This study also demonstrated that electrospun fibers composed of poly(1,4-butanediol diacrylate-co-4,4′-trimethylenedipiperidine) (4P) and PCL can be combined with 447-based multilayers to form PBAE-based surfaces that can enable gene transfer. PBAEs are biodegradable via hydrolytic cleavage of their ester groups. Polymers 447 and 4P used in this study showed they could be applied in different ways to achieve their desirable effect as gene delivery coatings and surfaces. Electrospinning and dipping into solutions are effective methods of depositing PBAE polymer onto substrates, which allows for DNA loading and release over 24 hrs. Combining the two methods can enhance DNA release when evaluated in vitro with a GB 319 human glioblastoma cell line. Ultimately, the released DNA performed as well as fresh DNA in transfection studies, and polymer 447 mediated highly effective gene delivery to human brain cancer cells. Used for layering as well as for a DNA transfection agent, 447 was able to achieve 60–80% transfection efficacy with DNA released from the LbL films with <20% loss in cell viability. Cells seeded directly onto LbL films could be transfected with <20% loss in viability when films were fabricated with an extra top layer of 447, and cells seeded onto films and incubated with additional 447 were transfected with <10% loss in viability. Electrospun PBAEs and their combination with polyelectrolyte PBAE multilayers are promising for DNA release and delivery from a surface.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health (1R01EB016721-01). CL acknowledges the Global-COE program (Kyushu University, Japan) foundation for fellowship support. SYT acknowledges the National Science Foundation (Arlington, VA, USA) for fellowship support.
References
- 1.Green JJ, Langer R, Anderson DG. Acc Chem Res. 2008;41(6):749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sunshine J, Green JJ, Mahon K, Yang F, Eltoukhy A, Nguyen DN, Langer R, Anderson DG. Adv Mater. 2009;21(48):4947–4951. doi: 10.1002/adma.200901718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzeng SY, Guerrero-Cazares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ. Biomaterials. 2011;32(23):5402–5410. doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhise NS, Shmueli RB, Gonzalez J, Green JJ. Small. 2012;8(3):367–373. doi: 10.1002/smll.201101718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green JJ. Ann Biomed Eng. 2012;40(7):1408–1418. doi: 10.1007/s10439-012-0550-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sunshine JC, Akanda MI, Li D, Kozielski KL, Green JJ. Biomacromolecules. 2011;12(10):3592–3600. doi: 10.1021/bm200807s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim BS, Smith RC, Poon Z, Hammond PT. Langmuir. 2009;25(24):14086–14092. doi: 10.1021/la9017618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shmueli RB, Anderson DG, Green JJ. Expert Opin Drug Del. 2010;7(4):535–550. doi: 10.1517/17425241003603653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vazquez E, Dewitt DM, Hammond PT, Lynn DM. J Am Chem Soc. 2002;124(47):13992–13993. doi: 10.1021/ja026405w. [DOI] [PubMed] [Google Scholar]
- 10.Wood KC, Zacharia NS, Schmidt DJ, Wrightman SN, Andaya BJ, Hammond PT. Proc Natl Acad Sci U S A. 2008;105(7):2280–2285. doi: 10.1073/pnas.0706994105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond PT. Curr Opin Colloid Interface Sci. 1999;4(6):430–442. [Google Scholar]
- 12.Hammond PT. Adv Mater. 2004;16(15):1271–1293. [Google Scholar]
- 13.Tan AR, Ifkovits JL, Baker BM, Brey DM, Mauck RL, Burdick JA. J Biomed Mater Res A. 2008;87A(4):1034–1043. doi: 10.1002/jbm.a.31853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li D, Xia YN. Adv Mater. 2004;16(14):1151–1170. [Google Scholar]
- 15.Shmueli RB, Sunshine JC, Xu Z, Duh EJ, Green JJ. Nanomedicine. 2012;8(7):1200–1207. doi: 10.1016/j.nano.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzeng SY, Hung BP, Grayson WL, Green JJ. Biomaterials. 2012;33(32):8142–8151. doi: 10.1016/j.biomaterials.2012.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzeng SY, Yang PH, Grayson WL, Green JJ. Int J Nanomed. 2011;6:3309–3322. doi: 10.2147/IJN.S27269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang JT, Chua LS, Lynn DM. Langmuir. 2004;20(19):8015–8021. doi: 10.1021/la048888i. [DOI] [PubMed] [Google Scholar]
- 19.Tzeng SY, Green JJ. Adv Healthcare Mater. 2012;2(3):468–480. doi: 10.1002/adhm.201200257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tzeng SY, Higgins LJ, Pomper MG, Green JJ. J Biomed Mater Res A. 2013;101A:1837–1845. doi: 10.1002/jbm.a.34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parlow A, Weiske C, Gasteiger J. J Chem Inf Model. 1990;30(4):400–402. [Google Scholar]
- 22.Akinc A, Lynn DM, Anderson DG, Langer R. J Am Chem Soc. 2003;125(18):5316–5323. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- 23.Shukla A, Fleming KE, Chuang HF, Chau TM, Loose CR, Stephanopoulos GN, Hammond PT. Biomaterials. 2010;31(8):2348–2357. doi: 10.1016/j.biomaterials.2009.11.082. [DOI] [PubMed] [Google Scholar]
- 24.Wang XF, Ji J. Langmuir. 2009;25(19):11664–11671. doi: 10.1021/la9013575. [DOI] [PubMed] [Google Scholar]
- 25.Ariga K, McShane M, Lvov YM, Ji QM, Hill JP. Expert Opin Drug Del. 2011;8(5):633–644. doi: 10.1517/17425247.2011.566268. [DOI] [PubMed] [Google Scholar]
- 26.McGirt MJ, Than KD, Weingart JD, Chaichana KL, Attenello FJ, Olivi A, Laterra J, Kleinberg LR, Grossman SA, Brem H, Quinones-Hinojosa A. J Neurosurg. 2009;110(3):583–588. doi: 10.3171/2008.5.17557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tambralli A, Anderson JM, Andukuri A, Lim DJ, Dean DR. Am Ceram Soc Bull. 2012;91(3):21–21. [Google Scholar]
- 28.Dhandayuthapani B, Yoshida Y, Maekawa T, Kumar DS. Int J Polym Sci. 2011:1–19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.