Abstract
Assisted reproductive technologies (ART) offer revolutionary infertility treatments for millions of childless couples around the world. Currently, ART accounts for 1 to 3% of annual births in industrialized countries and continues to expand rapidly. Except for an increased incidence of premature births, these technologies are considered safe. However, new evidence published during the past decade has suggested an increased incidence of imprinting disorders in children conceived by ART. Specifically, an increased risk was reported for Beckwith-Wiedemann syndrome (BWS), Angelman syndrome (AS), Silver-Russell syndrome, and retinoblastoma. In contrast, some studies have found no association between ART and BWS, AS, Prader-Willi syndrome, transient neonatal diabetes mellitus, and retinoblastoma. The variability in ART protocols and the rarity of imprinting disorders complicate determining the causative relationship between ART and an increased incidence of imprinting disorders. Nevertheless, compelling experimental data from animal studies also suggest a link between increased imprinting disorders and ART. Further comprehensive, appropriately powered studies are needed to better address the magnitude of the risk for ART-associated imprinting disorders. Large longitudinal studies are particularly critical to evaluate long-term effects of ART not only during the perinatal period but also into adulthood. An important consideration is to determine if the implicated association between ART and imprinting disorders is actually related to the procedures or to infertility itself.
Keywords: Epigenetics, genomic Imprinting, methylation, imprinting disorders, ART
Infertility was classified as a disease by the World Health Organization in 2009.1 According to a recent estimation, 1 in 10 people of reproductive age are involuntarily infertile.2 Consequently, there is a great demand for infertility treatments suchas assisted reproductive technologies (ART). Since the birth of the first test tube baby, Louise Brown in 1978, ART procedures such as in vitro fertilization (IVF), intracytoplasmic sperm injection (ICSI), embryo culture, and embryo cryopreservation have rapidly evolved and opened a new era in the treatment of infertility for millions of childless couples around the world. Currently, ART accounts for 1 to 3% of annual births in industrialized countries3 and continues to expand rapidly. Except for an increased incidence of premature births, these technologies are considered safe.4 However, studies published since 2002 suggest a relatively high incidence of birth defects such as Beckwith-Wiedemann syndrome (BWS), Angelman syndrome (AS), and Silver-Russell syndrome (SRS) among children conceived by ART.5–15 Similarly, experimental evidence obtained using different animal models suggests that ART may induce aberrant epigenetic changes potentially leading to various disorders.16–23 This article first briefly explains the overall mechanisms of epigenetic gene regulation and then reviews the current status of imprinting disorders linked to ART.
Epigenetic Gene Regulation
It has long been recognized that phenotype is a result of genotype and environment. However, the molecular mechanism(s) beyond some of the environmental effects has remained poorly understood. Nevertheless, studies published over the last 2 decades revealed that environment may contribute to the regulation of gene expression by inducing various chemical modifications to DNA (e.g., DNA methylation) and histones (e.g., acetylation, methylation, phosphorylation, sumoylation, and ubiquitination of histone tails) without changing DNA sequence.24–28 Such chemical modifications represent another layer of gene regulation in addition to a given gene sequence and are defined by the term epigenetics, which literally means on top of genetics (epi: “over, above”). Originally, the word epigenetics was introduced by Waddington in the 1940s to refer to a new discipline known today as developmental biology.29 Nowadays, the term epigenetics is typically used more narrowly to define heritable changes in gene expression without altering the underlying DNA sequence, although different definitions have also been proposed.30–32 The principal mechanisms of epigenetic gene regulation include DNA methylation, histone modifications, and non-coding RNAs, all of which are involved in chromatin remodeling and thus in the regulation of a transcriptionally permissive/nonpermissive state.
DNA Methylation
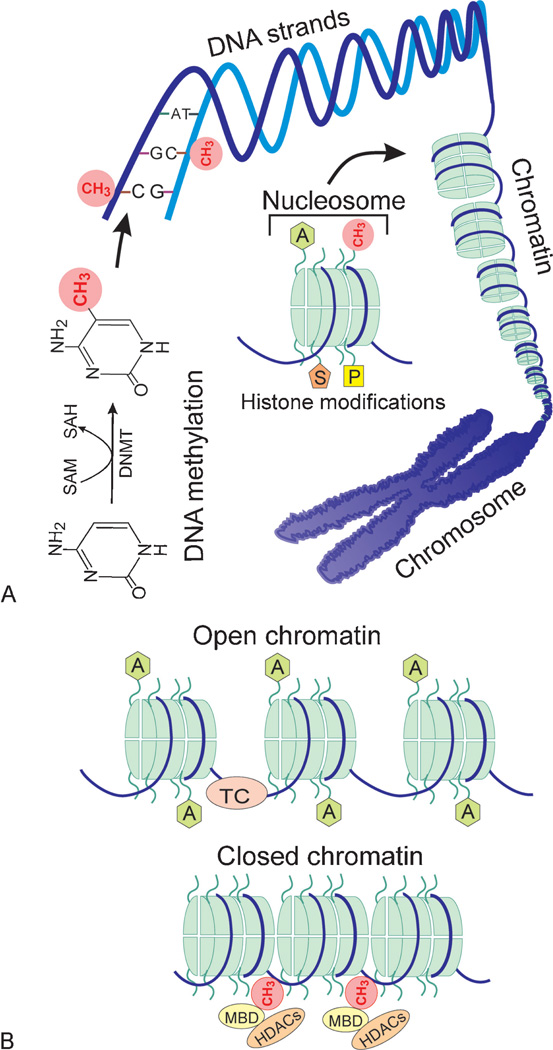
Among the epigenetic modifications just mentioned, DNA methylation is best characterized and involves the addition of a methyl group from S-adenosylmethionine (SAM) to the carbon-5 position of a cytosine base (Fig. 1A). This covalent chemical modification is catalyzed by DNA methyltransferases (DNMTs) that include DNMT1, DNMT3A, and DNMT3B.33 Whereas DNMT3A and DNMT3B primarily establish methylation marks by de novo methylation during early development, DNMT1 maintains methylation patterns during DNA replication by adding methyl groups to hemimethylated sites.34–37
Figure 1.
Epigenetic gene regulation. (A) The overall organization of DNA in relation to epigenetic modifications. DNA methylation is mediated through DNA methyltransferases (DNMTs) by transferring a methyl group from S-adenosylmethionine (SAM) to the carbon-5 position of a cytosine base (shown as a red circle). DNA methylation typically occurs on cytosines (C) preceding guanines (G) at both strands of the double helix. DNA strands are wrapped around histone octamers to form nucleosomes that are building blocks of chromatin. The chromatin in turn is organized into a chromosome. The specific structure of nucleosomes facilitates epigenetic gene regulation. Histone tails protruding from each nucleosome can undergo numerous posttranslational modifications such as acetylation (A in a green hexagon), methylation (CH3 in red circles), sumoylation (S in a brown pentagon), and phosphorylation (P in a yellow rectangle). Such modifications along with DNA methylation can influence chromatin remodeling. (B) The role of chromatin remodeling in gene expression. The open and closed structure of chromatin is regulated by interactions of DNA methylation, numerous histone modifications, and noncoding RNAs. Only shown are DNA methylation (CH3 in red circles) and acetylation of histone tails (A in green hexagons) for a simplified view. Typically, demethylation of CpG dinucleotides (absence of red circles) and acetylation of histone tails (green hexagons) result in an open chromatin structure, which allows binding of the transcription complex (TC as a pink ellipse) to a specific sequence leading to transcription. In contrast, upon methylation of cytosine residues (red circles) by DNA methyltransferases, methyl binding domain proteins (MBD as yellow ellipses) bind to the methylated DNA and recruit histone deacetylases (HDACs as brown ellipses). These events induce deacetylation of histone tails, compaction of the chromatin, and thus transcriptional silencing due to inability of the TC to bind to DNA.
In contrast to DNA methylation, demethylation of the established marks is less well understood and seems to occur through both passive and active mechanisms.38 Passive DNA demethylation may occur by suppression of maintenance methyltransferase activity (i.e., addition of methyl groups to cytosine bases during DNA replication). According to recent studies, active demethylation seems to occur through successive oxidation of 5-methylcytosine (5mC) by ten eleven translocation (Tet) proteins into first 5-hydroxymethylcytosine (5hmC), then 5-formylcytosine (5fC), and finally 5-carboxylcytosine (5caC).39–41 The latter appears to be excised by thymine-DNA glycosylase (TDG) and then repaired by the base excision repair (BER) pathway into an unmethylated cytosine.41 In mammals, DNA methylation overwhelmingly occurs on cytosines (C) preceding guanines (G). These CpG dinucleotides, where “p” indicates a phosphate group, are nonuniformly distributed in the mammalian genome and usually enriched in the promoter region of ~60% of genes.42–44 The CpG-rich regions consisting of 1000 to 2000 base pairs with a CG content >55% are also known as CpG islands and generally remain unmethylated with some exceptions.44–46 DNA methylation is typically associated with silencing of the targeted genes.44,46,47 Therefore, aberrant de novo methylation of CpG islands may lead to developmental disorders.
Histone Modifications
The role of histone modifications in epigenetic gene regulation was recognized early on.48 In eukaryotic cells, core histone proteins (i.e., H2A, H2B, H3, and H4) serve as scaffolds for the packaging of DNA into chromatin by assembling into octamers (Fig. 1A). The nucleosomes, the basic packaging unit of chromatin, are formed by the wrapping of 147 base pairs of DNA around each histone octamer. The resulting nucleosomes are then organized into chromatin, the building block of a chromosome. The extent of chromatin compaction is dynamic and can be influenced by modification of histone tails along with DNA methylation/demethylation.49 In general, highly compacted chromatin is inaccessible to transcription factors, and thus genes are “off” while the open form of chromatin allows binding of transcription factors and hence gene expression (Fig. 1B).50
The NH2-terminal tail of each core histone may undergo numerous covalent posttranslational modifications including acetylation of lysines, methylation of lysines and arginines, ubiquitination and sumoylation of lysines, and phosphorylation of serines and threonines.50 Acetylation of histone tails is catalyzed by histone acetyltransferases (HATs) and generally associated with open chromatin domains and active gene transcription.48 The removal of acetyl groups from lysine residues by histone deacetylases (HDACs) results in condensed chromatin and the silencing of gene transcription.51 Compared with histone acetylation, methylation of histone tails is more complex and can occur in different forms. Lysine residues can be modified with mono-, di-, and trimethyl groups, whereas arginine residues can be either mono- or dimethylated by histone methyltranferases. Depending on modified residues and timing, methylation of histone tails may activate (e.g., H3K4, H3K36, and H3K79) or repress (e.g., H3K9, H3K27, and H4K20) gene expression.50 Considering numerous other histone modifications and cross talk between them, the so-called histone code gets highly complex and is beyond the scope of this review. For a detailed discussion on this subject, readers are referred to specific review articles.50,52,53
Noncoding RNAs
According to high-throughput transcriptomic analyses, up to 90% of the eukaryotic genome seems to be transcribed.54 Whereas ~1 to 2% of these transcripts encode proteins, the overwhelming portion of the genomic DNA is transcribed into functional noncoding RNAs (ncRNAs) that are not further translated. In addition to well-known transfer RNAs and ribosomal RNAs, ncRNAs include micro RNAs (miRNAs), Piwi-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), long noncoding RNAs (lncRNAs), enhancer RNAs (eRNAs), and promoter-associated RNAs (PARs).55,56
In recent years, the involvement of several noncoding RNAs in epigenetic gene regulation was demonstrated.56–60 For example, miRNAs are small single-stranded molecules (~22 nucleotides long) that downregulate gene expression mostly by binding to the 3′ untranslated region (UTR) of messenger RNAs and subsequently causing their destabilization and degradation.61 By targeting enzymes responsible for histone modifications and DNA methylation, miRNAs contribute to epigenetic gene regulation.57–59 SiRNAs are also small but double-stranded RNA molecules (20 to 24 nucleotides long) that are involved in both posttranscriptional and direct sequence-specific transcriptional gene silencing by increasing epigenetic marks.62,63 LncRNAs (>200 nucleotides long) represent most of the non-protein-coding transcripts (~80%)55,64 and seem to regulate gene expression through modulation of the chromatin state. Recent studies suggest that a subgroup of lncRNAs, called large intergenic noncoding RNAs (lincRNAs), guide chromatin-modifying complexes to specific genomic loci to establish cell type–specific epigenetic states.65,66 The inactivation of one of the X chromosomes in female cells is a good example of the involvement of lncRNAs in epigenetic gene regulation. Experimental data suggest that X-inactive specific transcript (Xist) RNA coats the X-chromosome in cis and recruits histone modifications and DNA methylation, which lead to a transcriptionally inactive chromatin state.67,68 In recent years, the diverse functions of ncRNAs in epigenetic gene regulation have been increasingly recognized as discussed in detail in several recent reviews.56,69,70 Taken together, epigenetic gene regulation consists of highly complex processes and requires coordinated interactions of DNA methylation/demethylation, histone modifications, ncRNAs, and various nonhistone proteins to ensure timely expression/repression of genes. Therefore, any disturbance to this complex system may result in developmental disorders.
Genomic Imprinting
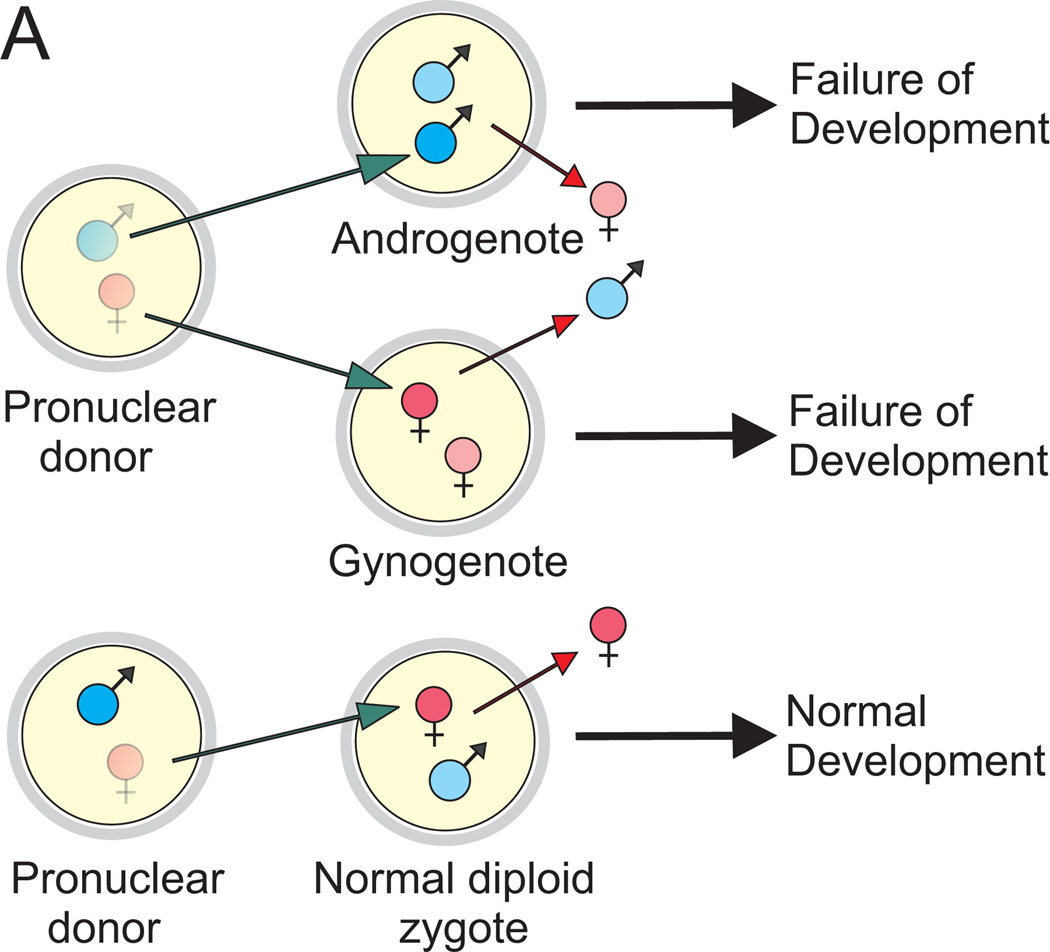
Genomic imprinting is an epigenetic form of gene regulation that results in expression of certain genes depending on their parental origin. In mammals, genomic imprinting was uncovered in the early 1980s as a result of pronuclear transplantation and chromosome translocation (uniparental disomy) experiments and led to the revision of classical Mendelian genetics.71–75 Until then, it was assumed that both maternal and paternal alleles of each gene were expressed (Fig. 2), although observations through the centuries provided hints on differential behavior of paternal and maternal genomes.76 As opposed to diploid mouse embryos containing one maternal and one paternal pronucleus, the combination of two maternal (gynogenote) or two paternal (androgenote) pronuclei in a mouse embryo by micromanipulation resulted in developmental failure despite diploidy and clearly indicated that maternal and paternal genomes are not functionally equivalent and both are required for normal development (Fig. 3). The maternal genome seems to be more important for fetal development, whereas the paternal genome is necessary for placental development. In humans, this is exemplified by two conditions: a molar pregnancy (placental overgrowth and underdeveloped fetal structures with two copies of the paternal genome) and a benign ovarian teratoma (an ovarian cyst manifested by an attempt at fetal development—hair, teeth, cartilage, bone, etc., without placental components, with two copies of the maternal genome). The discovery of the first imprinted genes in the early 1990s further substantiated the impact of genomic imprinting on human health and diseases.77–81 It has been postulated that genomic imprinting has evolved in mammals to regulate the dosage of developmentally important genes. Several other hypotheses including avoidance of parthenogenesis, the parental conflict hypothesis, the complementation hypothesis, and the ovarian time bomb hypothesis have also been proposed, as discussed in a recent review.82
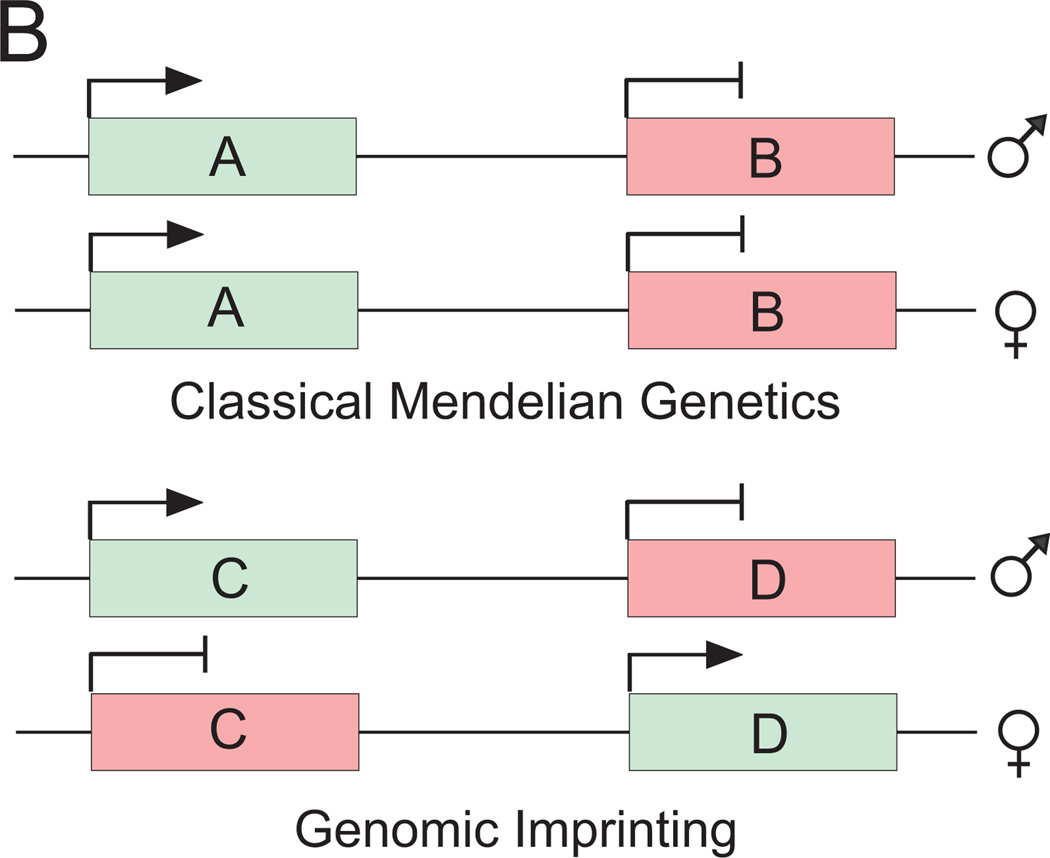
Figure 2.
Expression of imprinted and nonimprinted genes. According to classical genetics, genes are expressed from either both alleles (gene A) or none of them (gene B). This is true for most of the genes that are not imprinted. However, a subset of genes are imprinted; that is, their imprinting control regions are methylated on either the maternal or paternal allele allowing expression of only the paternal allele (gene C) or the maternal allele (gene D), respectively. This sex-specific parent-of-origin allelic methylation mark is inherited from one generation to another. Nonexpressed genes are indicated by red boxes with blunt arrows; expressed genes are depicted with green boxes with an arrowhead.
Figure 3.
Pronuclear transplantation studies demonstrating experimental evidence for genomic printing. Through sophisticated micromanipulation techniques, maternal or paternal pronuclei from normal mouse zygotes were removed and replaced with reciprocal ones resulting in diploid zygotes containing either two paternal pronuclei (androgenote) or two maternal pronuclei (gynogenote). Such androgenotes and gynogenotes were transferred to pseudopregnant recipients to study their subsequent development. Neither type of the reconstituted mouse zygotes was viable, whereas mouse zygotes reconstituted with paternal and maternal pronuclei normally developed to term. The gynogenotes displayed some fetal growth but poorly developed placentas. In contrast, androgenetic development was rather extraembryonic. These results suggested that both maternal and paternal genomes are necessary for normal development. Further, these experiments indicated that paternally expressed genes contribute to placental development, whereas maternally expressed ones are involved in embryonic development.
The mechanism of imprinting is complex and not fully understood. However, methylation of CpG-rich domains is a key part of this phenomenon. To date, at least 100 imprinted genes have been identified, and it has been estimated that there could be several hundred.83 The vast majority of imprinted genes are found in clusters, probably to share epigenetic regulatory elements.26 Such clusters are similarly organized in humans and mice, and they may include both paternally and maternally imprinted genes, nonimprinted genes, and ncRNAs.83–85 They also contain CpG-rich regions up to several kilobase pairs, which are differentially methylated in paternal and maternal alleles and known as differentially methylated regions (DMRs).26 DMRs differ in their function. Some DMRs acquire their methylation marks early in germ cells and serve as imprinting control regions (ICRs) to regulate monoallelic expression in cis. Deletion of ICRs (also known as imprinting centers or imprinting control elements in the literature) results in loss of monoallelic expression in the linked genes, indicating the critical role of ICRs in the imprinting mechanism.86–89 Whereas methylation marks in some DMRs remain stable throughout development and are maintained in all tissues, other DMRs experience considerable changes during development and acquire tissue-specific methylation marks.26,90
Imprinted genes play a critical role in regulating fetal and placental growth and development, as well as in neurological pathways and behavior.91 Therefore, consequences of aberrant genomic imprinting include phenotypic developmental abnormalities and neurological disorders such as Prader-Willi syndrome, BWS, AS, SRS, Albright hereditary dystrophy, and transient neonatal diabetes mellitus. It has also been recognized that aberrant genomic imprinting is involved in the development of both childhood (e.g., Wilms' tumor, retinoblastoma, and neuroblastoma) and adult (e.g., bladder, breast, cervical, colorectal, ovarian, prostate, uterine) tumors.92 As explained later, genomic imprinting occurs during gametogenesis and embryogenesis, and thus its timing coincides with the use of ART. Consequently, it is possible that techniques used in ART could cause aberrant genomic imprinting and thus imprinting disorders.
Transgenerational Cycle of Imprinting Marks
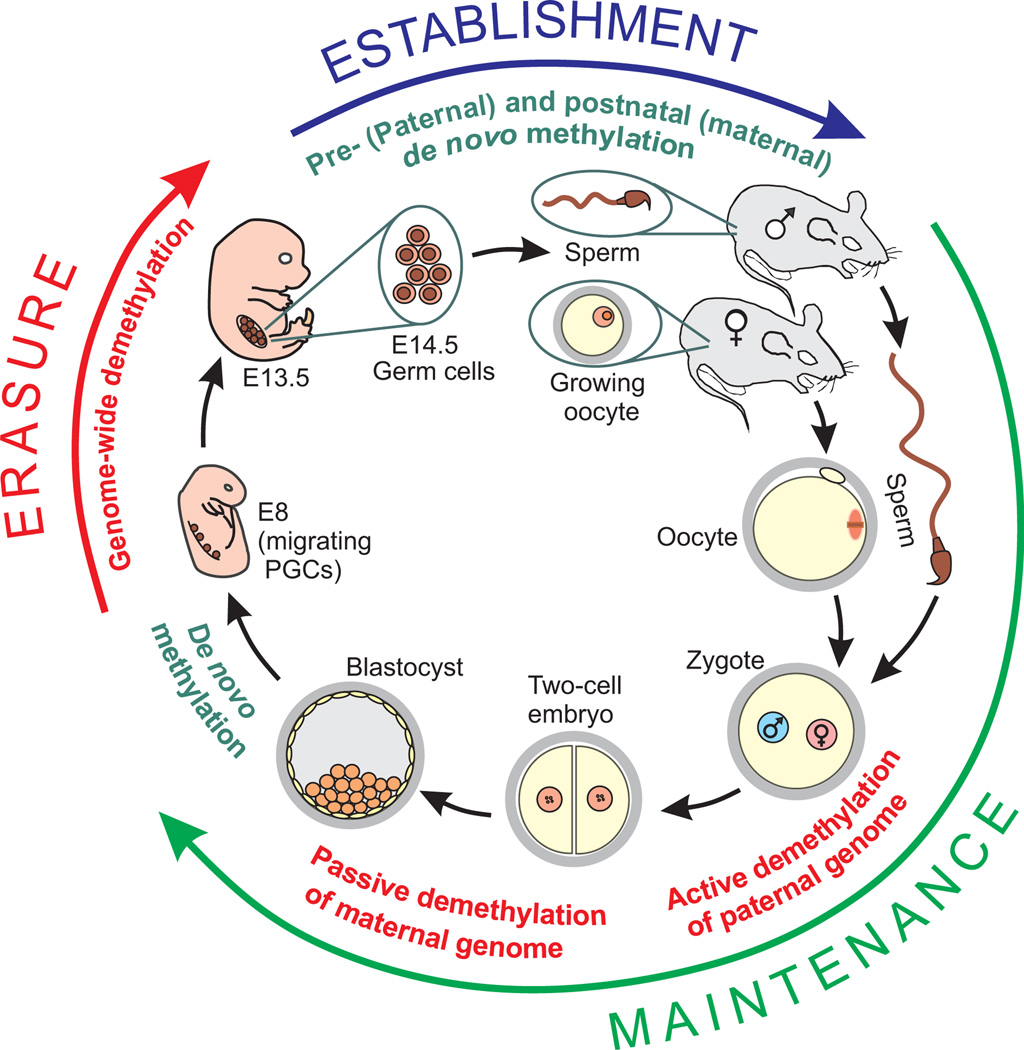
Genomic imprinting requires resetting sex-specific imprints in every generation. This process, which has been characterized by studying methylation marks mostly in the mouse and is also referred to as epigenetic reprogramming, goes through cyclic events consisting of erasure, establishment, and maintenance stages of imprinting marks26 (Fig. 4).
Figure 4.
Transgenerational cycle of imprinting marks in mouse. Generational inheritance and epigenetic reprogramming occur through a cycle of erasure, establishment, and maintenance of imprinting marks as illustrated here by successive demethylation and methylation events in the paternal and maternal genome. All methylation marks are erased in primordial germ cells (PGCs) by a genome-wide methylation event between embryonic day 8 (E8) and E13.5. Next, the imprinting marks are established by de novo methylation starting first in the male germ cells around E14.5 and then in growing oocytes after birth. The established sex-specific imprints are maintained until the appearance of the next-generation PGCs; however, further epigenetic reprogramming events occur to direct embryonic development and tissue-specific gene expression in nonimprinted genes. While the paternal genome undergoes active demethylation at fertilization, the maternal genome is passively demethylated during successive cleavages of the early embryo. Finally, another de novo methylation takes place at the time of implantation.
The erasure of the existing imprints occurs in primordial germ cells (PGCs). In the mouse embryo, PGCs first appear in the proximal epiblast on embryonic day 7 (E7) and then migrate along the genital ridge on E8. They colonize the developing gonads around E10.5. The erasure stage starts with a decrease in DNA methylation during migration of PGCs around E893,94 and is completed after migration by E13.5D with a possibly active wave of genome-wide demethylation.95–97 This epigenetic reprogramming event is probably required to restore totipotency.98,99
In the next cycle stage, imprinting marks are reestablished during development of germ cells into sperm or oocytes through de novo methylation.100,101 In the male germ cells, de novo methylation of DMRs starts in mitotically arrested prospermatogonia (gonocytes) around E14.5, and the paternal methylation imprints are entirely established in perinatal prospermatogonia.100,102,103 In the female germ line, imprinting marks are asynchronously acquired after birth during the growth phase of oocytes.104,105 Some germ-line DMRs including Snrpn, Peg3, Igf2r, and p57KIP2 are methylated during the primary and secondary follicle stages; others such as peg1/mest acquired their methylation marks later during the tertiary and antral follicle stages.105,106
The third stage of the cycle basically refers to maintenance of the imprinting marks during embryonic and fetal development, as well as during adulthood in somatic cells. Further epigenetic reprogramming occurs during embryonic development. Following fertilization, the second wave of demethylation takes place. First, the male pronucleus undergoes active demethylation.107,108 This is followed by passive demethylation of the maternal genome during successive cleavages. Both parental genomes are equally demethylated by the 16-cell stage. However, methylation marks on imprinted genes remain protected from this demethylation wave to maintain parental imprints.109 At the time of implantation, another de novo methylation occurs in a lineage-specific pattern.110 These waves of methylation/demethylation events regulate timely expression of embryonic and tissue-specific genes, and thus they ensure normal development.
The timing of epigenetic reprogramming in humans is not well characterized. Unlike the mouse, methylation marks in the human male germ line seem to be established later during spermatogonial differentiation in the adult testis,111 whereas human female germ cells appear to progressively acquire methylation marks before112 and during oocyte maturation113,114 or after fertilization.115 Taken together, major imprinting events take place in gametes and embryos when ART is used. Consequently, imprinting defects in the course of assisted reproduction may occur during the establishment of genomic imprinting (ovarian hyperstimulation, in vitro maturation of gametes, cryopreservation of gametes) as well as its maintenance (embryo culture, embryo cryopreservation).
Experimental Evidence Supporting Link Between ART and Imprinting Disorders
Because the establishment and maintenance of genomic imprints occur during both gametogenesis and embryogenesis, it has been suggested that in vitro culture of oocytes and embryos may alter methylation status and thus expression of imprinted genes. Indeed, studies in ruminants such as sheep and cattle have documented a particular overgrowth syndrome (known as large offspring syndrome [LOS]) after in vitro culture of embryos. It was shown that sheep with LOS displayed both lack of expression and abnormal methylation of the Igf2r gene.16 Furthermore, studies using mouse models have clearly shown that embryo culture is associated with altered methylation and expression of imprinted genes.17–19 Doherty et al18 investigated the effects of two culture media on the expression of H19 and Snrpn after culturing two-cell embryos to the blastocyst stage. The culture of mouse embryos in Whitten's medium resulted in demethylation of the paternal allele and biallelic expression of the H19 gene, which is normally expressed from the maternal allele, while embryo culture in KSOM containing amino acids showed normal methylation and expression of the H19 gene. Expression of the Snrpn gene remained unaffected in both media. Using a mouse model, Khosla et al17 showed that the addition of fetal bovine serum to M16 medium can alter methylation and expression of imprinted genes (i.e.,H19,Igf2, Grb7,and Grb10) leading to reduced fetal weight. Moreover, a recent study showed that culturing mouse embryos in a commonly used human embryo culture medium (i.e., human tubal fluid [HTF]) also results in aberrant imprinting of H19.20 Other recent studies further support the view that suboptimal culture of embryos may have serious consequences later on.19,21,116 Furthermore, experimental studies on ovarian stimulation in the human and mouse revealed that hormonal induction of superovulation affects expression and DNA methylation of imprinted genes.22,23
Incidence of ART-Related Congenital Anomalies
In general, ART is considered safe.4 However, recent studies suggest that there may be links between ART and increased risks for congenital anomalies.5–8,11,12,117–121 It is known that ART is associated with high rates of low birthweight. This can be partially attributable to the increased rates of multiple gestations due to transfer of two or more embryos. However, Schieve et al117 reported that the rate of low birthweight in singletons conceived with ART is 2.6 times higher than that in the general population. Furthermore, the risks remained high after restricting analyses to subgroups conceived with presumably healthy gametes (e.g., oocytes from egg donors and sperm from a partner without male-factor infertility).122 Because imprinted genes are involved in the regulation of embryonic and fetal growth, as well in placental growth and function,123 these findings might be related to inappropriate imprinting, although the role of parental factors also needs to be clarified.
Another retrospective study attempted to determine the risk for major birth defects when IVF and ICSI were used to achieve a pregnancy.118 The authors evaluated data from registries in Western Australia for 1993 to 1997 and found that 26 of the 301 infants conceived through ICSI (8.6%) and 75 of the 837 infants conceived through IVF (9%) had major birth defects (musculoskeletal, metabolic, cardiovascular, urogenital, gastrointestinal, central nervous system) diagnosed by 1 year of age, compared with 4.2% reported for natural conceptions. However, findings from this study also did not clarify the role of preexisting infertility in the increased prevalence of birth defects.
Incidence of ART-Related Imprinting Disorders
Normal genomic imprinting is characterized by the expression of either the maternal or paternal allele (but not both) Disruption of normal imprinting results in clinically identifiable disorders in humans and animals. In general, three mechanisms could cause imprinting disorders124: (1) deletion or mutation in known imprinted genes or imprinting control regions, (2) large deletions or duplications of chromosomal regions containing imprinted genes, or (3) uniparental disomy (both members of a chromosome pair for a particular chromosome come from one parent). Imprinting disorders that result in clinically recognized syndromes in humans occur principally on chromosomes 6,7,11, 14, 15, and 20 (Table 1). As more small deletions/duplications are identified throughout the genome (referred to as copy number variations, or CNVs), it is likely that additional phenotypes will be added to this list. Most of the data regarding imprinting disorders and ART come from studies of BWS and AS. BWS is the imprinting disorder studied more completely than any others.
Table 1.
Human Imprinting Disorders and Uniparental Disomy
| Chromosome | 6 | 7 | 11 | 14 | 15 | 20 | |||
|---|---|---|---|---|---|---|---|---|---|
| Paternal | Maternal | Paternal | Maternal | Paternal | Maternal | Paternal | Maternal | Paternal | |
| Phenotype | TNDM | SRS | BWS | Pre-postnatal growth restriction, dimorphisms; Early puberty; Developmental delay |
Polyhydramnios; Pre-postnatal growth restriction; bell-shaped thorax; muscular hypotonia |
PWS | AS | Pre-postnatal growth restriction; microcephaly; psychomotor developmental delay |
Pseudo- hypoparathyroidism type 1b |
| Prevalence UPD |
40% | 6–10% | 20% | 50 cases | 30 cases | 25% | 2% | ? | ? |
| Gene |
PLAGL, HYMAI |
GRB10, MEST? |
IFGF2; miR483, miR675, CDKN1C |
DLK1, RTU | DLK1, RTU | UBE3A |
SNRPN, snoR116 |
GNAS cluster | GNAS |
The consequences of uniparental disomy and copy number neutral loss-of-heterozygosity during human development and cancer. (Modified from Lapunzina P and Monk D. Biol Cell 2011;103:303–317.142) TNDM, transient neonatal diabetes mellitus; SRS, Silver-Russell syndrome; BWS, Beckwith-Wiedemann syndrome; PWS, Prader-Willi syndrome; AS, Angelman syndrome.
Beckwith-Wiedemann Syndrome
BWS is an overgrowth disorder characterized by pre- and postnatal overgrowth, neonatal hypoglycemia, macroglossia, macrosomia, and an increased risk of embryonal tumors such as Wilms' tumor, rhabdomyosarcoma, and hepatoblastoma.125,126 BWS affects ~1 in 13,700 children126 with most of the patients having an imprinting defect at the maternal allele of one of two DMRs on chromosome 11p15. Imprinting Center 1 (IC1), also known as DMR1, regulates the genes H19 and IGF2; and IC2, also known as DMR2 and in some studies KvDMR1, regulates the genes CDKN1C, KCNQ1OT1, and KCNQ1.124,126
In seven different studies out of nine, BWS cases were identified and the number having ART was ascertained (Table 2A). When these are combined, 53 of 656 (5.9%) BWS cases were the product of an ART procedure compared with ~0.007% in general population in most countries that perform IVF. Investigators then tried to determine if IVF or ICSI were used (some also were ovulation stimulation only or intrauterine insemination). Through the comparison of the percentage of BWS through ART versus the general population, these authors suggested that BWS was significantly increased in mothers undergoing ART.
Table 2.
Studies on Beckwith-Wiedemann Syndrome and Angelman’s Syndrome with Respect to Assisted Reproduction Techniques and Methylation Analysis Imprinting Disorder
| Locus and Genes |
Reference | Study Location |
Study Type | Total No. of Cases in Series |
No. of Cases and ART Type |
Cases Studied for Imprinting | Expected Etiology | |
|---|---|---|---|---|---|---|---|---|
| A. Studies published regarding BWS with respect to ART and methylation analysis | ||||||||
| BWS | 11p15.5 ICF2 and H19in IC1 (DMR1); CDKN1C KCNQ10T1 and KCNQ1 in IC2 (DMR2) |
1. DeBaun et al, 20035 | USA | Case series | 3/65 (4.6%) BWS cases had IVF; 4 BWS cases from another registry (not able to determine prevalence of IVF) |
2 IVF/5 ICSI | 6 studied; 5 had hypomethylation of IC2; 1 of these had both hypomethylation of IC2 and hypermethylation of IC1 |
Maternal hypomethylation of IC2 (50%); Maternal gain methylation of IC1 (5%); Paternal UPD (20%); Mutation of maternal CDKN1C allele (5%); Unknown (20%) Microdeletion (1%); Translocation/ inversion (1%); Duplication (<1%) |
| 2. Maher et al, 20036 | UK | Case series | 6/149 (4%)BWS cases had IVF |
3 IVF/3 ICSI | 4 studied: 0/4 had UPD; 2/4 hypomethylation of IC2 |
|||
| 3. Gicquel et al, 20037 | France | Case series | 6/149 (4%) with IVF | 4 IVF/2 ICSI | 6/6 studied had hypomethylation of IC2 |
|||
| 4. Halliday et al, 20048 | Australia | Case-control | 4/37 (10.8%) of BWS cases had IVF vs. 1/148 controls had IVF |
3 IVF/1 ICSI | 3/3 had hypomethylation of IC2 (1 BWS in non-IVF was not studied) |
|||
| 5. Chang et al, 20059 | USA | Case series | 19/341 (5.6%) BWS had IVF (12 available) |
5 IVF/5 ICSI; 1 CC/IUI; 1 IUI |
ND | |||
| 6. Rossignol et al, 200610 | France | Case series | 40 BWS with demethylation of IC2 (11 had IVF and 29 did not) |
NA | 3/11 (27%) IVF and 7/29(24%) had abnormal methylation at other loci than IC1 and IC2 |
|||
| 7. Sutcliffe et al, 2006120 | UK | Case series | 213 BWS; 83 replied (4 familial); 11/79(13.9%) had ART |
5 ICSI; 1IVF; 5 OI |
8/8 had hypomethylation of IC2 (not specified which had IVForOI) |
|||
| 8. Doornbos 2007143 | Dutch | Case series | 138 BWS; 75 responded and 4/76(1.3%) |
4 IVF; 1 OI; 1 IUI |
6/6 tested had hypomethylation of IC2 |
|||
| 9. Lim 2009144 | UK | Case series | 25 BWS referred compared with 87 BWS known to have IC2 hypomethylation |
12 IVF; 13 ICSI |
24/25 had hypomethylation of IC2; Loss of maternal allele methylation in other DMRs in 37.5% of ART and 6.4% of non-ART BWS IC2 defect cases |
|||
| Total: 656 BWS available | Total: 53 (5.9%) IVF |
Total: 46/50 (92%) | ||||||
| Locus and Genes |
Reference | Study Location |
Study Type | Total No. of Cases in Series |
No. of Cases and ART Type |
Cases Studied for Imprinting | Expected Etiology | |
|---|---|---|---|---|---|---|---|---|
| B. Studies published regarding AS with respect to ART and methylation analysis | ||||||||
| AS | 15q11.2-q13 SNRPN and UBE3A |
1. Ludwig et al 200513 | German | Case Series | 270 AS requested; 79 replied (30%); 16/79(20.2%) had infertility |
3 ICSI 5 OI 8 untreated |
1 of 3 hypomethylation 1 of 5 hypomethylation 2 of 8 hypomethylation |
Maternal 6–7Mb 15q11.2-q13 deletion (~68%) Upd(15)(7%) IC 6- to 200-kb deletions (3%) UBE3A mutations (11%) Deletion/ duplication analysis UBE3A (rare) |
| 2. Sutcliffe et al, 2006120 | UK | Case series | 75 AS cases; 3/75 (4%) had infertility |
2 donor IUI 1 IVF |
2 had deletions 1 had imprinting defect (donor IUI had hypomethylation) |
|||
| 3. Doornbos 2007143 | Dutch | Case series | 135 AS cases; 98 responded and 4/98(4.1%) had infertility |
3 OI 1 donor IUI |
2 of 4 had deletion 2 others unknown |
|||
| 23/252 AS patients (9.1%) had infertility |
4/23 (17.4%) | |||||||
ART, assisted reproduction techniques; IVF, in vitro fertilization; ICSI, intracytoplasmic sperm injection; OI, ovulation induction; IUI, intrauterine insemination; ND, not determined; NA, not applicable.
Table 2A: Studies 1–5, 7, and 8 were used to calculate frequencies of ART in BWS; study 5 did not examine methylation status (ND = not determined); study 6 could not be used for ART prevalence because it only enrolled 40 BWS patients with hypomethylation of IC2; studies 6 and 9 showed additional loci (other than 11 pi 5.5) that had imprinting abnormalities.
Furthermore, some investigators have studied the molecular basis of BWS who had ART to determine if BWS was due to an imprinting defect. For interpretation of the results of these data, it is important to know the prevalence of each mechanism for all cases of BWS. As can be seen in Table 2A, hypomethylation of IC2 accounts for ~50% of all BWS cases, whereas hypermethylation of IC1 occurs in ~5% of BWS. Paternal uniparental disomy of 11p15, annotated as upd (11)pat, is found in 20% of BWS, followed by unknown causes (20%), mutation of the maternally imprinted CDKN1C gene in IC2 (5%), and rarely structural changes or CNVs (3%). Therefore, ~75% of all BWS patients have an epigenetic etiology. In studies done to date, the epigenetic status was analyzed by testing for hypomethylation of IC2 or hypermethylation of IC1. No upd(11)pat has been reported, but it has not been addressed in most studies. Given this information, ~55% of BWS cases would be expected to demonstrate epigenetic changes. However, 46 of 50 of BWS patients (92%) exposed to ART had an imprinting defect of IC1 or IC2, which is considerably more frequent than the estimated 55%.
However, there are some problems determining the prevalence of ART in BWS patients versus the 1 to 3% prevalence of ART in the general population. Ascertainment bias is very likely to be involved in families who have a child with a congenital anomaly. In addition, the appropriate control group should be infertile couples because these disorders may be increased in patients with infertility, regardless of whether they have IVF or ICSI. In fact, cases of BWS with methylation abnormalities have been described in patients who underwent ovulation induction or intrauterine insemination without IVF/ICSI (Table 2A). In addition, in all of the studies cited here, the BWS patient cohort unexposed to ART was not studied for methylation abnormalities. Also, several additional studies reported a very low incidence (either 0 or 1 case) of BWS among the children conceived by ART.127–129 However, even with these methodological problems, the prevalence of imprinting abnormalities in 92% of BWS patients exposed to ART is concerning and requires further study.
Angelman Syndrome
AS, characterized by severe mental retardation, absence of speech, ataxia, seizures, and hyperactivity, was first recognized in 1965. Maternal deletions of 15q11.2-q13 were found to cause AS, and interestingly, paternal deletions of the same region result in Prader-Willi syndrome. This region contains the SNRPN and UBE3A genes, which appear to be imprinted. Three decades later, it was found that single gene mutations in UBE3A could cause AS.130 Interestingly, the UBE3A gene is imprinted in the human brain and only expressed from the maternal copy while the paternal copy of the gene is silent. There are five major genetic causes of AS: (1) maternal chromosome 15q11.2-q13 deletions (68%), (2) paternal uniparental disomy 15 (7%), (3) UBE3A gene mutations (11%), (4) imprinting defects (3%), and (5) partial or whole gene deletions of UBE3A (rare). It is important to emphasize that imprinting defects account for ~3% of AS patients.131
To date, a total of six ART-conceived AS patients with imprinting defects have been reported.11–13,120 However, the evidence is less compelling for AS than BWS. Initially, several case reports of AS who had ICSI were reported.11,12 In the first, two unrelated patients had affected AS children with hypomethylation of the maternal SNRPN/EBEA3 region on 15q11.2-q13.11 Another single case was also reported with hypomethylation of the maternal allele.12 In all three cases, upd(15)pat was excluded. Because AS with imprinting defects is very rare, these authors suggested the possible involvement of ART in AS.
Of three studies that ascertained AS patients exposed to ART, 23 of 252 AS parents (9.1%) had infertility. As shown in Table 2B, 4 of 23 (17.4%) had hypomethylation at the AS IC. Importantly, only three IVF patients were studied, and one of the three had an imprinting etiology. The remaining cases were all infertility, and 3 of 20 (15%) had hypomethylation. The frequency of imprinting in all AS patients approximates 3%, but the numbers with ART are too low to really characterize the effects of ART on imprinting disorders. Nevertheless, the rarity of AS in the general population has prompted some investigators to express concern that ART could be causative. From the data presented in Table 2B, it seems that infertility with or without treatment, rather than strictly ART, could be involved, but this requires future study. As reviewed by Amor and Halliday,124 considering (1) the rarity of AS in the general population (~1 in 15,000), (2) its occurrence as a result of imprinting defects (~3% of cases), and (3) the overall contribution of ARTs to annual births (1 to 3%), the coincidental occurrence of these three events by chance is ~20 million births, which has prompted the suggestion of a link between AS and ART.
Silver-Russell Syndrome
SRS is a disorder characterized by intrauterine and postnatal growth retardation, craniofacial abnormalities in addition to variable learning disabilities. It affects ~1 in 100,000 children. Up to 60% of SRS patients have an imprinting defect (hypomethylation) on the paternal allele of DMR1 at 11p15.132,133 Recently, several studies reported a total of nine SRS patients conceived with ART.14,15,119,134 Of nine patients, six (67%) were confirmed to have an imprinting defect (hypomethylation) in DMR1 on 11p15,14,15 which is not sufficient to provide evidence for or against an association between ART and imprinting defects in SRS.
Retinoblastoma
Retinoblastoma is a malignant tumor of the retina with an incidence of ~1 per 17,000 live births.135 Retinoblastoma usually occurs as a result of a mutation of the tumor suppressor gene RB1; however, hypermethylation of the RB1 gene can also cause retinoblastoma by inactivating its tumor suppressor function.136,137 In 2001, the first case of retinoblastoma was reported in a child born through IVF.138 An additional five cases of retinoblastoma were reported by a Dutch study in 2003.121 Of these five cases, one appeared to be caused by a mutation; the imprinting status of the RB1 gene was not determined in the remaining four children. Based on these cases, the investigators of this study estimated an increased risk of 4.2 to 6.7 for IVF-born infants to develop retinoblastoma. In 2004, another case of retinoblastoma was reported in a child conceived by IVF.139 In contrast, a survey study found no retinoblastoma cases among 176 children conceived by IVF.140 When the usual incidence of retinoblastoma (1 per 17,000 live births) is considered, the number of the evaluated children in the latter study is too small to draw any definitive conclusion. Taken together, further studies with the imprinting status of the RB1 gene are needed to confirm or rule out an association between ART and retinoblastoma.
Conclusions and Future Directions
Until recently, mutations in DNA sequence have mostly been the focus for understanding the genetic background of various diseases, including birth defects. In recent years, compelling evidence has been presented that epigenetic modifications not only regulate gene expression during development but also play a critical role in pathogenesis of complex diseases such as cancer and neurodegenerative disorders.141
Considering major epigenetic reprogramming events during gametogenesis/embryonic development and timing of ART, it is possible that suboptimal conditions in ART may induce aberrant epigenetic modifications leading to abnormal development and imprinting disorders. Due to the variability in ART protocols and the rarity of imprinting disorders, it is difficult to determine reliably the causative relationship between an increased risk for imprinting disorders and ART exposure. Nevertheless, despite some conflicting results, both human and animal studies suggest a possible link between ART and imprinting disorders, most convincingly for BWS and less so for AS, but the magnitude of the risk is still unclear. Further comprehensive studies are needed to better address the risk for ART-associated imprinting disorders. Although not always realistically possible, it would be helpful to minimize ascertainment bias by studying all affected individuals, perform similar molecular analyses on both those patients who had affected children from ARTversus those that did not. Perhaps, most importantly, the prevalence of these imprinting disorders in patients experiencing infertility should be compared with those having ART. Evidence from AS in particular suggests that infertility could be a risk factor. Nonhuman primates may serve as excellent experimental models to tackle some questions that cannot be addressed in humans due to ethical reasons. To minimize the risk, further optimization of ART with respect to imprinting events is of significance. Particularly, large longitudinal studies are critical to evaluate long-term effects of ART during the perinatal period and also into adulthood.
Acknowledgments
This work was supported by an extramural grant from the National Institutes of Health (Grant No. R01HD049537 from the NICHD) to Ali Eroglu and an intramural award from Georgia Health Sciences University (IRP00003) to Ali Eroglu and Lawrence Layman.
References
- 1.Zegers-Hochschild F, Adamson GD, de Mouzon J, et al. International Committee for Monitoring Assisted Reproductive Technology; World Health Organization. International Committee for Monitoring Assisted Reproductive Technology (ICMART) and the World Health Organization (WHO) revised glossary of ART terminology, 2009. Fertil Steril. 2009;92(5):1520–1524. doi: 10.1016/j.fertnstert.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Evers JL. Female subfertility. Lancet. 2002;360(9327):151–159. doi: 10.1016/S0140-6736(02)09417-5. [DOI] [PubMed] [Google Scholar]
- 3.Gosden R, Trasler J, Lucifero D, Faddy M. Rare congenital disorders, imprinted genes, and assisted reproductive technology. Lancet. 2003;361(9373):1975–1977. doi: 10.1016/S0140-6736(03)13592-1. [DOI] [PubMed] [Google Scholar]
- 4.Koulischer L, Verloes A, Lesenfants S, Jamar M, Herens C. Genetic risk in natural and medically assisted procreation. Early Pregnancy. 1997;3(3):164–171. [PubMed] [Google Scholar]
- 5.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72(1):156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maher ER, Brueton LA, Bowdin SC, et al. Beckwith-Wiedemann syndrome and assisted reproduction technology (ART) J Med Genet. 2003;40(1):62–64. doi: 10.1136/jmg.40.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gicquel C, Gaston V, Mandelbaum J, Siffroi JP, Flahault A, Le Bouc Y. In vitro fertilization may increase the risk of Beckwith-Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am J Hum Genet. 2003;72(5):1338–1341. doi: 10.1086/374824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halliday J, Oke K, Breheny S, Algar E, J Amor D. Beckwith-Wiedemann syndrome and IVF: a case-control study. Am J Hum Genet. 2004;75(3):526–528. doi: 10.1086/423902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang AS, Moley KH, Wangler M, Feinberg AP, Debaun MR. Association between Beckwith-Wiedemann syndrome and assisted reproductive technology: a case series of 19 patients. Fertil Steril. 2005;83(2):349–354. doi: 10.1016/j.fertnstert.2004.07.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossignol S, Steunou V, Chalas C, et al. The epigenetic imprinting defect of patients with Beckwith-Wiedemann syndrome born after assisted reproductive technology is not restricted to the 11p15 region. J Med Genet. 2006;43(12):902–907. doi: 10.1136/jmg.2006.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox GF, Bürger J, Lip V, et al. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71(1):162–164. doi: 10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ørstavik KH, Eiklid K, van der Hagen CB, et al. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic semen injection. Am J Hum Genet. 2003;72(1):218–219. doi: 10.1086/346030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludwig M, Katalinic A, Gross S, Sutcliffe A, Varon R, Horsthemke B. Increased prevalence of imprinting defects in patients with Angelman syndrome born to subfertile couples. J Med Genet. 2005;42(4):289–291. doi: 10.1136/jmg.2004.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bliek J, Terhal P, van den Bogaard MJ, et al. Hypomethylation of the H19 gene causes not only Silver-Russell syndrome (SRS) but also isolated asymmetry or an SRS-like phenotype. Am J Hum Genet. 2006;78(4):604–614. doi: 10.1086/502981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakeling EL, Amero SA, Alders M, et al. Epigenotype-phenotype correlations in Silver-Russell syndrome. J Med Genet. 2010;47(11):760–768. doi: 10.1136/jmg.2010.079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young LE, Fernandes K, McEvoy TG, et al. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet. 2001;27(2):153–154. doi: 10.1038/84769. [DOI] [PubMed] [Google Scholar]
- 17.Khosla S, Dean W, Brown D, Reik W, Feil R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod. 2001;64(3):918–926. doi: 10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- 18.Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62(6):1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- 19.Mann MR, Lee SS, Doherty AS, et al. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131(15):3727–3735. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Vu TH, Ulaner GA, et al. IVF results in de novo DNA methylation and histone methylation at an Igf2-H19 imprinting epigenetic switch. Mol Hum Reprod. 2005;11(9):631–640. doi: 10.1093/molehr/gah230. [DOI] [PubMed] [Google Scholar]
- 21.Farin PW, Piedrahita JA, Farin CE. Errors in development of fetuses and placentas from in vitro-produced bovine embryos. Theriogenology. 2006;65(1):178–191. doi: 10.1016/j.theriogenology.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 22.Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod. 2007;22(1):26–35. doi: 10.1093/humrep/del316. [DOI] [PubMed] [Google Scholar]
- 23.Fortier AL, Lopes FL, Darricarrère N, Martel J, Trasler JM. Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet. 2008;17(11):1653–1665. doi: 10.1093/hmg/ddn055. [DOI] [PubMed] [Google Scholar]
- 24.Jablonka E, Lamb MJ. The inheritance of acquired epigenetic variations. J Theor Biol. 1989;139(1):69–83. doi: 10.1016/s0022-5193(89)80058-x. [DOI] [PubMed] [Google Scholar]
- 25.Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93(4):505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 26.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2(1):21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 27.Aguilera O, Fernández AF, Muñoz A, Fraga MF. Epigenetics and environment: a complex relationship. J Appl Physiol. 2010;109(1):243–251. doi: 10.1152/japplphysiol.00068.2010. [DOI] [PubMed] [Google Scholar]
- 28.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21(4):214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 30.Riggs AD, Martiennssen RA, Russo VEA. Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 31.Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 32.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23(7):781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 34.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 35.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279(46):48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 36.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10(11):805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ooi SK, O'Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122(Pt 16):2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carey N, Marques CJ, Reik W. DNA demethylases: a new epigenetic frontier in drug discovery. Drug Discov Today. 2011;16(15–16):683–690. doi: 10.1016/j.drudis.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103(5):1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13(4):1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 44.Illingworth RS, Bird AP. CpG islands—'a rough guide.'. FEBS Lett. 2009;583(11):1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 45.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99(6):3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen L, Kondo Y, Guo Y, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3(10):2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187(4173):226–232. [PubMed] [Google Scholar]
- 48.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 50.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 51.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 53.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 54.The ENCODE; ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306(5696):636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 55.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 56.Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90(3):430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Friedman JM, Jones PA, Liang G. The tumor suppressor microRNA-101 becomes an epigenetic player by targeting the polycomb group protein EZH2 in cancer. Cell Cycle. 2009;8(15):2313–2314. doi: 10.4161/cc.8.15.9168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noonan EJ, Place RF, Pookot D, et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28(14):1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 60.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61(5 Pt 2):24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 61.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian micro-RNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20(2):134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 65.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 68.Silva J, Mak W, Zvetkova I, et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4(4):481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 69.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457(7228):413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220(2):126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 71.Barton SC, Surani MA, Norris ML. Role of paternal and maternal genomes in mouse development. Nature. 1984;311(5984):374–376. doi: 10.1038/311374a0. [DOI] [PubMed] [Google Scholar]
- 72.McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37(1):179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- 73.Cattanach BM, Kirk M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature. 1985;315(6019):496–498. doi: 10.1038/315496a0. [DOI] [PubMed] [Google Scholar]
- 74.Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308(5959):548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 75.Mann JR, Lovell-Badge RH. Inviability of parthenogenones is determined by pronuclei, not egg cytoplasm. Nature. 1984;310(5972):66–67. doi: 10.1038/310066a0. [DOI] [PubMed] [Google Scholar]
- 76.Hunter P. The silence of genes. Is genomic imprinting the software of evolution or just a battleground for gender conflict? EMBO Rep. 2007;2007;8(5):441–443. doi: 10.1038/sj.embor.7400965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64(4):849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 78.Barlow DP, Stöger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349(6304):84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 79.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351(6322):153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 80.Nicholls RD, Knoll JH, Butler MG, Karam S, Lalande M. Genetic imprinting suggested by maternal heterodisomy in nondeletion Prader-Willi syndrome. Nature. 1989;342(6247):281–285. doi: 10.1038/342281a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Henry I, Bonaiti-Pellié C, Chehensse V, et al. Uniparental paternal disomy in a genetic cancer-predisposing syndrome. Nature. 1991;351(6328):665–667. doi: 10.1038/351665a0. [DOI] [PubMed] [Google Scholar]
- 82.Das R, Hampton DD, Jirtle RL. Imprinting evolution and human health. Mamm Genome. 2009;20(9–10):563–572. doi: 10.1007/s00335-009-9229-y. [DOI] [PubMed] [Google Scholar]
- 83.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21(8):457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 84.Thorvaldsen JL, Bartolomei MS. SnapShot: imprinted gene clusters. Cell. 2007;130(5):958. doi: 10.1016/j.cell.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 85.Royo H, Cavaillé J. Non-coding RNAs in imprinted gene clusters. Biol Cell. 2008;100(3):149–166. doi: 10.1042/BC20070126. [DOI] [PubMed] [Google Scholar]
- 86.Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389(6652):745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 87.Yang T, Adamson TE, Resnick JL, et al. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet. 1998;19(1):25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- 88.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12(23):3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Williamson CM, Turner MD, Ball ST, et al. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet. 2006;38(3):350–355. doi: 10.1038/ng1731. [DOI] [PubMed] [Google Scholar]
- 90.Feil R, Walter J, Allen ND, Reik W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development. 1994;120(10):2933–2943. doi: 10.1242/dev.120.10.2933. [DOI] [PubMed] [Google Scholar]
- 91.Falls JG, Pulford DJ, Wylie AA, Jirtle RL. Genomic imprinting: implications for human disease. Am J Pathol. 1999;154(3):635–647. doi: 10.1016/S0002-9440(10)65309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Paulsen M, Ferguson-Smith AC. DNA methylation in genomic imprinting, development, and disease. J Pathol. 2001;195(1):97–110. doi: 10.1002/path.890. [DOI] [PubMed] [Google Scholar]
- 93.Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278(2):440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 94.Hajkova P, Ancelin K, Waldmann T, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452(7189):877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hajkova P, Erhardt S, Lane N, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117(1–2):15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 96.Lee J, Inoue K, Ono R, et al. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129(8):1807–1817. doi: 10.1242/dev.129.8.1807. [DOI] [PubMed] [Google Scholar]
- 97.Yamazaki Y, Mann MR, Lee SS, et al. Reprogramming of primordial germ cells begins before migration into the genital ridge, making these cells inadequate donors for reproductive cloning. Proc Natl Acad Sci U S A. 2003;100(21):12207–12212. doi: 10.1073/pnas.2035119100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128(4):747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 99.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9(2):129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 100.Davis TL, Yang GJ, McCarrey JR, Bartolomei MS. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet. 2000;9(19):2885–2894. doi: 10.1093/hmg/9.19.2885. [DOI] [PubMed] [Google Scholar]
- 101.Obata Y, Kono T. Maternal primary imprinting is established at a specific time for each gene through out oocyte growth. J Biol Chem. 2002;277(7):5285–5289. doi: 10.1074/jbc.M108586200. [DOI] [PubMed] [Google Scholar]
- 102.Ueda T, Abe K, Miura A, et al. The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells. 2000;5(8):649–659. doi: 10.1046/j.1365-2443.2000.00351.x. [DOI] [PubMed] [Google Scholar]
- 103.Kato Y, Kaneda M, Hata K, et al. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16(19):2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- 104.Hiura H, Obata Y, Komiyama J, Shirai M, Kono T. Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells. 2006;11(4):353–361. doi: 10.1111/j.1365-2443.2006.00943.x. [DOI] [PubMed] [Google Scholar]
- 105.Lucifero D, Mann MR, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet. 2004;13(8):839–849. doi: 10.1093/hmg/ddh104. [DOI] [PubMed] [Google Scholar]
- 106.Obata Y, Kono T. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem. 2002;277(7):5285–5289. doi: 10.1074/jbc.M108586200. [DOI] [PubMed] [Google Scholar]
- 107.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403(6769):501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 108.Oswald J, Engemann S, Lane N, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10(8):475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 109.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241(1):172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 110.Rossant J, Sanford JP, Chapman VM, Andrews GK. Undermethylation of structural gene sequences in extraembryonic lineages of the mouse. Dev Biol. 1986;117(2):567–573. doi: 10.1016/0012-1606(86)90325-8. [DOI] [PubMed] [Google Scholar]
- 111.Kerjean A, Dupont JM, Vasseur C, et al. Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum Mol Genet. 2000;9(14):2183–2187. doi: 10.1093/hmg/9.14.2183. [DOI] [PubMed] [Google Scholar]
- 112.Geuns E, Hilven P, Van Steirteghem A, Liebaers I, De Rycke M. Methylation analysis of KvDMR1 in human oocytes. J Med Genet. 2007;44(2):144–147. doi: 10.1136/jmg.2006.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Khoueiry R, Ibala-Rhomdane S, Méry L, et al. Dynamic CpG methylation of the KCNQ1OT1 gene during maturation of human oocytes. J Med Genet. 2008;45(9):583–588. doi: 10.1136/jmg.2008.057943. [DOI] [PubMed] [Google Scholar]
- 114.Geuns E, De Rycke M, Van Steirteghem A, Liebaers I. Methylation imprints of the imprint control region of the SNRPN-gene in human gametes and preimplantation embryos. Hum Mol Genet. 2003;12(22):2873–2879. doi: 10.1093/hmg/ddg315. [DOI] [PubMed] [Google Scholar]
- 115.El-Maarri O, Buiting K, Peery EG, et al. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet. 2001;27(3):341–344. doi: 10.1038/85927. [DOI] [PubMed] [Google Scholar]
- 116.Ecker DJ, Stein P, Xu Z, et al. Long-term effects of culture of preimplantation mouse embryos on behavior. Proc Natl Acad Sci U S A. 2004;101(6):1595–1600. doi: 10.1073/pnas.0306846101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schieve LA, Rasmussen SA, Buck GM, Schendel DE, Reynolds MA, Wright VC. Are children born after assisted reproductive technology at increased risk for adverse health outcomes? Obstet Gynecol. 2004;103(6):1154–1163. doi: 10.1097/01.AOG.0000124571.04890.67. [DOI] [PubMed] [Google Scholar]
- 118.Hansen M, Kurinczuk JJ, Bower C, Webb S. The risk of major birth defects after intracytoplasmic sperm injection and in vitro fertilization. N Engl J Med. 2002;346(10):725–730. doi: 10.1056/NEJMoa010035. [DOI] [PubMed] [Google Scholar]
- 119.Svensson J, Björnståhl A, Ivarsson SA. Increased risk of Silver-Russell syndrome after in vitro fertilization? Acta Paediatr. 2005;94(8):1163–1165. doi: 10.1111/j.1651-2227.2005.tb02066.x. [DOI] [PubMed] [Google Scholar]
- 120.Sutcliffe AG, Peters CJ, Bowdin S, et al. Assisted reproductive therapies and imprinting disorders—a preliminary British survey. Hum Reprod. 2006;21(4):1009–1011. doi: 10.1093/humrep/dei405. [DOI] [PubMed] [Google Scholar]
- 121.Moll AC, Imhof SM, Cruysberg JR, Schouten-van Meeteren AY, Boers M, van Leeuwen FE. Incidence of retinoblastoma in children born after in-vitro fertilisation. Lancet. 2003;361(9354):309–310. doi: 10.1016/S0140-6736(03)12332-X. [DOI] [PubMed] [Google Scholar]
- 122.Schieve LA, Meikle SF, Ferre C, Peterson HB, Jeng G, Wilcox LS. Low and very low birth weight in infants conceived with use of assisted reproductive technology. N Engl J Med. 2002;346(10):731–737. doi: 10.1056/NEJMoa010806. [DOI] [PubMed] [Google Scholar]
- 123.Reik W, Constância M, Fowden A, et al. Regulation of supply and demand for maternal nutrients in mammals by imprinted genes. J Physiol. 2003;547(Pt 1):35–44. doi: 10.1113/jphysiol.2002.033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Amor DJ, Halliday J. A review of known imprinting syndromes and their association with assisted reproduction technologies. Hum Reprod. 2008;23(12):2826–2834. doi: 10.1093/humrep/den310. [DOI] [PubMed] [Google Scholar]
- 125.Junien C. Beckwith-Wiedemann syndrome, tumourigenesis and imprinting. Curr Opin Genet Dev. 1992;2(3):431–438. doi: 10.1016/s0959-437x(05)80154-6. [DOI] [PubMed] [Google Scholar]
- 126.Weksberg R, Shuman C, Smith AC. Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet. 2005;137C(1):12–23. doi: 10.1002/ajmg.c.30058. [DOI] [PubMed] [Google Scholar]
- 127.Lidegaard O, Pinborg A, Andersen AN. Imprinting diseases and IVF: Danish National IVF cohort study. Hum Reprod. 2005;20(4):950–954. doi: 10.1093/humrep/deh714. [DOI] [PubMed] [Google Scholar]
- 128.Källén B, Finnström O, Nygren KG, Olausson PO. In vitro fertilization (IVF) in Sweden: risk for congenital malformations after different IVF methods. Birth Defects Res A Clin Mol Teratol. 2005;73(3):162–169. doi: 10.1002/bdra.20107. [DOI] [PubMed] [Google Scholar]
- 129.Bowdin S, Allen C, Kirby G, et al. A survey of assisted reproductive technology births and imprinting disorders. Hum Reprod. 2007;22(12):3237–3240. doi: 10.1093/humrep/dem268. [DOI] [PubMed] [Google Scholar]
- 130.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15(1):70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 131.Clayton-Smith J, Laan L. Angelman syndrome: a review of the clinical and genetic aspects. J Med Genet. 2003;40(2):87–95. doi: 10.1136/jmg.40.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Abu-Amero S, Wakeling EL, Preece M, Whittaker J, Stanier P, Moore GE. Epigenetic signatures of Silver-Russell syndrome. J Med Genet. 2010;47(3):150–154. doi: 10.1136/jmg.2009.071316. [DOI] [PubMed] [Google Scholar]
- 133.Netchine I, Rossignol S, Dufourg MN, et al. 11p15 imprinting center region 1 loss of methylation is a common and specific cause of typical Russell-Silver syndrome: clinical scoring system and epigenetic-phenotypic correlations. J Clin Endocrinol Metab. 2007;92(8):3148–3154. doi: 10.1210/jc.2007-0354. [DOI] [PubMed] [Google Scholar]
- 134.Kagami M, Nagai T, Fukami M, Yamazawa K, Ogata T. Silver-Russell syndrome in a girl born after in vitro fertilization: partial hypermethylation at the differentially methylated region of PEG1/MEST. J Assist Reprod Genet. 2007;24(4):131–136. doi: 10.1007/s10815-006-9096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Moll AC, Kuik DJ, Bouter LM, et al. Incidence and survival of retinoblastoma in The Netherlands: a register based study 1862–1995. Br J Ophthalmol. 1997;81(7):559–562. doi: 10.1136/bjo.81.7.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Greger V, Passarge E, Höpping W, Messmer E, Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet. 1989;83(2):155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 137.Ohtani-Fujita N, Dryja TP, Rapaport JM, et al. Hypermethylation in the retinoblastoma gene is associated with unilateral, sporadic retinoblastoma. Cancer Genet Cytogenet. 1997;98(1):43–49. doi: 10.1016/s0165-4608(96)00395-0. [DOI] [PubMed] [Google Scholar]
- 138.Anteby I, Cohen E, Anteby E, BenEzra D. Ocular manifestations in children born after in vitro fertilization. Arch Ophthalmol. 2001;119(10):1525–1529. doi: 10.1001/archopht.119.10.1525. [DOI] [PubMed] [Google Scholar]
- 139.Lee I, Finger PT, Grifo JA, Rausen AR, Rebarber A, Barad DH. Retinoblastoma in a child conceived by in vitro fertilisation. Br J Ophthalmol. 2004;88(8):1098–1099. doi: 10.1136/bjo.2003.041160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bradbury BD, Jick H. In vitro fertilization and childhood retinoblastoma. Br J Clin Pharmacol. 2004;58(2):209–211. doi: 10.1111/j.1365-2125.2004.02109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schofield PN, Joyce JA, Lam WK, et al. Genomic imprinting and cancer; new paradigms in the genetics of neoplasia. Toxicol Lett. 2001;120(1–3):151–160. doi: 10.1016/s0378-4274(01)00294-6. [DOI] [PubMed] [Google Scholar]
- 142.Lapunzina P, Monk D. The consequences of uniparental disomy and copy number neutral loss-Of-heterozygosity during human development and cancer. Biology of the cell/under the auspices of the European Cell Biology Organization. Bio Cell. 2011;103:303–317. doi: 10.1042/BC20110013. [DOI] [PubMed] [Google Scholar]
- 143.Doornbos ME, Maas SM, McDonnell J, Vermeiden JP, Hennekam RC. Infertility, assisted reproduction technologies and imprinting disturbances: a Dutch study. Human Reproduction. 2007;22:2476–2480. doi: 10.1093/humrep/dem172. [DOI] [PubMed] [Google Scholar]
- 144.Lim D, Bowdin SC, Tee L, et al. Clinical and molecular genetic features of Beckwith-Wiedemann syndrome associated with assisted reproductive technologies. Human reproduction. 2009;24:741–747. doi: 10.1093/humrep/den406. [DOI] [PubMed] [Google Scholar]