Abstract
Mechanical forces direct a host of cellular and tissue processes. Although much emphasis has been placed on cell-adhesion complexes as force sensors, the forces must nevertheless be transmitted through the cortical cytoskeleton. Yet how the actin cortex senses and transmits forces and how cytoskeletal proteins interact in response to the forces is poorly understood. Here, by combining molecular and mechanical experimental perturbations with theoretical multi-scale modeling, we decipher cortical mechanosensing from molecular to cellular scales. We show that forces are shared between myosin II and different actin crosslinkers, with myosin having potentiating or inhibitory effects on certain crosslinkers. Different types of cell deformations elicit distinct responses, with myosin and α-actinin responding to dilation, and filamin mainly reacting to shear. Our observations show that the accumulation kinetics of each protein may be explained by its molecular mechanisms, and that protein accumulation and the cell's viscoelastic state can explain cell contraction against mechanical load.
Cells are the ultimate smart material, being capable of self-renewal, self-repair and self-defense through mechanisms that include the regulation of the cells’ physical properties1. To accomplish these features, cells must be able to sense and respond to mechanical inputs. Tremendous effort has been invested in understanding how they sense mechanical cues from substrates through focal adhesions2–4. However, not all mechanosensation is mediated through focal adhesions. As an active material, the actin cytoskeleton is a highly dynamic network, which senses mechanical stimuli, remodels its own microstructures and activates associated signaling pathways5, 6. These properties are essential for many cellular events including cell division, differentiation, migration, morphogenesis, and stem cell fate determination3, 7–12. In in vitro assembled actin networks, the evolution of the microstructures is a result of active forces due to actin polymerization and myosin II contractility13. The force-dependent behaviors of the major cytoskeletal proteins have been well characterized in single molecule assays14, and the mechanical properties of in vitro assembled actin networks with different concentrations of cytoskeletal proteins have been systematically explored15–21. However, the kinetic mechanisms of the mechanosensory behaviors of the proteins, the quantitative links between the different hierarchical levels (from molecules to cells to tissues), and how these networks sense forces in living cells are largely unknown. These unresolved issues not only limit our understanding of mechanosensing at different scales but also hamper our ability to design smart materials using cellular components.
Here, by combining molecular and mechanical experimental perturbations of the social amoeba Dictyostelium discoideum with multi-scale modeling, we identify a complex molecular landscape for force sensing and transmission through the cortical cytoskeleton. We demonstrate experimentally that the magnitude of myosin II mechanosensitive accumulation is modulated by the presence of actin crosslinkers, which we interpret as force sharing between myosin II and the crosslinkers. Furthermore, we reveal that myosin II, α-actinin, and filamin react to different deformations, and demonstrate how their molecular mechanisms account for the cellular-scale response of these proteins. Finally, we show how mechanosensitive accumulation of myosin and actin-crosslinking proteins and the cell’s viscoelastic properties account for the dynamics of monotonic and oscillatory contractility in cells, which could explain the oscillatory contractile behaviors observed in some tissues22, 23.
The actin cytoskeleton is composed of myosin II motors, actin filaments and actin crosslinkers (ACs), which are physically linked to the membrane by anchoring proteins. To determine the spectrum of force transmission in the cell cortex in the absence of focal adhesions, we analyzed 37 proteins, each tagged with a fluorescent protein (FP), for changes in their localization in response to mechanical stress (Supplementary Table 1). To apply this stress, we primarily used micropipette aspiration, which allows precisely controlled forces to be applied to specific regions along the cell surface24. Micropipette aspiration has proven to be a useful approach for studying myosin II mechanosensitive accumulation in several systems, including Dictyostelium7, 25–27, Drosophila embryos28; and mammalian stem cells (D. E. Discher, personal communication). Twenty-three of these proteins, most of which are components of the cortical cytoskeleton-membrane composite (Fig. 1a), were identified as being of interest for further characterization in WT and selected genetic mutants. This analysis also uncovered several key features of cellular mechanosensitivity, which we then evaluated theoretically.
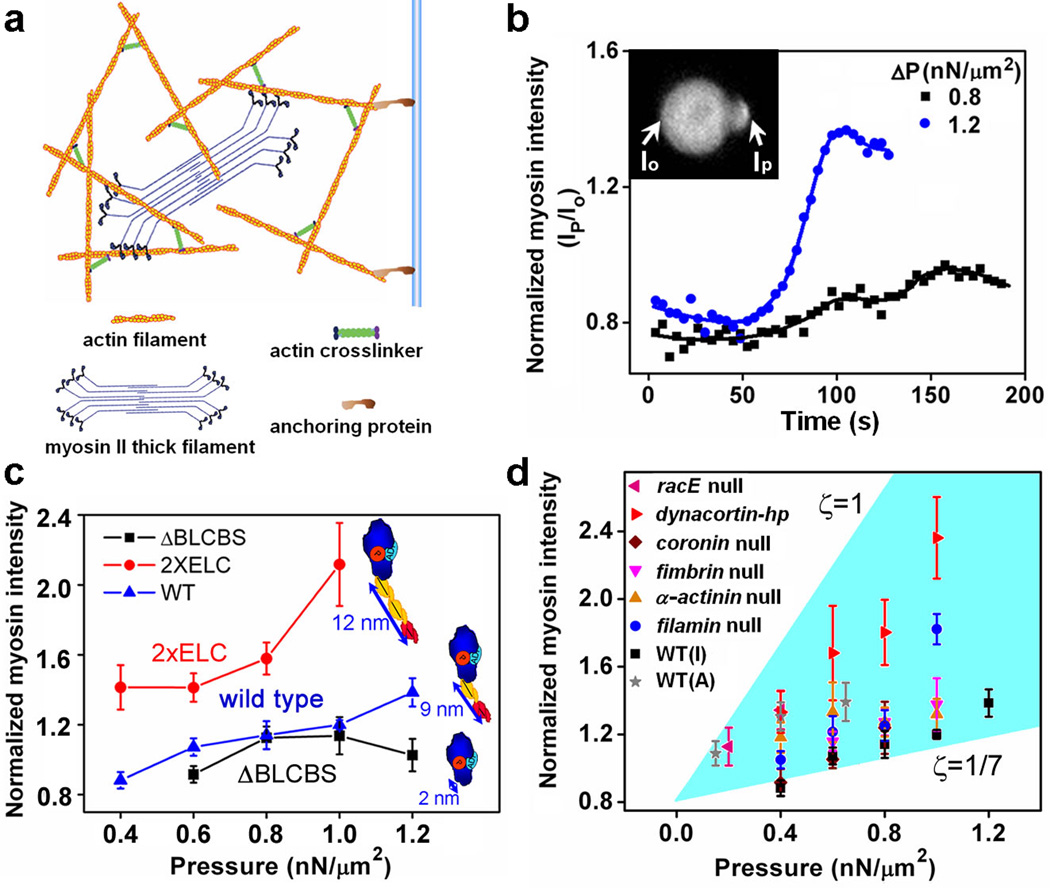
Figure 1. Mechanosensitivity of myosin II in interphase Dictyostelium cells.
a, Schematic diagram of cytoskeleton-membrane complex. b, Time course of myosin II accumulations during micropipette aspiration. Displayed data are from one representative cell at two different pressures. c, Force dependency of the accumulation for myosin II motors with WT and altered lever arm lengths. In this and other panels, normalized intensity is taken at its peak. Dot plots of all data are provided in Supplementary Fig. 2. d, Normalized intensity of the aspirated region for various Dictyostelium strains. The shaded region is bounded by the lines where ζ = |F⃗myosin|/|F⃗internal| equals 1 and 1/7. "I" and "A" represent interphase and anaphase, respectively. Dot plots of all data are provided in Supplementary Fig. 3. A schematic graph for force-sharing and force-transmission is shown in Supplementary Fig. 5. Error bars in c and d correspond to SEM and are based typically on n ≥ 10 at each point.
We established a baseline by characterizing the mechanosensitive response of myosin II in interphase WT cells and comparing it to that in mitotic cells7, 26. When interphase cells were aspirated by a micropipette, we observed that myosin II accumulated in the tip inside the pipette, where the highest dilation of the cortex occurs (Supplementary Movie S1)29. Under constant aspiration pressure, myosin II accumulation accelerated until reaching its peak (Fig. 1b), suggesting the presence of cooperativity in this type of mechanosensory response. Greater applied pressures also led to higher levels of accumulation (Fig. 1c). Moreover, at the same pressures, the accumulation of myosin II increased with increased lever arm lengths for engineered myosins (Fig. 1c). The lever arm length, not motor speed, was the critical determinant of mechanosensitivity. This is demonstrated by the fact that the S456L uncoupler motor (a mutant with shorter step size, slower ADP-release, and 10-fold slower unloaded actin-filament sliding velocity, but normal ATP hydrolysis and lever arm length) showed strong mechanosensitive accumulation (Supplementary Fig. 2). Finally, only intact myosin II accumulated since neither the myosin motor domain (myosin S1) nor the long coiled-coil tail domain were sufficient, confirming that actin binding and bipolar thick filament (BTF) assembly are both essential for mechanosensitive accumulation of myosin (Supplementary Fig. 2). These results, especially the myosin lever arm length dependency, demonstrate that mechanical stress is directly felt by the cortex where it promotes myosin II accumulation, rather than acting through a signaling pathway that is activated by mechanical stress acting on the plasma membrane.
The force-dependent accumulation of non-muscle myosin II at the cellular scale may be explained by using a molecular level catch-bond model. This class of models has been proposed as a simplification of the actin-myosin interaction during the myosin cross-bridge cycle30–33. In the model, the effective off-rate of the myosin head from the actin filament, koff, is a function of force applied to the head, where is the off-rate in the absence of force, f is the force applied on myosin, and Δx is the bond length. Smaller values of koff imply that myosin heads bind longer to F-actin leading to greater myosin accumulation. Thus, the relation Δx2×ELC > ΔxWT > ΔxΔBLCBS between the bond lengths of these myosin proteins with different lever-arm lengths34 indicates that 2×ELC (extra essential light chain binding site) myosin should have the highest accumulation of these motors, while ΔBLCBS (deletion of both light chain binding sites) myosin should have the lowest accumulation, provided that the applied force is the same for all three myosin II motors. This prediction matched the experimental observations (Fig. 1c). Though the mechanosensitive accumulation of myosin II in interphase and mitotic cells is qualitatively similar7, 26, interphase cells required a higher pressure range (see Supplementary Fig. 2a for comparison).
The difference in the mechanosensitive response between interphase and mitotic cells could be a result of the depletion of actin crosslinkers in the polar regions of mitotic cells35. Consistent with this view, we further observed that myosin II accumulation in various mutants, especially dynacortin and racE mutants, in which ACs were deleted was higher than that in WT cells over a wide force range (Fig. 1d). In contrast, the mechanosensitive accumulation of myosin II was reduced in mutants where the physical link between the actin cortex and the plasma membrane was disrupted. To this end, we studied cells in which anchoring proteins (cortexillin I, myosin I D, E, and F isoforms, or enlazin, which is the Dictyostelium ezrin-radixin-moesin-family protein) were deleted, or cells missing PTEN, which catalyzes the formation of the lipid, phosphatidylinositol 4,5-bisphosphate (Fig. 1a). These data suggest that the applied force is transmitted by anchoring proteins from the plasma membrane to the actin cortex where it is shared among myosin II and the cortical ACs. The force sharing here is analogous to that proposed for cell adhesion31, 36, 37.
To test this force sharing further, we plotted the myosin II mechanosensitive response for a number of strains (Fig. 1d; Supplementary Figs. 3a, 5). All the data fell in a region that is bounded by two lines that correspond to different assumptions regarding the fraction (ζ) of the total internal force in the actin cytoskeleton borne by myosin II. These two lines were obtained by solving reaction-diffusion equations describing the mechanosensitive accumulation of myosin II thick filaments25 (Supplementary Material). In the graph, WT cells fall near the line corresponding to ζ = 1/7 consistent with the fact that myosin II contributes ~10–20% of the cortical tension of WT cells (Supplementary Fig. 1c). At the other extreme, if ζ = 1, then 100% of the force would be carried by myosin II and the ACs would bear no force. This would correspond to a hypothetical mutant in which all ACs are inactivated or deleted. The dynacortin-depleted cells and racE null cells, which have reduced levels of several ACs at the cortex38, come close to this limit (Fig. 1d).
These data suggest a simplified picture where the cytoskeleton-membrane composite can be viewed structurally as an assembly of dynamic elastic components (Supplementary Fig. 5): force is applied to the membrane-anchoring proteins which function in series to an array in which myosin II and cortical ACs share forces in a parallel fashion. Thus, disruption of the link between the membrane and the cytoskeleton reduced force-transmission and hence myosin II experienced less force, leading to lower mechanosensitive accumulation. On the other hand, depletion of any ACs redistributed the force that would be borne by that AC onto the remaining proteins, resulting in greater myosin II mechanosensitive accumulation. Consistent with this observation, a plot of the myosin mechanosensitive accumulation as a function of the measured cortical tensions for each of the AC mutant cell-lines revealed a negative correlation between these parameters (Supplementary Figs. 1c, 3b).
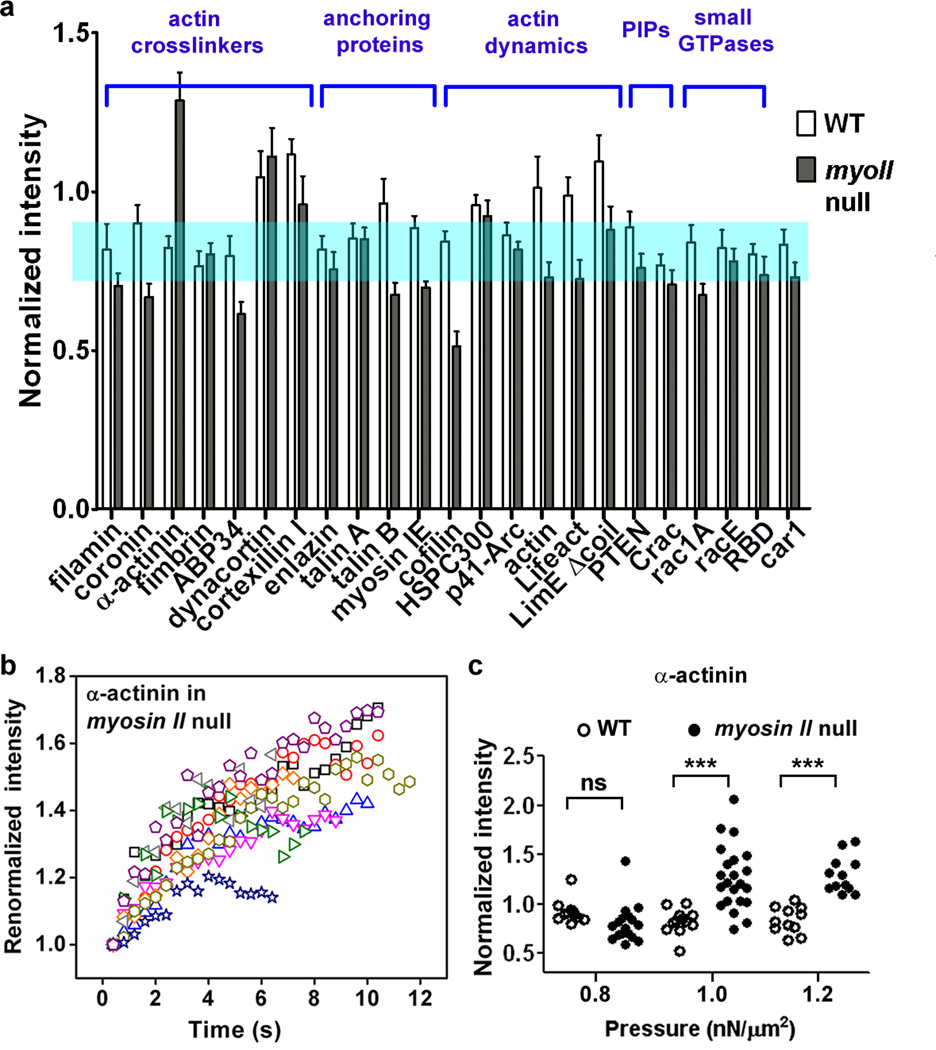
Having established that ACs modulate myosin II’s response to external force, we next studied the effect of myosin II on the mechanosensory responses of ACs. Cells expressing a number of FP-labeled proteins in the presence and absence of myosin II were aspirated at a fixed applied pressure of 1.0 nN/µm2 and the protein accumulation at the tip was quantified (Fig. 2a; Supplementary Fig. 6). These data showed that myosin had different effects on the accumulation at the tip of various AC proteins. For example, cortexillin I accumulation correlated with and depended on myosin II, as previously observed in mitotic cells (Supplementary Fig. 7). However, myosin II was antagonistic with α-actinin as the latter only accumulated in the absence of myosin II (Supplementary Movie S4). Further, α-actinin accumulation increased with time (Fig. 2b) and with increasing applied force (Fig. 2c). Most ACs showed weak myosin II dependency (Supplementary Material; Supplementary Fig. 6). The accumulations of anchoring proteins, such as talin B and myosin IE, were dramatically affected by myosin II. We also imaged actin polymers using FP-labeled actin, Lifeact, and LimE-Δcoil, all of which displayed transient, low-level accumulations in WT cells. These accumulations were more dynamic and did not correlate with the myosin II increase but were instead similar to the patterns of actin waves documented by others39 (Supplementary Movies 2, 3). These probes failed to accumulate in myoII null cells, indicating that actin accumulation by itself is not mechanosensitive to the applied pressure and does not provide the driving force for the myosin II accumulations in the various mutants described here.
Figure 2. The responses of different actin associated proteins to aspiration pressure.
a, Different actin-associated proteins have distinctive mechanosensitive accumulations in the presence and absence of myosin II (n>10 cells for each measurement; details in Supplementary Fig. 6). The shaded region shows the mean ±SEM of a soluble GFP volume marker8. b, The kinetics of α-actinin accumulation in myosin II null cells. c, The pressure-dependence of α-actinin accumulation. The “ns” and “***” denote “not significantly” different and p<0.005, respectively.
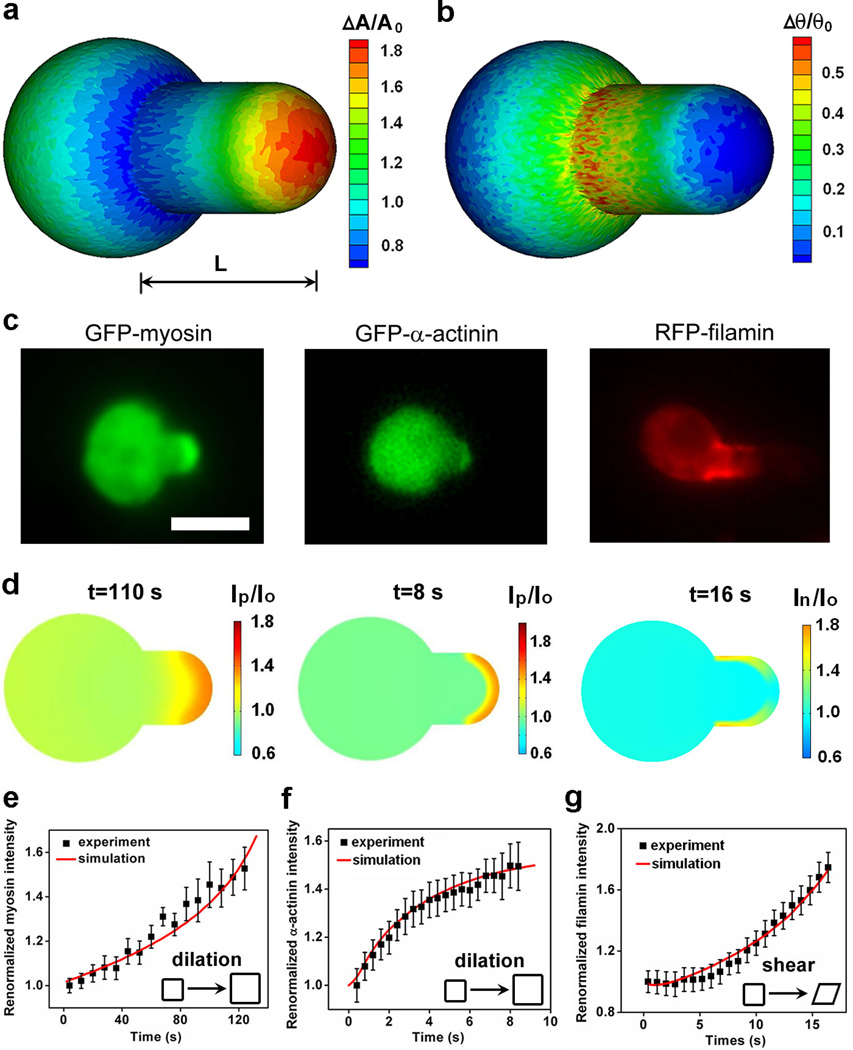
So far, we have described protein accumulations only in the tip region of the aspirated cortex. However, as we analyzed the accumulation of proteins in other mutant strains, we found that in racE null cells, filamin alone (of 20 proteins tested) localized along the neck of the pipette (Supplementary Movie S5; Supplementary Figs. 9, 10; Fig. 3). Filamin also accumulated at the neck in cells depleted of dynacortin using a RNAi hairpin plasmid (dyn-hp cells38; data not shown), but not in the other tested Dictyostelium strains. We conjectured that the different accumulations could be due to responses to varying types of deformations. To test this, we calculated the strain field of a deformed cell in a micropipette using coarse-grained molecular simulations (Supplementary Material; Supplementary Fig. 8). The deformation due to dilation had its highest value at the tip region (Fig. 3a) while the deformation due to shear is greatest at the neck region adjacent to the entrance of the micropipette (Fig. 3b). Thus, the spatial accumulation patterns of myosin II (in various cell lines) and that of α-actinin in myosin II null cells coincided with the dilation strain field suggesting that myosin II and α-actinin were sensitive to dilation strain/stress (Fig. 3c). On the other hand, filamin localization along the neck of the pipette suggests that filamin is sensitive to shear deformation. Because multiple actin crosslinking and bundling proteins (including dynacortin) are down-regulated from the cortex in racE null cells40 and dynacortin depletion itself allowed filamin to accumulate at shear-strained domains, these results suggest that the actin network structure and composition in these mutant cells is more conducive for filamin accumulation in response to shear stress19, 20, 41. We further tested the hypothesis that these proteins respond to specific deformations using a cell compression assay. As expected, we observed that myosin II and α-actinin tracked the dilated regions while filamin accumulated at the sheared regions (Supplementary Fig. 11).
Figure 3. Deformations and corresponding protein accumulation during micropipette aspiration.
Panels a and b show the dilation and angle-change of the cytoskeleton-membrane complex, respectively, calculated using coarse-grained molecular mechanics. Panels c and d show the experimentally observed and simulated accumulations of myosin II, α-actinin, and filamin, respectively. Panels e, f, and g show the comparisons between the measured kinetics of the accumulations and the ones simulated using the corresponding force-dependent binding models for these proteins (Supplementary Material). Insets in e–g show the type of deformation to which each protein responds. Scale bar in c, 10 µm. Model parameters may be found in Supplementary Tables 2 and 3.
To test quantitatively the hypothesis that different proteins are sensitive to different types of cellular deformations, we identified different mechanisms for the force-dependent binding of myosin II (Fig. 3e; Supplementary Fig. 12 and Ref25), α-actinin, and filamin to F-actin. We then carried out simulations in a 3D-deformed cell shape based on experimentally measured kinetic values for diffusion, binding and assembly of their functional units (Supplementary Material). The functional unit of myosin II is the assembled bipolar thick filament (BTF), which generates contractile force, while the actin crosslinkers α-actinin and filamin form rod-shaped42 and V-shaped dimers43, respectively. In these simulations, the dilation and shear strain/stress profiles (Fig. 3a,b) obtained from coarse-grained simulations were used as inputs. Previously, we demonstrated that the catch-bond model together with a model of myosin BTF assembly can account for force-dependent myosin II accumulation25. Here, to account for the force-dependent accumulations of α-actinin and filamin, we tested four types of bond formation: force-independent bonds, slip bonds, simplified catch bonds, and structural cooperativity (Supplementary Materials; Supplementary Figs. 13–17). We found that a simplified catch-bond model based on measured force-dependent bond parameters for α–actinin44 reproduced the accumulation levels and accounted for the deformation-type specificity (Fig. 3f; Supplementary Fig. 13). For filamin accumulation, we found that structural cooperativity resulting in increased on-rates during binding was required along with the catch-bond behavior observed in single molecule studies (Fig. 3g; Supplementary Fig. 16f,g)44. The structural cooperativity proposed here could result from the V-shape of the filamin dimer (Supplementary Materials). The simplified catch bond is sufficient for these simulations because our experimental approach only probes low- to mid-range forces. However, a model (Supplementary Materials) similar to those used in studies of muscle myosin II45 and selectin46, 47 that incorporates a catch-slip transition and accounts for the rupturing of bonds at higher force regimes, also reproduces the mechanosensitive accumulation observed in our experimental data (Supplementary Figs. 15, 17).
Our experiments and simulations showed that myosin II and α-actinin accumulate in the pipette tip while filamin accumulates in the neck region (Fig. 3d; Supplementary Movies S6–S8). More importantly, both the magnitude and kinetics obtained from simulations are in good agreement with experimental observations (Fig. 3e–g), indicating that different proteins are indeed sensitive to different types of cytoskeletal deformation (Fig. 3a). The proposed molecular mechanisms for dilation and shear (dilation could occur through the sliding between parallel and anti-parallel actin-filaments, while shear could involve angle changes between actin filaments) might be responsible for the force-induced accumulations. Because the catch-bond properties and actin binding domains associated with these proteins are relatively similar44, 48, 49, the observed deformation specificity of these proteins likely comes from the overall crosslinker architecture (anti-parallel rods vs. V-shaped).
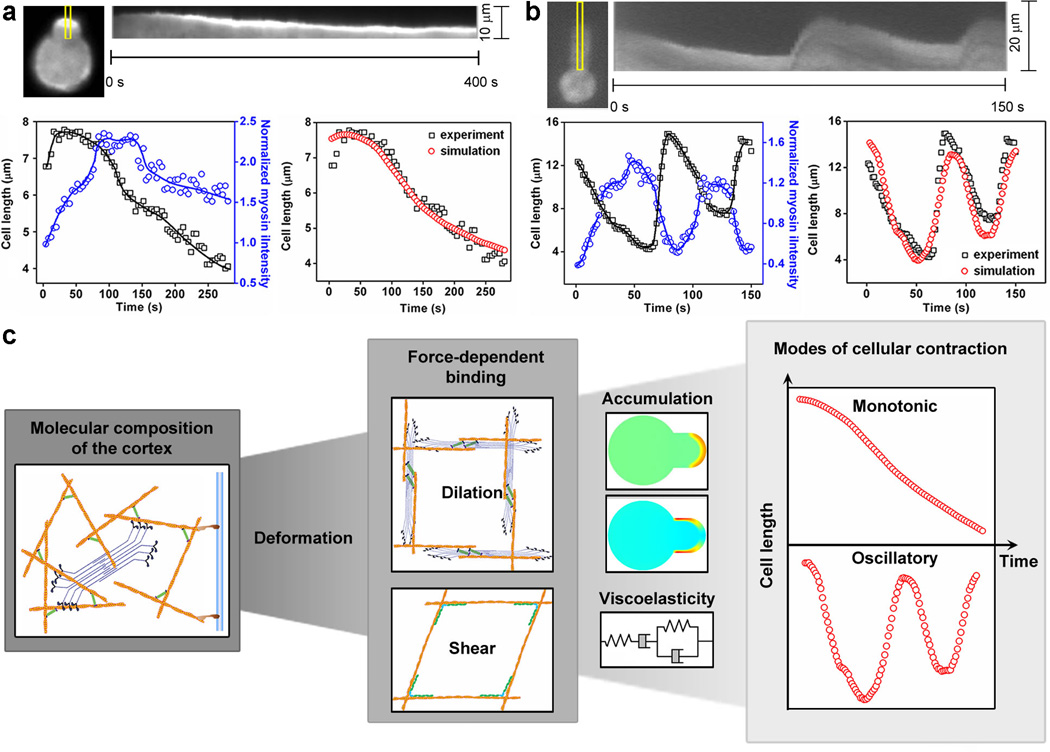
One consequence of the force-induced accumulation of these proteins is the increase of local cortical tension and therefore the resistance to further deformation. Myosin II, in particular, provides local contractility and generates active force to counteract external force. When the contractile force is large enough, cells pull themselves back from the micropipette without forming blebs50 (Fig. 4). We tested this model of contractility-driven cell retraction using simulations (Supplementary Material). We used the experimentally measured myosin II intensities to account for forces. These forces, along with the viscoelastic properties of each particular cell-type, led to simulated cell lengths that matched the monotonic contractility in WT and most mutant cell-types (Fig. 4a; Supplementary Figs. 18, 19) as well as the myosin II-driven oscillations observed in racE null cells (Fig. 4b; Supplementary Fig. 18; Supplementary Movie S9). Although other cytoskeletal proteins such as α-actinin also accumulated in the tip region, their accumulation in the absence of myosin II did not cause noticeable retractions (Supplementary Movie S4), indicating that myosin II is necessary for cell retraction.
Figure 4. Retraction of cells due to the accumulation of cytoskeletal proteins.
Retractions of filamin null (a), and racE null (b) cells, respectively. Kymographs show the accumulation of myosin II along the marked region. Lower left panels show the cell length (squares) and myosin II intensity (circles). The lines depict the trends. Lower right panels compare the experimentally measured aspiration length to that calculated from the viscoelastic model using the measured myosin II intensity as an input. c, The flow diagram summarizes the linking of the hierarchical levels of cell shape change. From the molecular (composition and properties of cortical cytoskeletal proteins) to network (types of deformation and force-dependent binding) to cellular scale (protein accumulation and viscoelastic state), the modes and kinetics of cellular contractility may be explained quantitatively. Model parameters may be found in Supplementary Tables 2–5.
By combining molecular, genetic, and mechanical perturbations with multi-scale modeling, we accounted for the cell’s response to mechanical deformation based on molecular mechanisms (Fig. 4c). We found that some cytoskeletal proteins accumulate to deformation sites in response to mechanical stimuli, and these different proteins are responsive to specific types of deformation. Both cooperative and antagonistic interactions exist between myosin II and ACs, and these mechanosensitive protein accumulations can be predicted quantitatively by their corresponding force-dependent binding reactions.
Our data suggest that the crosslinking proteins may be separated based on three different categories of behaviors. These categories are the types of deformation (dilation vs. shear) to which they respond (e.g. α-actinin vs. filamin), whether they show myosin II dependency in their mechanosensitive accumulation (e.g. α-actinin vs. cortexillin I), and the magnitude of impact they have on cortical tension and myosin II’s mechanosensitive accumulation (e.g. dynacortin vs. fimbrin). One can also envision that different isoforms of a given crosslinker might have considerably different actin affinities, which might translate into further variations in a cell’s mechanosensitivity49. Thus, actin crosslinking proteins provide considerable richness in how cells can respond to mechanical stress inputs, and much work will be required to discern how the specific crosslinker structures and regulation tune these features.
Further, the antagonistic interactions between myosin and α-actinin might help explain segregation of myosin II from α-actinin in self-organized actin structures such as stress fibers and sarcomeres51, 52. Moreover, in addition to filamin GAP, which is a downstream effector of filamin and is sensitive to shear stress6, our results demonstrate that filamin itself can accumulate in response to shear stress when other major crosslinking proteins are absent. Because the proteins studied here are important for the differentiation and migration of Dictyostelium discoideum as well as other organisms5, 6, 11, we expect that these mechanisms will greatly impact the understanding of these biological systems. Finally, the deformation specificities of α-actinin and filamin follow the different structures of their dimers (rod-shaped vs. V-shaped), suggesting a potential strategy for designing novel strain sensors for filamentous biomaterials such as collagen and fibronectin.
Overall, we have demonstrated how protein accumulations proceed and contractility is generated in an active material, the actin cytoskeleton-membrane composite, based on molecular mechanisms and the local microstructures of the cell cortex. We can now directly interpret how the active force generated by the cell cortex and the viscoelasticity of the cells themselves govern the dynamics of cell shape changes during many other biological events, such as cell division8, cell-cell adhesive interactions in tissues22, 23, and epithelial tube formation8, 53. The molecular mechanisms of cellular mechanosensing revealed here not only confer on the cell a self-defense mechanism for rejecting unwanted deformations, but may also allow for the artificial tuning of mechanosensitivity by genetic and pharmacological manipulation for medical purposes. From a materials research perspective, this quantitative understanding may provide guidance for the design of smart materials and strain sensors using cellular components.
Methods
Cell culture and plasmids
WT and mutant Dictyostelium strains were grown at 22°C in Hans’ Enriched HL-5 media. Plasmids were transformed using electroporation and transformants were selected using the appropriate selection medium, which included either G418 or hygromycin. A complete list of fluorescent protein (FP) expression plasmids may be found in Supplementary Table 1. The strains used in this study are: WT, which includes Ax3(replicase orf+) and myosin II null rescued with myosin II; myosin II null (mhcA−; strain ID DBS0236379); cortexillin I null (ctxA−; strain ID DBS0235598); filamin null (abpC−; strain ID DBS0236167); α-actinin null (abpA−; strain ID DBS0235459); fimbrin null (fimA−; strain ID DBS0236175); coronin null (corA−; strain ID DBS0236174); racE null (3G1; strain ID DBS0235415); pten null (pten−; strain ID DBS0349884); myosin I DEF null (myoI D−E−F−54).
Measurements of mechanosensory response of proteins using micropipette aspiration
Micropipette aspiration was performed as described previously7, 26. In short, to apply aspiration pressure, the pressure difference was generated by adjusting the height of a motor-driven water manometer. The Dictyostelium cells expressing desired FPs were loaded into the observation chamber filled with sterile filtered MES buffer (50 mM MES at pH 6.5, 2 mM MgCl2, 0.2 mM CaCl2). The images were collected using an Olympus IX81 microscope equipped with Metamorph software and analyzed using ImageJ (http://rsbweb.nih.gov/ij/). After background correction, the fluorescence intensity at the accumulation sites inside the micropipette was normalized against the opposite cortex in each frame to account for photobleaching. The fluorescence signals are assumed to be linearly proportional to the concentrations of the corresponding protein.
Compression assay using agar-overlay
Compression assay was conducted as described previously8. The images were collected on an Olympus IX81 microscope or a Zeiss 510 Meta confocal microscope.
Statistical analysis
Statistical analysis was performed with software Graph Prism (www.graphpad.com). Mann-Whitney test was used for the non-parametric comparisons for different data sets. In the figures, the asterisks *, ** and *** indicate that the calculated p-value was less than 0.05, 0.01 and 0.005, respectively. Samples not significantly different are denoted with the abbreviation “ns”. Data sets were also analyzed by ANOVA with a Fisher’s Least Significant Difference comparison, which led to nearly identical conclusions.
Supplementary Material
Acknowledgements
We thank Dr. Peter Devreotes, Dr. Miho Iijima and their lab members and members of the Robinson lab for reagents and discussions. We thank Takanari Inoue, Ron Rock, Rob Jensen and Robinson lab members for comments on the manuscript. We thank dictyBase (www.dictybase.org), Dr. David Knecht, Dr. Margaret Titus, Dr. Günther Gerisch, Dr. Thomas Egelhoff and Dr. Paul Steimle for reagents. We thank Vasudha Srivastava for help with confocal imaging. This work is supported by the National Institutes of Health grants GM066817 (to D.N.R.) and GM086704 (to D.N.R. and P.A.I.).
Footnotes
Author contributions
T.L. and D.N.R. conceived, designed and wrote the paper. T.L. performed the experiments and analyzed the data. T.L. conducted the coarse-grained molecular simulations. K.M. and P.A.I. carried out the continuum simulations. T.L., K.M., P.A.I. and D.N.R. proposed the strain-specific molecular mechanisms for different cytoskeletal proteins and the 1D retraction model. All authors discussed the results and commented on the manuscript.
Additional information
Supplementary information is available in the online version of the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.LeDuc PR, Robinson DN. Using lessons from cellular and molecular structures for future materials. Adv. Mat. 2007;19:3761–3770. [Google Scholar]
- 2.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell. Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 4.Tamada M, Sheetz MP, Sawada Y. Activation of signaling cascade by cytoskeleton stretch. Dev. Cell. 2004;7:709–718. doi: 10.1016/j.devcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 5.Chowdhury F, et al. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat. Mater. 2010;9:82–88. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrlicher AJ, Nakamura F, Hartwig JH, Weitz DA, Stossel TP. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature. 2011;478:260–263. doi: 10.1038/nature10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Effler JC, et al. Mitosis-specific mechanosensing and contractile protein redistribution control cell shape. Curr. Biol. 2006;16:1962–1967. doi: 10.1016/j.cub.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kee Y-S, et al. A mechanosensory system governs myosin II accumulation in dividing cells. Mol. Biol. Cell. 2012;23:1510–1523. doi: 10.1091/mbc.E11-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson CP, Tang H-Y, Carag C, Speicher DW, Discher DE. Forced Unfolding of Proteins Within Cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell. Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 12.Raab M, et al. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J. Cell Biol. 2012;199:669–683. doi: 10.1083/jcb.201205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Köhler S, Schaller V, Bausch AR. Structure formation in active networks. Nat. Mater. 2011;10:462–468. doi: 10.1038/nmat3009. [DOI] [PubMed] [Google Scholar]
- 14.del Rio A, et al. Stretching Single Talin Rod Molecules Activates Vinculin Binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardel ML, et al. Elastic behavior of cross-linked and bundled actin networks. Science. 2004;304:1301–1305. doi: 10.1126/science.1095087. [DOI] [PubMed] [Google Scholar]
- 16.Shin JH, Gardel ML, Mahadevan L, Matsudaira P, Weitz DA. Relating microstructure to rheology of a bundled and cross-linked F-actin network in vitro. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9636–9641. doi: 10.1073/pnas.0308733101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhuri O, Parekh SH, Fletcher DA. Reversible stress softening of actin networks. Nature. 2007;445:295–298. doi: 10.1038/nature05459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koenderink GH, et al. An active biopolymer network controlled by molecular motors. Proc. Natl. Acad. Sci. U.S. A. 2009;106:15192–15197. doi: 10.1073/pnas.0903974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner B, Tharmann R, Haase I, Fischer M, Bausch AR. Cytoskeletal polymer networks: The molecular structure of cross-linkers determines macroscopic properties. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13974–13978. doi: 10.1073/pnas.0510190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmoller KM, Lieleg O, Bausch AR. Cross-linking molecules modify composite actin networks independently. Phys. Rev. Lett. 2008;101:118102. doi: 10.1103/PhysRevLett.101.118102. [DOI] [PubMed] [Google Scholar]
- 21.Murrell M, et al. Spreading Dynamics of Biomimetic Actin Cortices. Biophys. J. 2011;100:1400–1409. doi: 10.1016/j.bpj.2011.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, Wang X, Tang HL, Montell DJ. Tissue elongation requires oscillating contractions of a basal actomyosin network. Nat. Cell. Biol. 2010;12:1133–1142. doi: 10.1038/ncb2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solon J, Kaya-Çopur A, Colombelli J, Brunner D. Pulsed Forces Timed by a Ratchet-like Mechanism Drive Directed Tissue Movement during Dorsal Closure. Cell. 2009;137:1331–1342. doi: 10.1016/j.cell.2009.03.050. [DOI] [PubMed] [Google Scholar]
- 24.Bao G, Suresh S. Cell and molecular mechanics of biological materials. Nat. Mater. 2003;2:715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 25.Luo T, et al. Understanding the cooperative interaction between myosin II and actin cross-linkers mediated by actin filaments during mechanosensation. Biophys. J. 2012;102:238–247. doi: 10.1016/j.bpj.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren Y, et al. Mechanosensing through cooperative interactions between myosin II and the actin crosslinker cortexillin I. Curr. Biol. 2009;19:1421–1428. doi: 10.1016/j.cub.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uyeda TQP, Iwadate Y, Umeki N, Nagasaki A, Yumura S. Stretching Actin Filaments within Cells Enhances their Affinity for the Myosin II Motor Domain. PloS one. 2011;6:e26200. doi: 10.1371/journal.pone.0026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez-Gonzalez R, Simoes Sde M, Roper JC, Eaton S, Zallen JA. Myosin II dynamics are regulated by tension in intercalating cells. Dev. Cell. 2009;17:736–743. doi: 10.1016/j.devcel.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Discher DE, Mohandas N. Kinematics of red cell aspiration by fluorescence-imaged microdeformation. Biophys. J. 1996;71:1680–1694. doi: 10.1016/S0006-3495(96)79424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans E. Probing the relation between force-life-time-and chemistry in single molecular bonds. Ann. Rev. Biophy. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 31.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 32.Veigel C, Molloy JE, Schmitz S, Kendrick-Jones J. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat. Cell Biol. 2003;5:980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- 33.Thomas WE, Vogel V, Sokurenko E. Biophysics of catch bonds. Ann. Rev. Biophy. 2008;37:399–416. doi: 10.1146/annurev.biophys.37.032807.125804. [DOI] [PubMed] [Google Scholar]
- 34.Uyeda TQ, Abramson PD, Spudich JA. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl. Acad. Sci. U.S. A. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson DN, Kee YS, Luo T, Surcel A. 7.5 Understanding how dividing cells change shape. In: Edward HE, editor. Comprehensive Biophysics. Amsterdam: Elsevier; 2012. pp. 48–72. [Google Scholar]
- 36.Seifert U. Rupture of multiple parallel molecular bonds under dynamic loading. Phys. Rev. Lett. 2000;84:2750–2753. doi: 10.1103/PhysRevLett.84.2750. [DOI] [PubMed] [Google Scholar]
- 37.Erdmann T, Schwarz US. Stability of adhesion clusters under constant force. Phys. Rev. Lett. 2004;92:108102. doi: 10.1103/PhysRevLett.92.108102. [DOI] [PubMed] [Google Scholar]
- 38.Girard KD, Chaney C, Delannoy M, Kuo SC, Robinson DN. Dynacortin contributes to cortical viscoelasticity and helps define the shape changes of cytokinesis. EMBO J. 2004;23:1536–1546. doi: 10.1038/sj.emboj.7600167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerisch G, et al. Mobile actin clusters and traveling waves in cells recovering from actin depolymerization. Biophy. J. 2004;87:3493–3503. doi: 10.1529/biophysj.104.047589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson DN, Spudich JA. Dynacortin, a genetic link between equatorial contractility and global shape control discovered by library complementation of a Dictyostelium discoideum cytokinesis mutant. J. Cell Biol. 2000;150:823–838. doi: 10.1083/jcb.150.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J. Cell. Biol. 2007;179:1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer RK, Aebi U. Bundling of actin filaments by alpha-actinin depends on its molecular length. J. Cell Biol. 1990;110:2013–2024. doi: 10.1083/jcb.110.6.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hock RS, Condeelis JS. Isolation of a 240-kilodalton actin-binding protein from Dictyostelium discoideum. J. Biol. Chem. 1987;262:394–400. [PubMed] [Google Scholar]
- 44.Ferrer JM, et al. Measuring molecular rupture forces between single actin filaments and actin-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9221–9226. doi: 10.1073/pnas.0706124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo B, Guilford WH. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9844–9849. doi: 10.1073/pnas.0601255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereverzev YV, Prezhdo OV, Forero M, Sokurenko EV, Thomas WE. The two-pathway model for the catch-slip transition in biological adhesion. Biophys. J. 2005;89:1446–1454. doi: 10.1529/biophysj.105.062158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans E, Leung A, Heinrich V, Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc. Natl. Acad. Sci. USA. 2004;101:11282–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gimona M, Djinovic-Carugo K, Kranewitter WJ, Winder SJ. Functional plasticity of CH domains. FEBS Lett. 2002;513:98–106. doi: 10.1016/s0014-5793(01)03240-9. [DOI] [PubMed] [Google Scholar]
- 49.Yao NY, et al. Stress-enhanced gelation: A dynamic nonlinearity of elasticity. Phys. Rev. Lett. 2013;110:018103. doi: 10.1103/PhysRevLett.110.018103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maugis B, et al. Dynamic instability of the intracellular pressure drives bleb-based motility. J. Cell Sci. 2010;123:3884–3892. doi: 10.1242/jcs.065672. [DOI] [PubMed] [Google Scholar]
- 51.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedrich BM, Friedrich EF, Gov NS, Safran SA. Sacromeric pattern formation by actin cluster coalescence. PLoS Comp. Biol. 2012;8:e1002544. doi: 10.1371/journal.pcbi.1002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dickinson DJ, Robinson DN, Nelson WJ, Weis WI. α-Catenin and IQGAP regulate myosin localization to control epithelial tube morphology in Dictyostelium morphogenesis. Dev. Cell. 2012;23:533–546. doi: 10.1016/j.devcel.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C-L, Wang Y, Sesaki H, Iijima M. Myosin I Links PIP3 Signaling to Remodeling of the Actin Cytoskeleton in Chemotaxis. Sci. Signal. 2012;5:ra10. doi: 10.1126/scisignal.2002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.