Abstract
Purpose
Amplification of MYC is one of the most common genetic alterations in lung cancer, contributing to a myriad of phenotypes associated with growth, invasion and drug resistance. Murine genetics has established both the centrality of somatic alterations of Kras in lung cancer, as well as the dependency of mutant Kras tumors on MYC function. Unfortunately, drug-like small-molecule inhibitors of KRAS and MYC have yet to be realized. The recent discovery, in hematologic malignancies, that BET bromodomain inhibition impairs MYC expression and MYC transcriptional function established the rationale of targeting KRAS-driven NSCLC with BET inhibition.
Experimental Design
We performed functional assays to evaluate the effects of JQ1 in genetically defined NSCLC cells lines harboring KRAS and/or LKB1 mutations. Furthermore, we evaluated JQ1 in transgenic mouse lung cancer models expressing mutant kras or concurrent mutant kras and lkb1. Effects of bromodomain inhibition on transcriptional pathways were explored and validated by expression analysis.
Results
While JQ1 is broadly active in NSCLC cells, activity of JQ1 in mutant KRAS NSCLC is abrogated by concurrent alteration or genetic knock-down of LKB1. In sensitive NSCLC models, JQ1 treatment results in the coordinate downregulation of the MYC-dependent transcriptional program. We found that JQ1 treatment produces significant tumor regression in mutant kras mice. As predicted, tumors from mutant kras and lkb1 mice did not respond to JQ1.
Conclusion
Bromodomain inhibition comprises a promising therapeutic strategy for KRAS mutant NSCLC with wild-type LKB1, via inhibition of MYC function. Clinical studies of BET bromodomain inhibitors in aggressive NSCLC will be actively pursued.
Keywords: LKB1, KRAS, NSCLC, BET, MYC
INTRODUCTION
Approximately 15 – 30% of human non-small cell lung cancer (NSCLC) patients harbor an oncogenic KRAS mutation associated with resistance to conventional treatment regimens (1). Oncogenic activation of kras in the murine lung compartment produces adenocarcinoma, and several human lung cancer cell lines harboring mutant KRAS are dependent on KRAS activity for survival (2–4); therefore, oncogenic mutant KRAS is a highly attractive therapeutic target in NSCLC. However, targeting mutant KRAS with small molecule inhibitors has not proven successful (1).
KRAS is a strong activator of mitogen-activated protein kinase (MAPK) and phosphatydilinositol-3-kinase (PI3K) pathways, which together confer oncogene addiction (5). In a murine lung adenocarcinoma model driven by mutant Kras, the concurrent inhibition of MAPK and PI3K pathways demonstrates significant tumor regression (6). Based on these preclinical data, this strategy has translated to phase I clinical trials for NSCLC patients with KRAS mutation. Although exact mechanisms of cell death remain elusive, several reports suggest that the inhibition of MAPK and PI3K pathways converges at the level of transcription factor MYC (7). The inhibition of KRAS-dependent MAPK and PI3K pathways results in the dephosphorylation of MYC at Serine-62 and phosphorylation at Threonine-58 (7, 8). These events contribute to rapid degradation of MYC in a proteasome-dependent manner. Several lines of evidence support a critical function for MYC in enforcing the transcriptional growth pathway downstream of KRAS (9). Notably, research by Gerard Evan and colleagues has established the dependence of KRAS-mutated NSCLC on MYC by pharmacologic induction of a dominant-negative genetic construct, called OmoMyc (10, 11). These observations establish a pressing rationale to test therapies targeting the Myc transcriptional signaling network in KRAS mutant NSCLC models in vitro and in vivo.
MYC is a master regulatory transcription factor governing cellular growth, and deregulation of MYC in cancer contributes to proliferation, metabolic adaptation, tumorigenesis and resistance to apoptosis. Indeed, amplification of MYC is one of the most common genetic alterations in cancer genomes (12). MYC is a basic helix-loop-helix leucine zipper transcriptional activator and amplifier (13), which localizes to E-box binding sites at promoter and enhancer regulatory regions, as a heterodimer with Max. To date, efforts to create drug-like, direct-acting inhibitors of MYC have not been successful. This perhaps is due to the difficulty of targeting the extended protein-protein interface defined by the heterodimer and the absence of a defined ligand-binding domain (14). We have therefore undertaken an effort to impede MYC-dependent transcriptional signaling by optimizing and characterizing inhibitors of putative MYC co-activator proteins. Recently, we reported MYC-specific inhibitory activity of a highly selective inhibitor of BET bromodomains, JQ1, in models of multiple myeloma and acute leukemia (15, 16). These studies demonstrated selective inhibition of growth- and metabolism-associated transcriptional pathways characteristic of MYC function, without overt effects on other key transcriptional complexes such as AP-1 and NF-κB. In an index study of BET bromodomain inhibition, we observed marked efficacy of JQ1 in models of the rare and lethal, t(15;19) subset of squamous adenocarcinoma of the head, neck and lung known as NUT midline carcinoma (17, 18). Recent research from our group has identified an inhibitory effect of BET inhibition on N-MYC function as well, in translational models of neuroblastoma (19).
Based on the rationale that BET bromodomain inhibition may confer MYC-specific anti-proliferative effects downstream of KRAS signaling, we undertook a detailed study of JQ1 in NSCLC. We have recently demonstrated that genetically-engineered mouse (GEM) models provide valuable platforms to test clinically-relevant hypotheses, which can be immediately extended to the design and implementation of human clinical trials (20). To evaluate the therapeutic efficacy of JQ1 in a common, clinically relevant subset of NSCLC, we performed genetic and functional analyses to assess the impact of JQ1 treatment in mouse lung tumors.
MATERIALS AND METHODS
Cell lines and viability assays
NSCLC cell lines were obtained from the American Type Culture Collection (ATCC) and were maintained as specified. The cells lines were tested by a certified third party laboratory for authenticity (See Supplementary Methods). JQ1 was dissolved in cell culture grade dimethylsulfoxide (DMSO; Sigma). Cell viability assay was performed as described previously (21), and as detailed in Supplementary Methods.
Western blot analysis
Lysate preparation and Western blot were performed as described previously (22). Detailed methods and a list of antibodies used are available in Supplementary Methods.
Luminex Assay
Luminex-based multi-bead assays were carried out as described previously (21). Briefly, quantification of MYC and apoptosis were assayed using the Milliplex Pluripotent Stem Cell Assay and Milliplex Human Apoptosis Assay, respectively (Millipore).
shRNA constructs, lentiviral infection, and siRNA transfection
pLKO.1 short hairpin RNA (shRNA) constructs designed by the Harvard RNAi consortium and siRNA (Thermo) was used as described previously (22). shRNA sequences and procedures are provided in Supplementary Methods.
Analysis of Microarray Gene Expression Data
Gene expression profiling methods (Total RNA isolation and microarray processing) are available in Supplementary Methods.
Murine drug treatment studies
Genetically engineered mice harboring a conditional activating mutation (G12D) at the endogenous Kras locus, conditional Lkb1 knockout, and conditional p53 knockout have been described previously (21, 23). All animal treatment studies were reviewed and approved by the IACUC at the Dana-Farber Cancer Institute. Kras and Kras/Lkb1 mice were treated with 5 × 106 p.f.u. adeno-Cre (University of Iowa viral vector core) intranasally as previously described (20, 21, 23), and subjected to magnetic resonance imaging (MRI) to document tumor burden. After initial imaging, animals were subjected to treatment with JQ1 formulated in 10%DMSO, 90% 10%-Hyroxypropyl Beta Cyclodextrin and given IP route at 50 mg/kg or vehicle daily. Primary lung tumors were macrodissected and formalin fixed or frozen in liquid nitrogen and stored at −80°C until use.
Imaging with tumor volume measurement and immunohistochemistry
MRI measurements were performed as previously described (20), to determine the reduction in tumor volume after the indicated time of treatment. Using the RARE sequence scans, tumor volume measurements were generated using in-house custom software. 3D Slicer was used to reconstruct MRI volumetric measurements as reported previously (20). All murine FDG-PET/CT studies were performed with a preclinical small animal PET/CT system (Siemens Inveon) after injection with 14MBq of 18F-FDG as previously reported (20). Immunohistochemistry was performed as described previously (20). Details and a list of antibodies used is available in the Supplementary Methods.
Statistical analysis
Unless otherwise stated, comparisons of statistical significance were performed using the Student’s t-test. A p-value < 0.05 was considered statistically significant.
RESULTS
JQ1 is active in NSCLC cells of different genotypes, but concomitant alteration of KRAS and LKB1 confers resistance
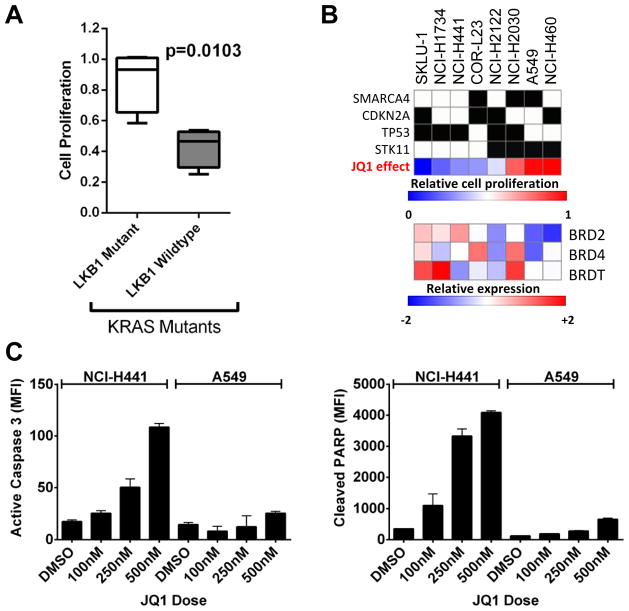
To evaluate the efficacy of BET bromodomain inhibition in NSCLC, human cell lines bearing mutations in KRAS were first screened for sensitivity to the prototype BET inhibitor, JQ1. Unlike hematologic malignancies, where uniform activity of JQ1 is observed, 24 NSCLC cell lines displayed clear dose-dependent sensitivity or resistance to bromodomain inhibition (Supplementary Fig. 1A and 1B). Anti-proliferative activity of BET inhibition was observed in all genetically-defined subtypes of NSCLC, but closer inspection of common genetic alterations revealed reduced efficacy when LKB1 and KRAS were both altered (Fig. 1A). In total, 8 out of 24 cells lines tested harbor KRAS mutation and 3 of 4 cell lines with concurrent KRAS and LKB1 mutation are found to be resistant to JQ1. In contrast, the 4 out of 4 KRAS mutated cell lines without LKB1 mutation responded to JQ1 treatment. Among the 24 cell lines tested, no predictive correlation was observed based on KRAS or EGFR mutational status (Supplementary Fig. 1C). Growth rates of cells under JQ1 treatment were compared to the expression levels of bromodomain and extra-terminal (BET) family proteins and genotype data extracted from the Cancer Cell Line Encyclopedia (CCLE) dataset (Fig. 1B). No significant correlation was found between growth inhibition by JQ1 and the expression level of BET genes (BRD2, BRD3, BRD4 and BRDT).
Figure 1. JQ1 treatment induces apoptosis in NSCLC cells with mutant KRAS but not KRAS/LKB1 mutant cells.
(A) NSCLC cells with KRAS/LKB1 mutations (Left, Open box) proliferate faster than NSCLC cells with KRAS mutation and wild-type LKB1 (Right, grey box) in the presence of JQ1. The proliferation rates of cells treated with 2.5 μM JQ1 were compared to that of cells in DMSO. Proliferation rates of the 24 cell lines tested are listed in Supplementary Fig. 1B. (B) Growth retardation by JQ1 treatment and genotypic characteristics of NSCLC cells harboring KRAS mutation. Percent growth with JQ1 in blue indicates small or no growth inhibition, while red indicates growth despite exposure to JQ1. In the heat map showing the expression level of BRD2, 4, and T, low relative expressions are represented by blue and high expression is indicated by red. (C) Exponentially growing NCI-H441 (KRASmut/LKB1WT) cells or A549 (KRASmut/LKB1mut) cells were treated with DMSO or indicated doses of JQ1 for 48 hours. Cells were harvested for Luminex-based active Caspase 3 (Left) and cleaved PARP (Right) assay for the detection of apoptosis. Dose dependent induction of activated Caspase-3 and cleaved PARP is evident in H441 cells but not in A549 cells. The results are from two independent experiments run with duplicate samples. Bars, S.D.
JQ1-induced apoptosis in KRAS mutant NSCLC cells is abrogated with concurrent LKB1 mutation
We next investigated whether BET bromodomain inhibition in mutant KRAS NSCLC promotes apoptosis. We employed Luminex-based apoptosis assays measuring activated (cleaved) caspase 3 and cleaved-PARP following 48 hours of JQ1 or DMSO (vehicle) treatment in NCI-H441 (mutant KRAS) and A549 (KRAS/LKB1 mutant) cells. These two cell lines showed markedly different apoptotic responses (Fig. 1C). Robust and dose-dependent increases in activated Caspase-3 and cleaved PARP were evident in H441 cells. In contrast, induction of apoptosis was subtle in A549 cells. Similarly, JQ1 treatment induced apoptosis in SK-LU-1 cells harboring mutant KRAS but not in NCI-H460 cells with KRAS/LKB1 mutation (Supplementary Fig. 1D). Cell viability assays confirmed the differences in apoptotic response to JQ1 treatment (Supplementary Fig. 1E).
MYC expression in KRAS positive NSCLC is BRD4 dependent but only in the absence of LKB1 mutation
To investigate the molecular determinants of differences observed between KRAS mutant and KRAS/LKB1 mutant cells, we treated NCI-H441 (KRASG12V), NCI-H1734 (KRASG13C), A549 (KRASG12S/LKB1mut), and NCI-H460 (KRASQ61K/LKB1mut) cells with JQ1 for 48 hours (Fig. 2). Among these cell lines, NCI-H441 and A549 human cell lines have been functionally validated as strongly dependent on KRAS-mediated signaling (4, 24). Proliferation assays demonstrated that H441 cells were sensitive to JQ1 with an IC50 of 0.06 μM while A549 cells were resistant to JQ1 with an IC50 higher than 10 μM (Supplementary Fig. 1A). Similarly, H460 cells harboring LKB1-mutant are resistant to JQ1 with IC50 around 10 μM (Supplementary Fig. 1E).
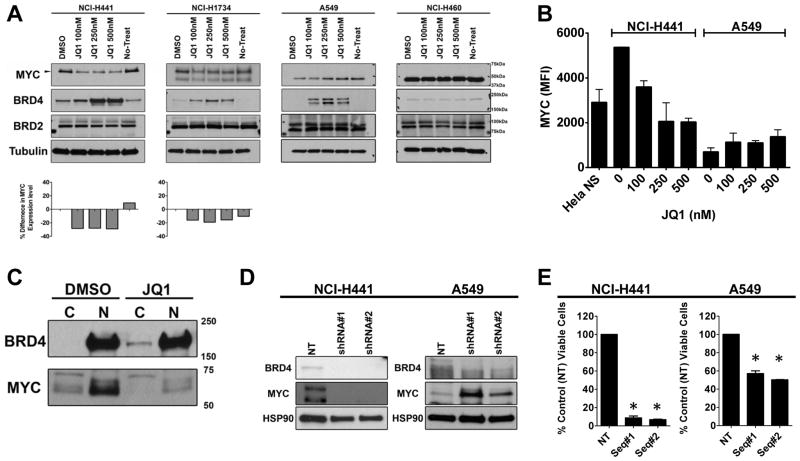
Figure 2. JQ1 promotes depletion of MYC in NSCLC cells with mutant KRAS.
(A) A panel of exponentially growing NSCLC cells (NCI-H441 and NCI-H1734 cells are with KRASmut/LKB1WT and A549 and NCI-H460 cells are with KRASmut/LKB1mut) were treated with indicated doses of JQ1 for 48 hr and lysates were prepared for Western blotting with the indicated antibodies and demonstrate dose dependent accumulation of BRD4. Note that JQ1 induced depletion of MYC (57kDa) is evident in KRASmut/LKB1WT cells. MYC (57kDa) was not detectable in KRASmut/LKB1mut cells with the film exposure time. (B) NCI-H441 or A549 cells were treated with DMSO or indicated doses of JQ1 for 48 hours. Cells were harvested for a Luminex assay to quantify the MYC abundance. Units are mean fluorescent intensity (MFI) and the number is proportional to the amount of protein in the cell. The results represent an average of two independent assays run in duplicate samples. (C) Exponentially growing NCI-H441 cells (KRASmut/LKB1WT) were treated with 500 nM JQ1 or vehicle for 48 hours and lysates were harvested. The cytosolic (C) and nuclear (N) fractions were prepared and the samples were subjected to Western blotting with the indicated antibodies, demonstrating the accumulation of BRD4 takes place in the cytosol and MYC depletion takes place in the nucleus. (D) A representative Western blot showing NCI-H441 or A549 cells transduced with lentiviruses encoding two shRNA sequences targeting BRD4 or a shRNA sequence control. Western blots were performed 5 days post-transduction; in each case, the control construct did not reduce expression of BRD4. In H441 cells, BRD4 knockdown promoted the decrease of MYC expression while BRD4 knockdown in A549 resulted in an increase of MYC expression. (E) Viability assays were performed 7 days after infection, and viability was normalized to cells infected with a lentivirus encoding a non-targeting shRNA. Columns represent the average of triplicate samples. The viability for H441 and A549 cells was significantly (*) reduced by BRD4 knockdown with the most profound reduction being observed in H441 cells. Results represent an average of two independent assays. Bars, SD.
We have demonstrated in highly sensitive hematologic malignancies that the observed, potent antiproliferative effect of BET bromodomain inhibition is variably associated with rapid downregulation of MYC (15). Therefore, we investigated how JQ1 treatment affects MYC expression across a panel of NSCLC cells. All cell lines tested expressed MYC and BRD4, with some variability in expression levels. As a technical note, we detected multiple electrophoretic bands by immunoblot with an anti-MYC antibody. To identify the correct MYC protein, we repeated the immunoblot using lysates harvested from A549, H441, and SK-LU-1 cells transduced with lentivirus coding for non-target and MYC (Supplementary Fig. 2A). As expected, a 57kDa reacting band was the only band depleted by MYC specific shRNA. Following 48 hours of JQ1 exposure, the 57kDa MYC was depleted in JQ1-sensitive H441 and H1734 cells. No effect was observed on MYC expression in resistant A549 and H460 cells (Fig. 2A). To quantify the expression of the 57kDa MYC protein in H441 and A549 cells upon JQ1 treatment, a sensitive Luminex-based assay was performed. The assay independently confirmed depletion of MYC in H441 cells in a dose-dependent manner, in contrast to resistant A549 cells (Fig. 2B). Time-dependent inhibition of MYC expression in sensitive H441 cells was demonstrated by immunoblot. H441 cells exposed to 250 nM of JQ1 begin to deplete MYC in 8 hours (Supplementary Fig. 2B).
Interestingly, exposure to JQ1 also promoted accumulation of BRD4 in cellular lysates within 2 hours (Supplementary Fig. 2B). As BRD4 and MYC primarily localize to the nucleus, we examined the compartment-specific enrichment of BRD4 and MYC in H441 cells upon JQ1 treatment (Fig. 2C). In H441 cells, JQ1 treatment promoted increased appearance of BRD4 in the cytosol and depletion of MYC in the nucleus, illustrating the effect of JQ1 on BET bromodomain displacement from chromatin and corroborating the marked effect on MYC expression.
BRD4 knockdown in H441 cells recapitulates effects of JQ1 in H441 cells
To understand the mechanism of MYC transcriptional inhibition, we assessed function of BET bromodomain proteins in enforcing MYC expression. H441 and A549 cells were transduced with two lentiviral shRNA vectors designed to target BRD4 or a control (non-target) shRNA, followed by puromycin selection. Within 96 hours post lentiviral transduction, BRD4 was significantly (>80%) depleted (Fig. 2D). RNA interference of BRD4 expression resulted in marked reduction in MYC by immunoblot in JQ1-sensitive H441 cells but not in JQ1-resistant A549 cells. In addition, the cell viability of H441 was significantly reduced by BRD4 shRNA at 7 days post-infection (p < 0.01), whereas only a minimal effect on A549 cell viability was observed (less than 50% reduction; Fig. 2E).
MYC transcriptional programs are arrested in JQ1-sensitive mutant KRAS positive NSCLC cells
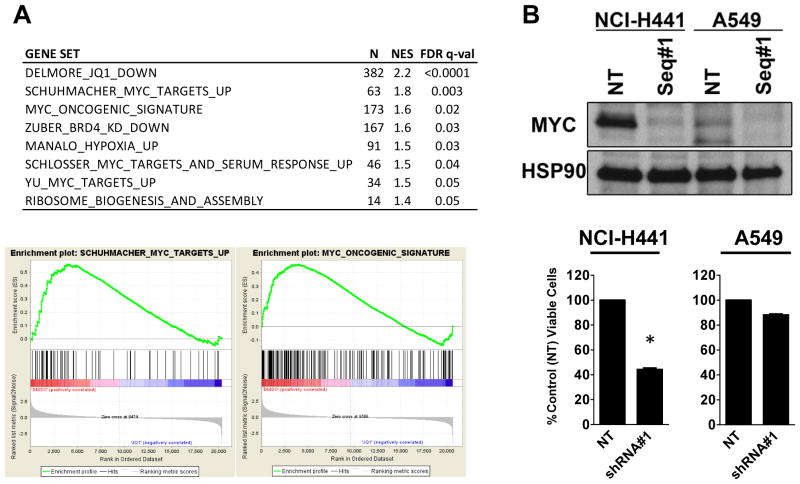
We have observed in highly sensitive hematologic malignancies that rapid downregulation of MYC expression is accompanied by suppression of the MYC-dependent transcriptional growth program (15). To investigate the effect of BET inhibition on NSCLC transcriptional pathways, we performed genome-wide transcriptional profiling of NSCLC cells treated with JQ1 or vehicle control. Unsupervised hierarchical clustering of replicate samples readily identified JQ1 versus vehicle treated samples. Effects on MYC-specific transcription were investigated using reported, functionally validated gene signatures correlating with deregulated MYC (15, 16, 25, 26). All MYC signatures were strongly correlated with downregulation of gene expression by JQ1 in NCI-H441 cells (Fig. 3A). To ascertain whether the H441 cells are MYC dependent, we depleted MYC using lentiviral shRNA (Fig. 3B). Five days post infection, the viability of H441 cells following MYC knockdown was significantly reduced compared to cells infected with virus expressing control shRNA (p < 0.01). In a sharp contrast, MYC depletion in JQ1-resistant A549 cells by lentiviral shRNA did not affect viability.
Figure 3. JQ1 suppresses the MYC transcriptional program and depletion of MYC phenocopies the JQ1 effect in H441 KRASmut/LKB1WT NSCLC cells.
(A) Table of gene sets enriched among genes downregulated by JQ1-treatment in NCI-H441 cells. N: the number of genes in gene set, NES: the normalized enrichment score, and FDR q-val: statistical significance of FDR test. MYC gene sets are significantly downregulated in JQ1-treated NCI-H441 cells. (B) NCI-H441 or A549 cells were transduced with lentiviruses encoding two different sequences of shRNAs targeting MYC or non-target shRNA (control). Western blots were performed 5 days post-transduction; in each case, the control construct did not reduce expression of MYC. In H441 cells, MYC knockdown promoted the significant (*, p < 0.01) decrease in cell viability while MYC depletion in A549 demonstrates no growth retardation. Bars, SD.
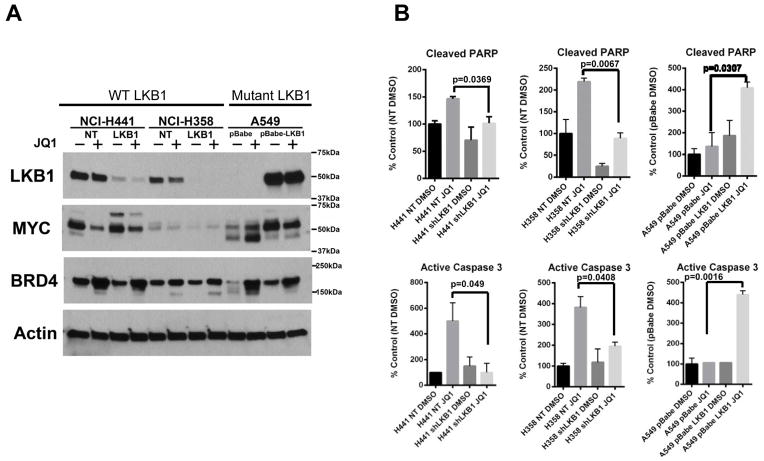
LKB1 mutational status determines sensitivity to JQ1 in mutant KRAS positive NSCLC cells
We have demonstrated that the efficacy of JQ1 and dependency on the BRD4-MYC axis is determined by the mutational status of LKB1 in mutant KRAS NSCLC cell lines. To confirm and extend this finding, we assessed whether loss of LKB1 alone impacts the cellular response to JQ1 treatment, using isogenic paired cell lines (NCI-H358 and NCI-H441) expressing shRNA to LKB1 (LKB1), a non-targeting shRNA (NT). Immunoblot analysis of cells treated with JQ1 (500 nM for 48 hours) or vehicle control demonstrated that LKB1 knock-down impairs the robust inhibition of MYC expression observed with JQ1 (Fig. 4A). We then explored the effect of forced expression of LKB1 in a LKB1 mutated, JQ1 resistant cell line. In contrast to vector-transduced, control A549 cells, LKB1 overexpressing A549 cells exhibited downregulation of MYC by JQ1 (Fig. 4A). After 48 hours of BET inhibition, LKB1 transduced A549 cells now exhibited induction of markers of late apoptosis, cleaved PARP and cleaved caspase 3 (Fig. 4B). The A549 cells transduced with vector control failed to undergo programmed cell death, compared to untreated controls (Fig. 4B).
Figure 4. The loss of LKB1 renders mutant KRAS NSCLC cells resistant to JQ1.
(A) Two paired LKB1 and control (non-target, NT) knockdown cells were generated in KRASmut/LKB1WT NSCLC cell lines, H358 (H358LKB1 and H358NT) and H441 (H441LKB1 and H441NT). Additionally, wild-type LKB1 was ectopically expressed in KRASmut/LKB1mut A549 cell line (A549 pBabe-LKB1). A549 cells were transduced with retrovirus coding for pBabe vector as a paired control (A549 pBabe). The paired cell lines were challenged with DMSO (−) or JQ1 (+) for 48 hours and lysates were subjected to Western blotting with the indicated antibodies. Note that JQ1-induced MYC depletion is modest in the mutant KRAS NSCLC cells with the loss of LKB1. (B) Exponentially growing paired NCI-H441 or NCI-H358 or A549 cells were treated with DMSO or JQ1 (500 nM) for 48 hours. Cells were harvested for Luminex-based active Caspase-3 (bottom) and cleaved PARP (top) assay for the detection of apoptosis. The induction of apoptosis markers is significantly more in cells with LKB1 than in LKB1 deficient cells. The results are from two independent experiments run with duplicate samples. Bars, S.D.
Efficacy of BET bromodomain inhibition in GEM models of KRAS mutated NSCLC
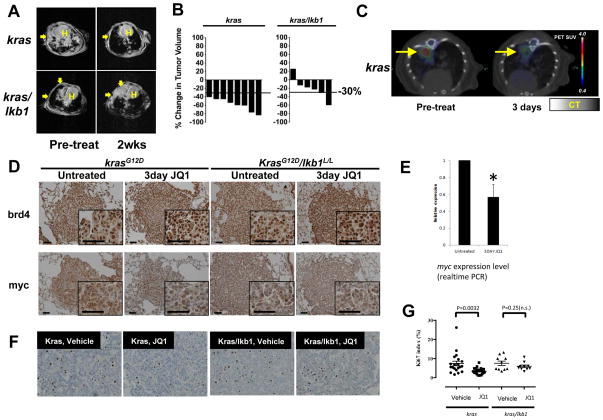
Having demonstrated that a subset of NSCLC cells harboring mutant KRAS are dependent on the BRD4-MYC transcriptional axis, we next explored the therapeutic relevance of BET bromodomain inhibition using genetically engineered mice (GEM) with either mutant kras or mutant kras/lkb1. Activation of mutant kras (G12D) and inactivation of lkb1 in the lung epithelium was initiated using nasal instillation of adenovirus encoding the CRE recombinase. Mice with established tumors, defined by evident and significant tumor burden by magnetic resonance imaging (MRI), were treated with 50 mg/kg JQ1 or vehicle control by intraperitoneal injection. Response to treatment was monitored by serial MRI imaging (Fig. 5A). Tumor volume in response to therapy was determined by previously validated, quantitative methods (20). Among mice with kras mutant tumors, single agent JQ1 provoked a partial response in 100 % of treated animals, defined by a greater than 30 % reduction in tumor volume (Fig. 5A and Fig. 5B). Mice bearing kras mutant tumors responded significantly better than mice with mutant kras tumors with concurrent lkb1 loss (Fig. 5A and Fig. 5B; Fisher’s exact test, p = 0.015).
Figure 5. JQ1 promotes BRD4 accumulation and MYC depletion in a mutant kras murine lung cancer model.
(A) Mice harboring mutant kras or kras/lkb1 were subjected to MRI to establish measurable tumor burden. Next, mice were treated daily with JQ1 (50 mg/kg) by intraperitoneal injection. Two weeks post-treatment, the mice were imaged to evaluate tumor burden. Arrows indicate tumors; H indicates Heart. (B) Volumetric measurements of tumor burden in kras and kras/lkb1 mice after two weeks of JQ1 treatment. kras mice exhibited an objective response (> 30%) to JQ1 treatment while kras/lkb1 tumors do not respond as favorably to JQ1-treatment. (C) Representative FDG-PET/CT images of kras mice at baseline and 3 days post-treatment. Baseline and post-treatment PET images are depicted with identical scales. The colored FDG-PET images are superimposed with the grey-scale cross-sectional CT images. (D) Immunohistochemical (IHC) staining of lung tumors shows JQ1-treatments are more efficacious in downregulating myc expression in mutant kras mice than in mutant kras/lkb1 mice. IHC for brd4 (Top), and myc (Bottom) of lung tumors from kras or kras/lkb1 mice. Mice were treated with 3 doses of JQ1 over 3 days. The third dose was given 3 hours prior to euthanasia, and lung sections were stained with indicated antibodies. Photos shown are representative fields in each group in low and high magnification. Scale bars measure 50 μm. (E) Quantitative RT-PCR analysis of total myc transcript isolated from untreated and JQ1-treated mutant kras mice tumors shows that effects on myc mRNA expression are significantly (p < 0.05) with evident decreases in tumor nodules harvested from kras mice treated with JQ1 for 3 days. Each sample was analyzed in triplicate for quantification of both total myc and β-actin transcripts. The endogenous mouse myc level from untreated mice was arbitrarily designated as 1. Data were analyzed by relative quantitation using the ΔΔCt method with normalization to β-actin. Error bars, S.D. (F) Representative pictures of Ki-67 IHC staining of lung tumors from mutant kras and kras/lkb1 mutant mice untreated or treated with JQ1. (G) Scoring of Ki-67 positive cells shows significant (p = 0.0032, Student’s t-test) reduction in Ki-67 positive cells in mutant kras tumors treated with JQ1 for 3 days. Five areas of each slide were counted (n = 4 each for mutant kras treated and untreated; n = 2 each for kras/lkb1 mutant treated and untreated). Error bars represent S.D.
To further explore early tumor metabolic changes following JQ1 therapy, FDG-PET/CT was performed on mutant kras mice at pre- and post- three days of JQ1 treatment. Treatment with JQ1 resulted in significant changes in tumor hyper-metabolism in mice with mutant kras tumors (Fig. 5C). Together, these results demonstrate that changes in tumor metabolism measured by FDG-PET/CT are in agreement with the volumetric analyses of tumor burden using MRI (Fig. 5A and 5C), and that both reveal tumor regressions in mutant kras tumors with single-agent BET bromodomain inhibition.
Histologic confirmation of radiographic responses to JQ1 treatment in mutant kras mice
To confirm the radiographic responses in kras mice, we performed a detailed immunohistochemical examination of tumor tissue isolated from treated animals. For these pharmacodynamic studies, mice were treated with three daily doses of vehicle or JQ1 prior to sacrifice and harvest of tumors for immunohistochemical analysis. In kras mice, JQ1 treatment resulted in increased staining for brd4 decreased staining for myc (Fig. 5D, left). In contrast, JQ1 treatment failed to deplete myc in mutant kras/lkb1 mice (Fig. 5D, right) while brd4 staining increased upon JQ1 treatment, consistent with our human cell line data (Fig. 2A). To confirm myc depletion in mutant Kras mice, total RNA was isolated from both JQ1 treated and vehicle treated tumor nodules and evaluated by real time PCR using probes specific to murine myc (Fig. 5E). Pharmacologic exposure to JQ1 significantly (p < 0.006) reduced transcription of myc in mutant kras mouse tumors.
Measurements of tumor volume confirmed that NSCLC tumors with mutant kras are considerably more responsive to JQ1 than tumors with combined alteration of kras and lkb1. Consistently, histopathological examination of tumors collected after 3 days of treatment revealed that BET inhibition produced a significant increase in apoptosis (p = 0.0075, Supplementary Fig. 4A) and reduction in proliferation (p < 0.05, Fig. 5F and G) in the mutant Kras tumors but not in Kras/Lkb1 mutant tumors.
DISCUSSION
While direct inhibition of KRAS remains a formidable challenge in the field of cancer drug discovery, the ultimate dependency of Ras signaling on MYC affords new opportunities for drug development within chromatin-dependent transcriptional signaling pathways. The emergence of BET bromodomain proteins as novel co-activators of MYC expression and function prompted consideration of this emerging class of investigational cancer agents as therapeutics for KRAS-dependent NSCLC.
Among 24 NSCLC cell lines representing the clinically apparent genotypes defining aggressive NSCLC, BET bromodomain inhibition confers a meaningful but highly variable response. Cytotoxic activity was not correlated with KRAS and EGFR mutational status; however, the increased fraction of resistant cell lines harboring both KRAS and LKB1 alterations prompted further mechanistic and translational interrogation. Interestingly, we observe that the downregulation of MYC expression and function by BET bromodomain inhibition is highly influenced by LKB1 status. We speculate that altered cell signaling in the context of LKB1 loss produces divergent enforcement of MYC expression.
The results with BRD4 knockdown (Fig. 2D and E) and MYC knockdown (Fig. 3B) suggest that MYC depletion following inhibition of BRD4 by JQ1 treatment is detrimental to NSCLC cells harboring mutant KRAS as evidenced by decreased cell viability and increased apoptosis. In contrast, JQ1 treatment in KRAS/LKB1 mutant cells does not promote the depletion of MYC (Fig. 2B). Similarly, the depletion of BRD4 in A549 cells did not result in the depletion of MYC (Fig. 2D), suggesting, but not proving, that the interactions between BRD4 and MYC in KRAS/LKB1 mutant cells are distinct from those in KRAS mutant cells.
Previously, we have reported that murine tumors with both kras and lkb1 mutations are highly metastatic and respond poorly to simultaneous inhibition of PI3K and MAPK pathways (6, 21). This observation suggests that LKB1 deletion impacts how mutant KRAS regulates PI3K and MAPK pathway activity. The differential activation of the PI3K and MAPK pathways by mutant KRAS in the absence of LKB1 might also promote cellular independence from MYC transcriptional activity, as PI3K and MAPK signaling activities have been shown to control MYC turnover (7, 9). Ongoing research in our laboratory using genome-wide chromatin immunoprecipitation will explore differential enhancer structure and function in the MYC gene desert.
Of note, murine NSCLC tumors harboring mutant kras were sensitive to BET bromodomain inhibition; however, not all human mutant KRAS NSCLC cell lines are sensitive to the inhibition. The discrepancy in the response to JQ1 may be due to the fact that the genetically engineered mouse model used in this study is kras-driven while not all human NSCLC cell lines harboring mutant KRAS are driven by mutant KRAS as reported previously (3, 4, 24, 27). The difference in cellular dependency on mutant KRAS may impact how the cells utilize myc function, ultimately influencing on how the cells respond to BET bromodomain inhibitors. Future work to investigate the impact of BET bromodomain inhibition on MYC-driven NSCLC cells with or without mutant KRAS will provide more mechanistic insights.
To confirm that mutational status of LKB1 determines the antitumoral activity of JQ1 in NSCLC harboring mutant KRAS, we generated a set of isogenic cell lines where LKB1 was restored (in mutated resistant cells) or enforced (in wild-type sensitive cells; Fig. 4). Together these studies confirmed the pronounced effect of LKB1 status on JQ1 sensitivity and JQ1-dependent MYC downregulation.
While these studies were being completed, Lockwood et al. reported the suppression of the oncogenic transcription factor FOSL1 by BET bromodomain inhibition, also using the JQ1 chemical probe from the Bradner laboratory (28). To confirm these observations, we treated JQ1 sensitive NCI-H441 with dose-ranging JQ1 for 48 hours (Supplementary Fig. 3A). Whole cell lysates were subjected to Western blot using MYC and FOSL1 antibodies. We observed dose-dependent depletion of MYC and FOSL1, though MYC exhibited comparatively increased sensitivity to BET inhibition.
To explore whether FOSL1 depletion by JQ1 explains the differential responses to JQ1 in mutant KRAS NSCLC cell lines with or without LKB1, whole cell lysates from isogenic paired cell lines treated with JQ1 or DMSO were subjected to immunoblot for FOSL1 (Supplementary Fig. 3B). In the paired H441 cell lines, the depletion of FOSL1 by JQ1 treatment did not differ between LKB1 expressing and deficient cells. In the paired resistant A549 cells, basal FOSL1 expression level was higher in the LKB1 reconstituted cells, but JQ1 treatment depleted FOSL1 in both sensitive and resistant cells. These data suggest that the suppression of FOSL1 may not entirely explain the differential effect of JQ1 in LKB1 deficient and proficient NSCLC cell lines harboring KRAS mutation. In vivo, JQ1 treatment did not promote fosl1 transcript depletion in murine mutant kras tumors (Data not shown). Further collaborative research will help to contextualize effects on these two BET-responsive master regulatory transcription factors.
Interestingly, the exposure to JQ1 promoted nuclear accumulation of BRD4 in all the NSCLC cell lines tested (Fig. 2A and 2C). The rapid increase in BRD4 staining by immunoblot (Supplementary Fig. 2B) may relate to competitive binding of JQ1 with chromatin, consistent with increased free BRD4 that could have significance for pharmacodynamic biomarker development.
Here, we provide a chemical strategy to inhibit MYC transcriptional signaling downstream of mutant KRAS in lung adenocarcinoma, a common and aggressive human malignancy. Single-agent efficacy of BET bromodomain inhibition in NSCLC has been demonstrated in GEM models of Kras mutant NSCLC. Use of multiallelic GEM models identifies a genetically-defined cohort of patients unlikely to respond, guiding patient selection. Together, these data and insights will guide immediate human clinical investigation of clinical BET inhibitors in early-stage cancer clinical trials.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Preclinical studies have played a key role in defining NUT midline carcinoma, multiple myeloma, and acute myeloid leukemia as diseases leveraged for sensitivity to bromodomain inhibition. Here, we provide a mechanistic rationale for the clinical study of bromodomain inhibitors in non-small cell lung cancer (NSCLC). Using validated models of NSCLC in vitro and in vivo, we provide experimental evidence that the prototype BET bromodomain inhibitor, JQ1, exerts remarkable anti-tumor efficacy in a subset of NSCLC cells harboring KRAS mutation. Sensitivity to bromodomain inhibition is highly influenced by LKB1 mutation status, as Kras/Lkb1 mutant tumor models are comparatively more resistant. Furthermore, the series of in vivo experiments using genetically engineered mouse lung cancer models described in this manuscript confirmed that single-agent bromodomain inhibition prompts bona fide tumor regression in mutant Kras mouse lung cancers. Consistent with cultured human cell lines, Kras/Lkb1 mutant tumors were less responsive to the bromodomain inhibitor. Sensitivity both in vitro and in vivo is mediated by down-regulation of MYC, a known master regulatory transcription factor downstream of the KRAS signaling pathway. The data presented in this manuscript identifies a genetically-defined cohort of patients unlikely to respond to JQ1 treatment, guiding patient selection in early-stage cancer clinical trials.
Acknowledgments
We thank Dr. Bjoern Chapuy for kindly providing pLKO.1 shRNA targeting BRD4. We thank Drs. Rick Wiese and Debra MacIvor at Millipore for providing invaluable technical assistance for Milliplex Pluripotent stem cell and apoptosis assays.
GRANT SUPPORT:
This work is supported by the National Institutes of Health (CA122794, CA140594, CA137181, CA137008, CA147940, CA137008-01, 1U01CA141576, Lung SPORE P50CA090578), United against Lung Cancer Foundation (to K.K.W.), American Lung Association (to K.K.W.), Susan Spooner Research Fund (to K.K.W.), the Damon-Runyon Cancer Research Foundation, the Broad Next Generation Award and a Smith Family Award (to J.E.B). This work is also supported by American Cancer Society Illinois Division Basic Sciences Grant #254563 (to T.S.).
Footnotes
On behalf of all authors on this manuscript, we disclose the following conflicts of interest. All authors have completed separate conflict of interest forms, and we have no additional conflicts of interest to report.
Kwok-Kin Wong
Conflict of interest disclosure statement:
Drug-like BET bromodomain inhibitors created by Drs. Bradner and Qi have been licensed by the Dana-Farber Cancer Institute to Tensha Therapeutics (Cambridge, MA) for therapeutic development.
Consultant/Advisory Board
Drs. Bradner (Major) and Qi (Minor) consult for Tensha Therapeutics to contribute to the clinical translation of drug-like BET bromodomain inhibitors.
Dr. Kimmelman is a consultant for Forma Therapeutics (Minor).
Ownership
Dr. Bradner and the Dana-Farber Cancer Institute have been allocated equity (minority) in Tensha Therapeutics.
Takeshi Shimamura, Zhao Chen, Margaret Soucheray, Julian Carretero, Eiki Kikuchi, Jeremy H. Tchaicha, Yandi Gao, Katherine A. Cheng, Travis J. Cohoon, Esra Akbay, Andrew L. Kung and Kwok-Kin Wong
No conflicts of interest
References
- 1.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–74. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–6. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 3.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 6.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–76. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 9.Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–18. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 10.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–83. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soucek L, Whitfield JR, Sodir NM, Masso-Valles D, Serrano E, Karnezis AN, et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–13. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional Amplification in Tumor Cells with Elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–9. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 15.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bauer DE, Mitchell CM, Strait KM, Lathan CS, Stelow EB, Luer SC, et al. Clinicopathologic Features and Long-term Outcomes of NUT Midline Carcinoma. Clin Cancer Res. 2012;18:5773–9. doi: 10.1158/1078-0432.CCR-12-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–23. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483:613–7. doi: 10.1038/nature10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;17:547–59. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimamura T, Li D, Ji H, Haringsma HJ, Liniker E, Borgman CL, et al. Hsp90 inhibition suppresses mutant EGFR-T790M signaling and overcomes kinase inhibitor resistance. Cancer Res. 2008;68:5827–38. doi: 10.1158/0008-5472.CAN-07-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–10. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 24.Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–34. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 25.Schlosser I, Holzel M, Hoffmann R, Burtscher H, Kohlhuber F, Schuhmacher M, et al. Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene. 2005;24:520–4. doi: 10.1038/sj.onc.1208198. [DOI] [PubMed] [Google Scholar]
- 26.Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci U S A. 2012;109:19408–13. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.