Abstract
The innate immune system senses viral DNA that enters mammalian cells, or in aberrant situations self-DNA, and triggers type I interferon production. Here we present an integrative approach that combines quantitative proteomics, genomics and small molecule perturbations to identify genes involved in this pathway. We silenced 809 candidate genes, measured the response to dsDNA and connected resulting hits with the known signaling network. We identified ABCF1 as a critical protein that associates with dsDNA and the DNA-sensing components HMGB2 and IFI204. We also found that CDC37 regulates the stability of the signaling molecule TBK1 and that chemical inhibition of the CDC37-HSP90 interaction and several other pathway regulators potently modulates the innate immune response to DNA and retroviral infection.
The innate immune system detects viral infection primarily by recognizing viral nucleic acids inside an infected cell1. In the case of retroviruses, which are RNA viruses that replicate via a DNA intermediate, genomic RNA can be recognized in endosomes of specialized innate immune cells using Toll-like receptor 7 (TLR7)2, whereas the reverse-transcribed DNA is believed to be recognized during entry into a host cell by a cytoplasmic DNA sensor (or sensors) that triggers type I interferon production3. This latter response has been observed in cells that lack the endoplasmic reticulum (ER)-associated 3′ to 5′ exonuclease, TREX1. When TREX1 is present, it can degrade the viral DNA before sensing occurs.
Self DNA from the host cell seems to have a similar fate: a deficiency in Trex1 leads to the accumulation of endogenous retroelements and genomic DNA in the cytoplasm, causing aberrant activation of the DNA-sensing pathway and subsequent initiation of autoimmunity4,5. Indeed, mutations in TREX1 are associated with human autoimmune disorders such as Aicardi-Goutières syndrome (AGS)6, familial chilblain lupus7 and systemic lupus erythematosus (SLE)6. Genetic ablation of DNA-sensing pathway components (Irf3 or Tmem173 (Sting)) or the type I interferon receptor can prevent disease onset in Trex1−/− mice (an animal model of AGS)4,5, demonstrating the key epistatic relationship between Trex1 and the interferon-stimulatory DNA (ISD)-sensing pathway, and linking the recognition of DNA viral and retroviral infection to the initiation of autoimmune disease. Identifying components of this pathway may facilitate the identification of therapeutics that modulate the DNA-sensing response to mitigate disease.
How cytosolic DNA elicits production of type I interferons is a long-standing question8 that has gained key insights recently. Specific DNA sequences, for example, (A+T)-rich DNA, are recognized by cytosolic RNA polymerase III, which transcribes the DNA ligand into an RNA product that is recognized by the cytosolic RNA-sensing receptor RIG-I9–11. The innate immune recognition of retroviruses, retroelements and other non–(A+T)-rich DNA does not involve RNA polymerase III or RIG-I3,5,12,13 but involves several of the downstream components of the RIG-I pathway. Recognition of cytosolic DNA relies on the intracellular transmembrane protein, STING14,15, which binds to the kinase TBK1 and the transcription factor IRF3 to allow IRF3 phosphorylation16. Phosphorylated IRF3 then dimerizes and translocates into the nucleus to induce Ifnb1 expression. In addition, certain HMGB family proteins as well as the AIM2-like receptor, IFI16, are believed to have at least a partial role in the cytosolic DNA response17,18. However, the underlying mechanisms remain poorly understood.
We describe here an integrative approach for identification of new components of this DNA-sensing pathway (referred to as the ‘ISD-sensing pathway’ here) and the innate immune response to retroviral infection. We combined unbiased quantitative proteomics with curation of candidates from existing proteomic, genomic and domain-based data sets, and functionally tested 809 of these candidates by high-throughput loss-of-function (RNA interference (RNAi)) screening. We then validated hits by chemical inhibition, cDNA rescue or targeted knockout, and mined existing protein-protein interaction data sets (host-host and host-viral) to form a network model of the ISD-sensing pathway.
RESULTS
Quantitative proteomics identifies candidate pathway components
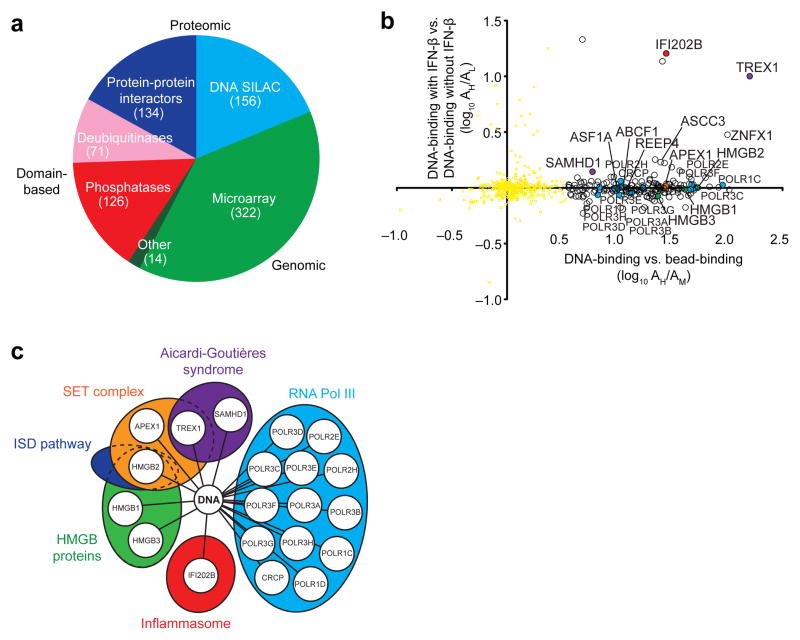
We generated 809 candidate genes from proteomic, genomic and domain-based data sets that we hypothesized contain unidentified ISD-sensing pathway components (Fig. 1a). First, we used protein-protein interaction data sets to nominate 36 candidate proteins that interacted with the known DNA-sensing signaling proteins STING14,15, TBK1 (refs. 13,16), IKKε13 and IRF3 (ref. 12) from a recent mass spectrometry study19 as well as 99 candidates from our own mass spectrometry–based list of putative STING-interacting proteins (Supplementary Table 1). Second, we selected 321 DNA-stimulated genes and interferon-stimulated genes (ISGs) from our own and existing microarray data sets (Online Methods) based on the hypothesis that a subset of components of this pathway are feedback-regulated17,20. Third, we focused on 126 annotated phosphatases (Gene Ontology (GO): 0004721) and 71 deubiquitinases (GO: 0004221 and ref. 21) as part of our pilot screen to identify regulators of the ISD-sensing pathway16,20.
Figure 1.
Generation of a candidate gene set by quantitative proteomics and curation. (a) Sources of the 809 ISD-sensing pathway candidates from proteomic, genomic and domain-based data sets. Data sets included protein-protein interactors of STING, TBK1, IKKε and IRF3 identified by mass spectrometry (Supplementary Table 1 and ref. 19); cytoplasmic DNA-interacting proteins identified by mass spectrometry in b; DNA-regulated and IFN-β–regulated genes found by microarray analysis; and annotated phosphatases and deubiquitinases. (b) Quantitative mass spectrometry analysis showing DNA-binding proteins precipitated from cytoplasmic extracts of MEFs; proteins were precipitated with biotinylated DNA immobilized on streptavidin beads with streptavidin beads alone used as a negative control; white, red, purple, blue, green and orange circles, DNA-interacting proteins with colors corresponding to pathways in c; yellow dots, nonsignificant precipitated proteins; A, abundance; H, M and L, isotope-labeled samples. Ratio of DNA-binding (DNA pull-down, +IFN-β; AH) to bead-binding (empty bead pulldown, +IFN-β; AM) per protein on the x axis is plotted against ratio of DNA binding with IFN-β prestimulation (DNA pulldown, +IFN-β; AH) to DNA binding without IFN-β prestimulation (DNA pull-down, –IFN-β; AL) per protein on the y axis. (c) Representation of SILAC assay-selected hits to indicate the involvement of identified proteins in known cytosolic DNA-sensing pathways.
Because there was no existing data set of cytoplasmic DNA-interacting proteins, we used quantitative proteomics to discover such proteins. We prepared cytoplasmic extracts from mouse embryonic fibroblasts (MEFs; Supplementary Fig. 1a) and added biotinylated 45-base-pair double-stranded DNA (‘ISD sequence’12) coupled to streptavidin beads as bait. We used three-state stable isotope labeling by amino acids in cell culture (SILAC) to label and quantify proteins using mass spectrometry22, with “medium MW (M)” isotope-labeled cells used as negative control (lysates incubated with beads alone), “light MW (L)” isotope-labeled cells for bead-DNA precipitation and “heavy MW (H)” isotope-labeled cells for bead-DNA precipitation preceded by IFN-β stimulation to upregulate pathway components (Supplementary Fig. 1b).
Although only a handful of bands were visually distinguishable by protein electrophoresis (Supplementary Fig. 1c,d), we identified 184 proteins with SILAC ratios that exhibited enrichment for DNA binding after mass spectrometry (Fig. 1b, Supplementary Table 2 and Supplementary Fig. 1e). Among the 184 proteins, 121 (64.2%) were classified by Gene Ontology as having nucleic acid binding function (P = 5.95 × 10−58; GO: 0003676), and others were components of DNA-binding complexes.
Twenty of the 184 identified proteins (10.9%) represent the majority of known proteins involved in the immune sensing of cytosolic DNA (Fig. 1c). We identified known components of DNA-sensing pathways including HMGB family proteins (HMGB1, HMGB2 and HMGB3)18, components of the AIM2 inflammasome (IFI202B and the HMGB proteins)18,23 and the cytosolic RNA polymerase III complex (POLR3A, POLR3B, POLR3C, POLR3D, POLR3E, POLR3F, POLR3G, POLR3H, POLR1C, POLR1D, POLR2E, POLR2H and CRCP). We also identified three members of the SET complex (TREX1, APEX1 and HMGB2) that regulate the ISD-sensing pathway as well as HIV-1 detection and infection3,4,18,24. In addition, we identified the proteins SAMHD1 and TREX1 (refs. 25,26), which are involved in regulating retroviral and retroelement detection and are responsible for AGS4,27. These results validate the utility of quantitative mass spectrometry for finding components of cytosolic DNA-sensing pathways.
RNAi screen defines components of the ISD-sensing pathway
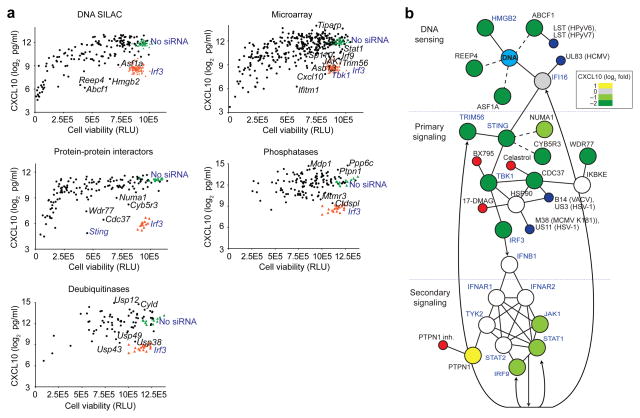
To functionally test the 809 proteomic, genomic and domain-based candidate genes described above (Fig. 1a), we developed a robust high-throughput screening assay, in which we knocked down genes with small interfering RNAs (siRNAs) in MEFs transfected with dsDNA and measured production of the interferon-inducible protein CXCL10 by enzyme-linked immunosorbent assay (ELISA; Supplementary Fig. 2a,b). We used CXCL10 as a readout for interferon-inducible genes because of its high induction in response to nucleic acids, its dependence on IRF3 and the availability of robust protein and RNA assays28. For selected genes, we confirmed by quantitative (q)RT-PCR that the siRNAs knocked down target gene expression (Supplementary Fig. 2c). The siRNA screen (Fig. 2a and Supplementary Table 3a–e) identified 15 genes that, upon knockdown, led to over 90% less CXCL10 production in response to ISD stimulation, including DNA interactors (for example, Abcf1 and Reep4) and protein-protein interactors (for example, Cdc37 and Wdr77); and five genes for which CXCL10 was upregulated more than fourfold after knockdown, including phosphatases (for example, Ppp6c and Mdp1), deubiquitinases (for example, Usp12 and Cyld) and regulated genes (for example, Tiparp). We investigated several of the siRNA screening hits in detail (see below).
Figure 2.
RNAi screen defines functional components of the ISD-sensing pathway. (a) siRNA screening assay showing CXCL10 concentration in supernatants of MEFs treated with siRNA and stimulated with DNA (ISD sequence), plotted against cell viability (in relative light units, RLU) after siRNA treatment to control for potential cytotoxic effects of the siRNAs. Black dots, siRNAs; orange triangles, siRNA targeting Irf3 (siIrf3) positive controls; green triangles, no siRNA; selected genes are labeled; control genes are labeled. Data represent averages of three biological replicates for each siRNA. (b) Representation of protein-protein interactions among known and new proteins involved in the ISD-sensing network based on published and our protein-protein interaction data sets. Blue text, known ISD-sensing pathway components; solid lines, interactions defined in other data sets; dashed lines, interactions defined in our data sets; arrows, transcriptional regulation; green, gray and yellow circles, screened by siRNA and colored by CXCL10 values relative to no siRNA control; white circles, toxic siRNA or not screened; blue circles, viral protein; red circles, chemical inhibitor.
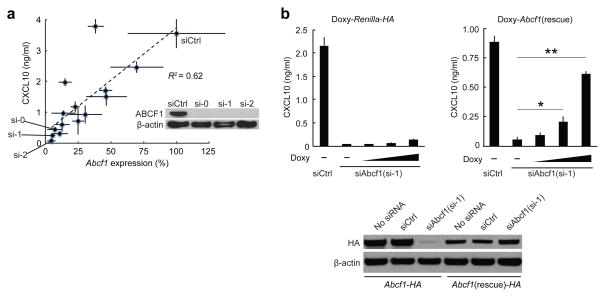
Several cytoplasmic DNA-interacting mass spectrometry hits had functional phenotypes in the siRNA screen (Supplementary Table 3a). HMGB2 regulates nucleic acid-sensing pathways (ref. 18 and Supplementary Fig. 3a) and interacts with DNA. ABCF1 is a cytosolic and ER-localized member of the ATP-binding cassette (ABC) family of transporters with a role in translational control29, but unlike other members of the ABC family, the Abcf subfamily proteins lack transmembrane domains. Although a role in DNA sensing had not been previously observed for ABCF1, there is evidence that human polyomavirus 6 and 7 proteins interact with ABCF1 (ref. 30 and Fig. 2b). We used 14 different siRNAs targeting Abcf1 to interfere with Abcf1 mRNA expression in MEFs and measured by ELISA the induction of CXCL10 in response to stimulation of the ISD-sensing pathway. Knockdown of Abcf1 correlated with loss of CXCL10 induction (R2 = 0.62), with three different siRNA screening pools (si-0, si-1 and si-2) that inhibited both Abcf1 mRNA and ABCF1 protein expression and CXCL10 induction most strongly (Fig. 3a). Knockdown of Abcf1 with the strongest Abcf1 siRNA (si-1) resulted in >93% less production of CXCL10 (Fig. 3b). Expression of an siRNA-resistant cDNA (Abcf1 rescue gene), but not of a Renilla luciferase cDNA control, significantly rescued this phenotype in a manner dependent on the amount of doxycycline used to titrate cDNA expression (Fig. 3b and Supplementary Fig. 3b,c).
Figure 3.
Validation of Abcf1 as a regulator of the ISD-sensing response. (a) ELISA of CXCL10 in MEFs treated with control siRNA (siCtrl) or 14 different siRNAs targeting Abcf1 and stimulated with DNA, plotted against qRT-PCR analysis of Abcf1 mRNA expression in corresponding siRNA-treated MEFs. Inset, immunoblot analysis showing knockdown efficiency of siCtrl or siRNAs si-0, si-1 and si-2 targeting mRNA encoding ABCF1 in MEFs; β-actin served as a loading control. (b) ELISA of CXCL10 in MEFs stably expressing doxycycline (Doxy)-inducible HA-tagged Renilla luciferase (Renilla-HA) or Doxy-inducible Abcf1-rescue cDNA treated with siRNA to Abcf1 (si-1) and doxycycline and stimulated with DNA. Bottom, immunoblot analysis showing knockdown efficiency of no siRNA, siCtrl or si-1 in MEFs stably expressing HA-tagged Abcf1 (Abcf1-HA) or HA-tagged siAbcf1(si-1)-knockdown-resistant Abcf1 (Abcf1(rescue)-HA). Data are from one experiment representative of three independent experiments (mean and s.d. in b). *P < 0.05, **P < 0.001 (Student’s t-test).
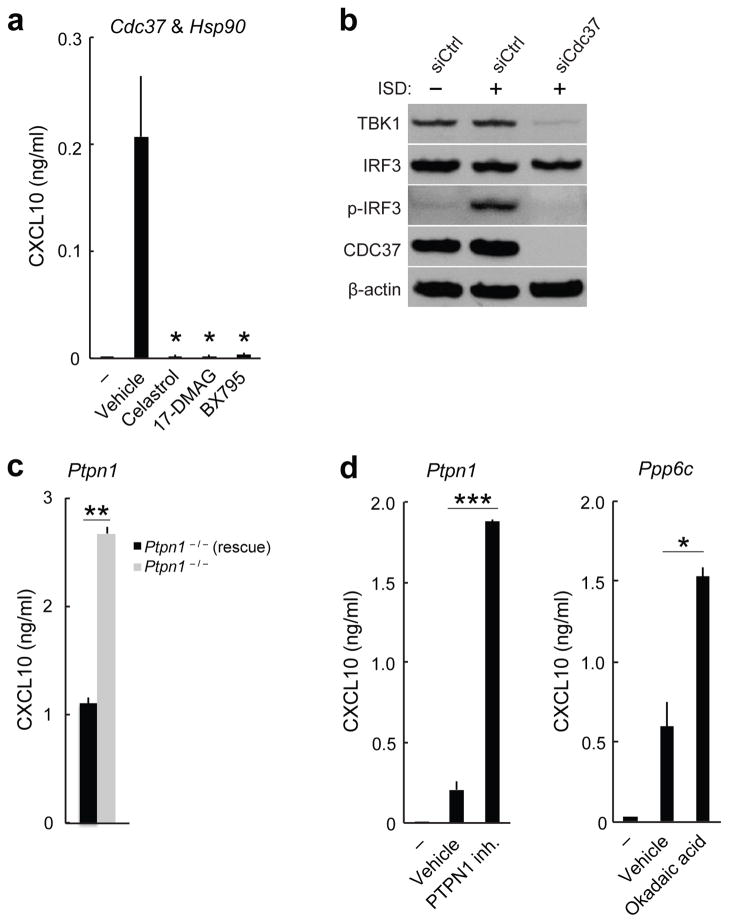
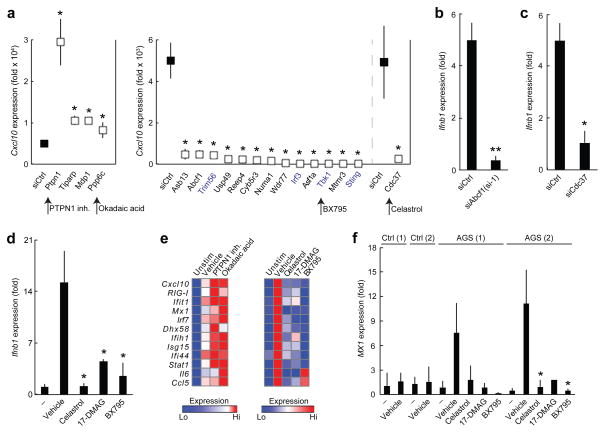
Known components of the ISD-sensing pathway such as Sting, Tbk1 and Irf3 represented strong hits in our siRNA screen (Fig. 2a). Knockdown of each of these genes resulted in over 90% less CXCL10 production in response to ISD stimulation (Supplementary Table 3b,c). Several siRNA screening hits, including Cdc37, Numa1 and Cyb5r3, whose protein products were found to interact with STING (Supplementary Table 1) or TBK1 (ref. 30) also resulted in loss of CXCL10 expression in the siRNA silencing screen (Fig. 2a,b and Supplementary Table 3c). siRNA-mediated knockdown of Cdc37 resulted in over 96% less CXCL10 production in response to ISD stimulation, which is comparable to the reduction induced by knockdown of Sting, Tbk1 or Irf3 (Supplementary Table 3c). In addition, treatment of primary mouse lung fibroblasts or human monocyte-derived dendritic cells (MoDCs) with celastrol, a small molecule inhibitor of the CDC37-HSP90 interaction31,32, potently decreased Ifnb1 and CXCL10 induction in cells stimulated with ISD (Fig. 4a and Supplementary Fig. 4a). CDC37 is a molecular co-chaperone that interacts with HSP90 to stabilize specific proteins, notably protein kinases32, and is a putative interacting partner of TBK1 (refs. 19,33). Thus we tested whether CDC37 regulates TBK1 expression. siRNA-mediated knockdown of Cdc37 in MEFs substantially decreased TBK1 protein expression (Fig. 4b). In addition, siRNA-mediated knockdown of Cdc37 in MEFs abrogated phosphorylation of IRF3 at Ser396, a modification known to occur during activation of the ISD-sensing pathway (Fig. 4b). Chemical inhibition of HSP90 (by 17-DMAG) or of TBK1 (by BX795) decreased ISD-stimulated Ifnb1 and CXCL10 production in mouse lung fibroblasts and human MoDCs, similar to the reductions in these cytokines caused by CDC37 inhibition (Fig. 4a and Supplementary Fig. 4a). Thus, targeting the members of this complex (Fig. 2b) with small molecules blocked the ISD-sensing response by potently inhibiting TBK1 protein stability or activity.
Figure 4.
Small molecule inhibitors modulate the ISD-sensing response. (a) ELISA of CXCL10 in human MoDCs treated with small-molecule inhibitors (500 nM celastrol, 75 nM 17-DMAG or 500 nM BX795) or vehicle control and stimulated with 0.3 μg/ml DNA (HIV gag-100) for 24 h or left unstimulated (–). (b) Immunoblot analysis of TBK1, IRF3, p-IRF3 or CDC37 in MEFs treated with control siRNA (siCtrl) or siRNA targeting Cdc37 (siCdc37) and stimulated for 4 h with DNA. (c) ELISA of CXCL10 in Ptpn1−/− MEFs and Ptpn1−/− MEFs rescued with Ptpn1 cDNA (Ptpn1−/− (rescue)) stimulated with DNA. (d) ELISA of CXCL10 in human MoDCs treated with 7.5 uM PTPN1 inhibitor, 1 nM okadaic acid or vehicle control and stimulated with 0.3 μg/ml DNA (HIV gag-100) or left unstimulated (–). Data are from one experiment representative of three independent experiments (mean and s.d. in a,c,d). *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-test).
Secondary signaling downstream of the interferon receptor is also important in the ISD-sensing response (Fig. 2b), and we identified known (for example, Irf9 and Stat1) and candidate mediators (for example, Ptpn1; ref. 34) of the secondary signaling network (Fig. 2b, Supplementary Fig. 4b and Supplementary Table 3b,d). MEFs deficient in the protein tyrosine phosphatase Ptpn1 produced 2.4-fold more CXCL10 in response to ISD stimulation than rescued MEFs did (Fig. 4c and Supplementary Fig. 4c). Consistent with this result, small molecule inhibition of PTPN1 increased CXCL10 production 9.1-fold in human MoDCs stimulated with ISD (Fig. 4d).
We also tested siRNA screening hits with unknown molecular interaction partners in the ISD-sensing pathway. SP110 is an interferon-regulated nuclear body protein. A natural deleterious mutation in Sp110 (ref. 35) resulted in over 30% less Ifnb1 induction in mouse conventional dendritic cells (cDCs) in response to ISD stimulation compared to wild-type cDCs (Supplementary Fig. 4d). The serine-threonine phosphatase PPP6C has been proposed to interact with IκB-ε33, but its target in the ISD-sensing pathway is unknown. Consistent with the increased CXCL10 production we observed in the siRNA screen, we found that small molecule inhibition of PPP6C by okadaic acid increased ISD-stimulated CXCL10 production in human MoDCs 2.6-fold (Fig. 4d). Taken together, our experiments identified regulators of the ISD-sensing pathway with roles in DNA sensing, primary signaling and secondary signaling.
ABCF1 interacts with HMGB2, IFI204 and the SET complex
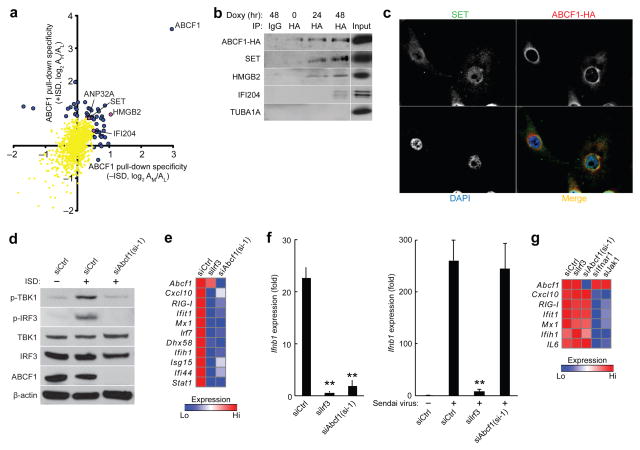
Because the role of ABCF1 in DNA sensing has not been established previously, we investigated the interaction partners of ABCF1. In an unbiased quantitative mass spectrometry–based approach (Supplementary Fig. 5a), we identified 53 proteins that significantly (P < 0.01) precipitated with hemagglutinin epitope (HA)-tagged ABCF1 (ABCF1-HA) in MEFs (Fig. 5a and Supplementary Table 4). Three of these proteins, SET, HMGB2 and ANP32A, are members of the ER-associated SET complex (Fig. 5a,b and Supplementary Fig. 5b) that also contains the DNA exonucleases TREX1 and APEX1, which we identified in ISD interaction experiments (Fig. 1b,c). None of these proteins were found upon pulldown of HA-tagged STING protein (STING-HA) (Supplementary Table 1), indicating specificity for ABCF1. By immunofluorescence staining of MEFs stably expressing ABCF1-HA, we found that a subset of ABCF1 localized with SET and the ER marker calreticulin (Fig. 5c and Supplementary Fig. 5c), consistent with previous reports that ABCF1 localizes to both ER and cytosolic compartments29. These results suggest that ABCF1 interacts with the SET complex at the ER.
Figure 5.
ABCF1 interacts with HMGB2, IFI204 and SET complex. (a) Quantitative mass spectrometry analysis of ABCF1-HA–interacting proteins precipitated from MEFs plotted against ABCF1-HA-interacting proteins precipitated from DNA-stimulated MEFs; proteins were precipitated with anti-HA antibody immobilized on Protein G beads. Blue and purple dots, ABCF1-HA-interacting proteins; yellow dots, nonsignificant precipitated proteins. (b) Immunoassay of association of ABCF1-HA with SET, HMGB2 and IFI204 in MEFs stably expressing doxycycline-inducible Abcf1-HA treated with doxycycline for 0 h, 24 h or 48 h; proteins were immunoprecipitated (IP) with antibody to HA or IgG control and analyzed by immunoblot; TUBA1A, negative control. (c) Immunofluorescence microscopy showing colocalization of ABCF1-HA and SET in MEFs stably expressing Abcf1-HA. Nuclei were stained with the DNA-intercalating dye DAPI. Original magnification X63. (d) Immunoblot analysis of p-TBK1, p-IRF3, TBK1, IRF3 or ABCF1 in MEFs treated with control siRNA (siCtrl) or si-1 and stimulated for 4 h with DNA. (e) qRT-PCR analysis of Abcf1 and ISG mRNA in MEFs treated with indicated siRNAs and infected with HSV-1 d109 (6 h). (f,g) qRT-PCR analysis of Ifnb1 (f) or Abcf1 and ISG mRNA (g) in Trex1−/− MEFs treated with indicated siRNAs and stimulated with 4 μg/ml DNA (HIV gag-100) for 5.5 h or infected with Sendai virus for 6 h (f) or stimulated with 300 U/ml IFN-β for 8 h (g). Data in b–g are from one experiment representative of three (mean in e,g; mean and s.d. in f). *P < 0.01 compared with stimulated siCtrl-treated cells (Student’s t-test).
In addition to its role in the SET complex, HMGB2 is thought to function as a co-ligand for nucleic acid sensors, though its precise role remains unclear18. In our mass spectrometry analysis of ABCF1-HA–interacting proteins (Fig. 5a), we found that ABCF1 interacted with HMGB2 as well as IFI204 (Fig. 5a,b), whose related human protein, IFI16, is a putative DNA sensor17. Consistent with the reported functions of Hmgb2 and Ifi16 (refs. 17,18), siRNA-mediated knockdown of Abcf1 suppressed TBK1 and IRF3 phosphorylation in MEFs stimulated with ISD (Fig. 5d and Supplementary Fig. 5d,e). siRNA-mediated knockdown of Abcf1 also significantly decreased Ifnb1 and ISG induction in MEFs after stimulation with dsDNA (HIV gag-100 sequence) or infection with HSV-1 d109 (Fig. 5e,f and Supplementary Fig. 5f,g) but had no significant effect on Ifnb1 or ISG induction by Sendai virus (which stimulates the RIG-I pathway) or by recombinant IFN-β (Fig. 5f,g). Taken together, these results suggest that ABCF1 interacts with HMGB2 and IFI204 and is a critical node in the DNA-sensing network (Supplementary Fig. 5b).
Identified factors regulate host responses to retrovirus
We further examined whether the ISD-sensing pathway regulators described above also regulate the innate immune response to retroviral infection. Infection by an HIV-based retrovirus induced type I interferon and ISG production in Trex1−/− MEFs but not in wild-type MEFs (ref. 3 and Supplementary Fig. 6a). Knockdown of four of the genes identified in our siRNA screen (Ptpn1, Tiparp, Mdp1 and Ppp6c) in Trex1−/− MEFs followed by infection with an HIV-based retrovirus significantly enhanced the ability of the retroviral infection to induce Ifnb1 and ISG production, whereas knockdown of ten of these genes (Asb13, Abcf1, Usp49, Reep4, Cyb5r3, Numa1, Wdr77, Asf1a, Mtmr3 and Cdc37) as well as of four known signal transduction components (Trim56, Sting, Tbk1 and Irf3) significantly abrogated the innate immune response (Fig. 6a–c and Supplementary Fig. 6b).
Figure 6.
Inhibition of identified regulators by RNAi or small molecules modulates the innate immune response to retroviral infection. (a) qRT-PCR analysis of Cxcl10 mRNA in Trex1−/− MEFs treated with indicated siRNAs and infected with retrovirus for 21.5 h. Known ISD-sensing pathway genes are shown in blue text; small molecule inhibitors of selected targets are labeled and marked with arrows. (b,c) qRT-PCR analysis of Ifnb1 mRNA in Trex1−/− MEFs treated with control siRNA (siCtrl), siRNA to Abcf1 (si-1) or siRNA to Cdc37 (siCdc37) and infected with retrovirus. (d,e) qRT-PCR analysis of Ifnb1 or ISG mRNA in Trex1−/− MEFs treated with vehicle alone or small molecule inhibitors (400 nM celastrol, 750 nM 17-DMAG, 500 nM BX795, 30 uM PTPN1 inhibitor or 10 nM okadaic acid) and infected with retrovirus. (f) qRT-PCR analysis of mRNA of the ISG, MX1, in TREX1 mutant (AGS patient 1 had an R114H substitution on one allele and an Asp201 insertion on the other allele, named ‘R114H,D201ins’, and AGS patient 2 had homozygous mutations that encoded proteins with R114H substitutions, called ‘R114H,R114H’) human fibroblasts and healthy control fibroblasts treated with vehicle alone or small molecule inhibitors (500 nM celastrol, 100 nM 17-DMAG and 500 nM BX795) and infected with retrovirus. Data are from one experiment representative of two (a) or three (b–f) independent experiments (mean and s.d. in a–d,f; mean in e). *P < 0.05, **P < 0.01 (Student’s t-test; after Benjamini-Hochberg correction for multiple testing in a).
Chemical inhibition of CDC37, HSP90 or TBK1 potently abrogated retroviral infection–induced Ifnb1 induction in Trex1−/− MEFs (Fig. 6d,e and Supplementary Fig. 6c); in contrast, chemical inhibition of PTPN1 or PPP6C increased ISG induction in response to retroviral infection in a dose-dependent manner (Fig. 6e and Supplementary Fig. 6c). We also tested several of these small molecules in human fibroblasts derived from two healthy individuals and from two patients with AGS that carry mutations in TREX1 (AGS patient 1: R114H substitution on one allele and a D201 insertion on the other allele, called ‘R114H/D201ins’; AGS patient 2: homozygous R114H, called ‘R114H/R114H’)25. The R114H variant is the most common mutation found in individuals with AGS and is also found in individuals with SLE6. Small-molecule targeting of CDC37 (celastrol), HSP90 (17-DMAG) and TBK1 (BX795) abrogated the induction of ISGs in the AGS fibroblasts (Fig. 6f and Supplementary Fig. 6d). Taken together, these results suggest that genetic or chemical inhibition of ISD-sensing pathway regulators we identified can modulate the innate immune response to retroviral infection in Trex1−/− MEFs and in fibroblasts from patients with AGS.
DISCUSSION
We described here the integration of complementary genomic and proteomic data sets to identify new components and physical interactions in the ISD-sensing signaling network. We generated and used DNA-protein interaction, protein-protein interaction and loss-of-function screening data sets to identify new ISD-sensing pathway components; validated several of the newly identified components, including Abcf1; demonstrated that a subset of these newly identified components have functional roles in the response to retroviral infection in Trex1−/− MEFs; and showed that small-molecule inhibitors of several of these components can modulate the innate immune response to dsDNA and retroviral infection in both mouse and human cells.
We identified SET complex members (SET, ANP32A and HMGB2) as interacting partners for ABCF1. The SET complex contains three DNA nucleases (TREX1, APEX1 and NME1), the chromatin-modifying proteins SET and ANP32A, and HMGB2, which functions as a co-receptor for nucleic acid receptors among other roles36. The observed interactions (whether direct or indirect) among dsDNA, ABCF1, HMGB2 and other SET complex members suggest that early steps in DNA recognition may occur at the ER-localized SET complex. Consistent with this hypothesis, the complex member TREX1 can prevent HIV-1 DNA detection, and its absence results in accumulation of retroelement DNA at the ER which drives an ISD-sensing response3,4,37. Furthermore, the complex members SET and NME1 also detect HIV-1 DNA and in turn regulate HIV-1 infectivity27. A recent model suggests that the SET complex may recognize viral DNA as damaged DNA, specifically via its base excision repair (BER) activity and/or its distorted structure (for example, HMGB2)38. Consistent with this model, the DNA interactors we identified included the SET and BER complex member APEX1 as well as nearly the entire BER complex (for example, PARP1, PARP2, POLB, LIG3, XRCC1, FEN1 and PCNA). Overall, these results suggest that the SET complex has a central role in DNA sensing and forms a coordinated system for detecting, modifying and degrading viral or retroelement DNA.
We also tested the impact of small-molecule inhibitors on the DNA-sensing response and found that inhibition of five components modulates the innate immune response to cytosolic DNA in human dendritic cells and to retroviral infection in TREX1 mutant fibroblasts. Targeting the two inhibitory phosphatases PTPN1 and PPP6C may serve as a way to enhance the immune response to DNA viruses and retroviruses or the immunogenicity of DNA vaccines15,39. In contrast, inhibition of CDC37, HSP90 or TBK1 may be useful in treating certain autoimmune conditions. Several of these small molecules have already been tested for other conditions in mice and humans. For example, 17-DMAG is being explored for the treatment of autoimmune disease and various cancers40, and extracts of the medicinal plant Tripterygium wilfordii, from which the natural product celastrol is derived, have been used as an anti-inflammatory41. Although current treatments for TREX1-dependent autoimmune disorders (including AGS, familial chilblain lupus and SLE) do not target the cause of these diseases, small molecules such as celastrol, 17-DMAG and BX795 that inhibit the ISD-sensing response may represent new therapeutics for this class of disorders.
Several concrete results and new lines of investigation have emerged from the integrative approach described here. First, the proteomic and functional RNA interference data sets provide a resource for immunologists to mine and identify factors involved in any aspect of the cellular response to cytosolic DNA. Second, the finding that ABCF1 interacts with the SET complex and other critical factors opens up a new direction for studies of the ISD-sensing pathway, focusing on the hypothesis that DNA degradation and sensing are coordinated. Finally, our list of putative factors provides a large target space for seeking small-molecule inhibitors, leading us to identify several compounds with potential utility in treating patients with an overactive ISD-sensing pathway, as observed for AGS and related disorders.
ONLINE METHODS
Cells, viruses and reagents
p53−/− MEFs were derived from p53−/− mice (gift from D.J. Kwiatkowski (Harvard Medical School) and D.M. Sabatini (MIT)). Trex1−/− MEFs and C57BL/6 wild-type control MEFs were a gift from T. Lindahl (London Research Institute). Ptpn1−/− MEFs and Ptpn1−/− MEFs rescued with Ptpn1 cDNA were a gift from B.G. Neel (Ontario Cancer Institute)42. Human AGS patient (TREX1 R114H/D201ins and TREX1 R114H/R114H) fibroblasts and healthy control fibroblasts were a gift from Y.J. Crow (University of Manchester). Primary murine lung fibroblasts were derived from lung tissue of 4–8 wk old female C57BL/6 mice. Mouse conventional dendritic cells (cDCs) were prepared from wild-type or B6.C3H-sst1 (Sp110 LoF) mice as previously described43,44. The 293T cells were obtained from the American Type Culture Collection. Cells were maintained in DMEM (Mediatech) with 10% FBS (Sigma). Human monocytes were isolated by negative selection (Life Technologies) from peripheral blood mononuclear cells, and differentiated into dendritic cells in GM-CSF (R&D) and IL-4 (R&D) in RPMI (Life Technologies) with 10% FBS (Life Technologies) for 7 d. Sendai virus was obtained from ATCC and used at a multiplicity of infection (MOI) of 1. HSV-1 d109 (ref 45) was obtained as a gift from N.A. DeLuca (University of Pittsburgh) and used at an MOI of 1. Self-inactivating minimal HIV-1 virus was produced in 293T cells using the vector pLX301 (TRC, Broad Institute), packaging construct psPAX2, and envelope plasmid pCMV-VSVG. ISD, HIV gag-100 and HSV60 dsDNA were annealed from oligonucleotides (IDT) as described3,12,17; sequences are listed in Supplementary Table 5a. In vitro transcribed RNA was synthesized as described46. Nucleic acids were mixed with Lipofectamine LTX (Life Technologies) at ratio of 1:3 (wt/vol) in Opti-MEM (Life Technologies), and added to cells at 1 μg/ml (DNA) or 0.1 μg/ml (RNA) unless otherwise indicated. Recombinant IFN-β was obtained from PBL Interferon Source; murine CXCL10 ELISA kit from R&D; NE-PER from Pierce; Coomassie blue (SimplyBlue SafeStain) from Life Technologies; EXPRESS35S Protein Labeling Mix from Perkin Elmer. Antibodies used were anti-p-TBK1 Ser172 (5483; Cell Signaling), anti-p-IRF3 Ser396 (4947; Cell Signaling), anti-TBK1 (3504; Cell Signaling), anti-IRF3 (4302; Cell Signaling), anti-CDC37 (4793; Cell Signaling), anti-ABCF1 (SAB2106638, Sigma), anti-HMGB2 antibody (ab67282, Abcam), anti-SET (sc25564, Santa Cruz Biotechnology), anti-IFI204 (SAB2105265, Sigma), anti-α-tubulin (T5168, Sigma), anti-β-actin (ab6276, Abcam), anti-HA (High Affinity 3F10; Roche), anti-SMARCB1 (H-300; Santa Cruz Biotechnology), anti-calreticulin (ab14234; Abcam) and rat IgG control (012-000-003 Jackson Laboratories). PTPN1 inhibitor (CAS 765317-72-4), okadaic acid and celastrol were obtained from Millipore; BX795 and 17-(Dimethylaminoethylamino)-17-demethoxygeldanamycin (17-DMAG) from Invivogen.
Plasmid construction
To create the tet-on lentiviral vector (pCW57d-P2AR), pLKO.1 (ref. 47) was modified as follows: U6 shRNA cassette was removed a tetracycline-responsive promoter element (TRE) with a multiple cloning site was inserted upstream of PGK promoter; rtTA was cloned 3′ of puroR (with 2A multicistronic cleavage site in between) with a woodchuck hepatitis virus posttranscriptional regulatory element (WPRE). To create the Abcf1-rescue construct, silent mutations were made in the siRNA to Abcf1 (si-1) targeting site using overlap extension PCR; the rescue cDNA was then cloned into pCW57d-P2AR. The Irf3-rescue construct was similarly cloned, with silent mutations made in the siRNA to Irf3 (siIrf3) targeting site. To generate the HA-tagged Abcf1 (Abcf1-HA) and HA-tagged Sting (Sting-HA) expression vectors, an HA tag–encoding sequence was added to C terminus of the cDNAs during PCR, and the constructs were cloned into pLX301. Primer sequences are listed in Supplementary Table 5b.
Identification of DNA-interacting proteins by SILAC mass spectrometry
p53−/− MEFs were grown for six cell doublings in DMEM depleted of L-arginine and L-lysine (Caisson Labs Inc.) and supplemented with 10% dialyzed FBS (Sigma) and either light MW (L; L-arginine and L-lysine), medium MW (M; L-arginine 13C6 and L-lysine D4), or heavy MW (H; L-arginine 13C6, 15N4 and L-lysine 13C6, 15N2) isotope-labeled amino acids (Sigma Isotec). L and H cells were stimulated with 1000 U/ml IFN-β for 18 h. Cells were pelleted and incubated in hypotonic lysis buffer (10 mM HEPES pH 7.4, 10 mM KCl, 1 mM EDTA; HLB) containing protease inhibitors (Roche) for 10 min on ice followed by lysis for 1 min in HLB + 0.3% Triton X-100. Nuclei and insoluble proteins were removed by centrifugation. 11 mg of H and L lysates were mixed with 1:1 mix of biotinylated DNA (‘ISD’ sequence is used throughout the study unless otherwise specificied) and DNA with a tetraethylene glycol arm between biotin and nucleic acid (IDT). No DNA was added to 11 mg of sample M. Streptavidin beads (Ultralink; Pierce) were added to all samples, which were then rotated for 2.5 h at 4 °C. Beads were pelleted and washed extensively with wash buffer (50 mM Tris-HCl pH 7.8, 150 mM NaCl, 1 mM EDTA, 0.75% NP-40, 0.175% sodium deoxycholate). All samples were mixed, cysteines were reduced by DTT and alkylated with iodoacetamide, and proteins eluted by heating in SDS sample buffer (Life Technologies) for 10 min before separation on a 4–12% gradient gel (NuPAGE; Life Technologies). Resolved proteins were divided into 13 fractions and subjected to trypsin proteolysis. Peptide extracts were cleaned up offline with C18 StageTips before 90 min nanoESI-LCMS analyses with gradient of 3–35% acetonitrile/0.1% formic acid. Protein and peptide identification and quantification was performed with MaxQuant (v1.0.12.31) using International Protein Index (IPI) mouse v3.52 as search database. Protein ratios were medians of ratios from at least two quantified peptides. Log(AH/AM) is the ratio of abundance of a protein when pulled down with DNA beads versus empty beads, and log(AH/AL) represents abundance of a protein when pulled down with DNA beads from cells stimulated with IFN-β versus from cells without IFN-β. To identify significant proteins, P values were calculated via Gaussian modeling of log(AH/AM) data, and significance threshold of P < 1 × 10−4 was used.
Identification of STING- and ABCF1-interacting proteins by SILAC mass spectrometry
p53−/− MEFs mock infected (L) or stably expressing Sting-HA (H) or Abcf1-HA (M and H) were grown in SILAC solutions (above) and then stimulated with 1,000 U/ml IFN-β for 18 h. H cells were transfected with DNA for 2.5 h. Cells were lysed in 50 mM Tris-HCl pH 7.8, 150 mM NaCl, 1 mM EDTA, 0.2% NP-40, and insoluble proteins were removed by centrifugation. We mixed 18 mg of each lysate with 1 μg/ml anti-HA antibody and Protein G beads (Pierce) and rotated the mixture for 2.5 h at 4 °C. Samples were then handled as above (with Sting-HA and Abcf1-HA cells handled separately) with following differences: peptides were analyzed with a 100 min acquisition method on a Thermo EASY-nLC 1000 UHPLC coupled to a Q Exactive mass spectrometer; MaxQuant (v1.2.2.5) and IPI mouse v3.68 were used. To identify significant interactions, P values were calculated via Gaussian modeling of the log2(AM/AL) and log2(AH/AL) data, and significance threshold of P < 0.01 was used.
RNA interference screen
750 p53−/− MEFs per well were seeded in 96-well plates in 60% DMEM and 40% Opti-MEM. siRNA (25 nM) was complexed with 0.5 μl Lipofectamine RNAiMax (Life Technologies) in Opti-MEM, incubated for 12 min and added to wells. Cells were transfected with DNA 72 h later. Supernatants were collected 26 h later, and CXCL10 was quantified by ELISA. Cell viability was estimated by CellTiter-Glo Luminescent Cell Viability Assay (Promega); CellTiter-Glo values below 3.75e5 were considered toxic. Dharmacon siGENOME SMARTpools from Harvard ICCB were used for screening. ON-TARGETplus Nontargeting Pool (Dharmacon) was used as negative control (siCtrl). Individual siRNAs are listed in Supplementary Table 5c.
cDNA rescue
p53−/− MEFs stably expressing cDNA in the pCW57d-P2AR vector were subjected to siRNA. 72 h later, doxycycline (Sigma) was added and cells were stimulated with ISD DNA.
Quantitative RT-PCR
Total RNA was extracted using RNeasy Mini kit (Qiagen). cDNA was synthesized using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time qPCR was performed using SYBR Green and LightCycler 480 system (Roche). The primers used for qPCR are listed in Supplementary Table 5d.
Co-immunoprecipitation assays
p53−/− MEFs stably expressing doxycycline-inducible Abcf1-HA were treated with 3 μg/ml doxycycline, and lysed in 10 mM Tris-HCl, 2 mM EDTA, 0.4% NP-40 with complete EDTA-free protease inhibitors (Roche). Anti-HA was cross-linked to Protein G beads (Roche) at 1 μg antibody per 20 μl beads using dimethyl pimelimidate dihydrochloride (Sigma). Cleared supernatants were incubated with antibody-bound beads, and rotated overnight at 4 °C. Rat IgG control bound to Protein G was used as IP control with 48 h doxycycline-treated lysates. Beads were washed extensively with wash buffer (10 mM Tris-HCl, 2 mM EDTA and 1% NP-40). Immunoprecipitates were eluted with 100 μl 2.5 M glycine (pH 3) and buffered to pH 7.5 by adding 25 μl of 2 M Tris. Samples were boiled in reducing Laemmli buffer for 10 min, separated by SDS-PAGE and immunoblotted.
Immunofluorescence assays
We grew 5 × 104 p53−/− MEFs stably transfected with doxycycline-inducible Abcf1-HA overnight on glass cover slips. Expression was induced by adding 3 μg/ml doxycycline for 48 h. Cells were fixed using 4% PFA for 15 min and permeabilized using PBS + 0.2% Triton X-100. Cells were treated with primary antibodies at concentration of 1/200 for 1 h, followed by treatment with fluorophore labeled secondary antibodies. Cells were mounted on glass sides using DAPI-containing Vectashield (Vector Laboratories) and visualized using 503 Platform (Intelligent Imaging Innovations) under ×40 and ×63 oil-immersion objective. Eight z stacks were taken per image at 1 μm per step; images were deconvolved using nearest neighbor algorithm.
DNA microarray analysis
(i) The 293T cells were stimulated with 1,000 U/ml IFN-β and lysed 12 h later. RNA was hybridized to Affymetrix Human U133 Plus 2.0 array by the Molecular Profiling Laboratory (MGH Center for Cancer Research). (ii) p53−/− MEFs were treated with siRNAs for 72 h and then transfected with DNA for 6 h (in biological duplicates). RNA was hybridized to Affymetrix GeneChip Mouse Gene 1.0 ST Array by Expression Analysis. (iii) Genes were curated as follows: >6.1-fold upregulation after poly(dA-dT)–poly(dT-dA) stimulation of MEFs for 4 h from GDS1773 (ref. 13); >3.9-fold upregulation after IFN-β stimulation of NIH3T3 cells for 4 h, and >6.25-fold upregulation after IFN-β stimulation of L929 cells for 4 h from GSE14413 (ref. 48); >1.3-fold upregulation after IFN-β stimulation of 293T cells from above-described arrays.
Network analysis
Network analysis was carried out using protein-protein interaction data from Ingenuity, the STRING database (http://string.embl.de), and protein-protein interactions found experimentally in published studies19,30,33,49,50 and in above-described SILAC experiments.
Statistics
Statistical significance was determined by paired Student’s t-test. P values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We are grateful to Y.J. Crow for AGS patient fibroblasts and control cells; T. Lindahl for Trex1−/− MEFs and wild type control MEFs; D. Sabatini and D. Kwiatkowski for p53−/− MEFs; N.A. DeLuca for HSV-1 d109 virus; C. Shamu (Institute of Chemistry and Cell Biology at Harvard Medical School) for the siRNA library and expert advice; The RNAi Consortium (TRC) at the Broad Institute for assistance with siRNA screening; J. Qiao for assistance with mass spectrometry; M. Rooney and C. Ye for advice about statistical analyses; D. Londono for help with microscopy; J. Kagan, N. Haining, L. Glimcher and M. Brenner for valuable discussions; and W.F. Pendergraft III and other members of the Hacohen laboratory for critical review of the manuscript. This work was supported by the US National Institutes of Health Director’s New Innovator award DP2OD002230 (N.H.). M.N.L. is supported by National Human Genome Research Institute grant HG005062 and a National Institutes of Health Medical Scientist Training Program fellowship; F.D. by a GlaxoSmithKline-Immune Disease Institute fellowship; and J.L. by NIAID grant AI102816.
Footnotes
AUTHOR CONTRIBUTIONS
M.N.L., M.R., S.O., P.M., W.L., F.D. and J.S. performed experiments and analyzed data; A.C.V. analyzed data; J.G.D., M.H.O. and I.K. provided reagents; D.M.K., J.L. and S.A.C. supervised experiments; N.H. designed and supervised the study. M.L. and N.H. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Supplementary information is available in the online version of the paper.
References
- 1.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 2.Altfeld M, Fadda L, Frleta D, Bhardwaj N. DCs and NK cells: critical effectors in the immune response to HIV-1. Nat Rev Immunol. 2011;11:176–186. doi: 10.1038/nri2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gall A, et al. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee-Kirsch MA, et al. Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 7.Lee-Kirsch MA, et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J Mol Med (Berl) 2007;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 8.Rotem Z, Cox RA, Isaacs A. Inhibition of virus multiplication by foreign nucleic acid. Nature. 1963;197:564–566. doi: 10.1038/197564a0. [DOI] [PubMed] [Google Scholar]
- 9.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 12.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 14.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanai H, et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 19.Li S, Wang L, Berman M, Kong YY, Dorf ME. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35:426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuchida T, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 23.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 24.Yan N, Cherepanov P, Daigle JE, Engelman A, Lieberman J. The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog. 2009;5:e1000327. doi: 10.1371/journal.ppat.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crow YJ, et al. Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 26.Rice GI, et al. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laguette N, Benkirane M. How SAMHD1 changes our view of viral restriction. Trends Immunol. 2012;33:26–33. doi: 10.1016/j.it.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okabe Y, Sano T, Nagata S. Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature. 2009;460:520–524. doi: 10.1038/nature08138. [DOI] [PubMed] [Google Scholar]
- 29.Paytubi S, et al. ABC50 promotes translation initiation in mammalian cells. J Biol Chem. 2009;284:24061–24073. doi: 10.1074/jbc.M109.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozenblatt-Rosen O, et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang T, et al. A novel Hsp90 inhibitor to disrupt Hsp90/Cdc37 complex against pancreatic cancer cells. Mol Cancer Ther. 2008;7:162–170. doi: 10.1158/1535-7163.MCT-07-0484. [DOI] [PubMed] [Google Scholar]
- 32.Gray PJ, Jr, Prince T, Cheng J, Stevenson MA, Calderwood SK. Targeting the oncogene and kinome chaperone CDC37. Nat Rev Cancer. 2008;8:491–495. doi: 10.1038/nrc2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouwmeester T, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 34.Myers MP, et al. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem. 2001;276:47771–47774. doi: 10.1074/jbc.C100583200. [DOI] [PubMed] [Google Scholar]
- 35.Pan H, et al. Ipr1 gene mediates innate immunity to tuberculosis. Nature. 2005;434:767–772. doi: 10.1038/nature03419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 38.Yan N, O’Day E, Wheeler LA, Engelman A, Lieberman J. HIV DNA is heavily uracilated, which protects it from autointegration. Proc Natl Acad Sci USA. 2011;108:9244–9249. doi: 10.1073/pnas.1102943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 40.Shimp SK, III, et al. HSP90 inhibition by 17-DMAG reduces inflammation in J774 macrophages through suppression of Akt and nuclear factor-kappaB pathways. Inflamm Res. 2012;61:521–533. doi: 10.1007/s00011-012-0442-x. [DOI] [PubMed] [Google Scholar]
- 41.Corson TW, Crews CM. Molecular understanding and modern application of traditional medicines: triumphs and trials. Cell. 2007;130:769–774. doi: 10.1016/j.cell.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klaman LD, et al. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol Cell Biol. 2000;20:5479–5489. doi: 10.1128/mcb.20.15.5479-5489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan H, et al. Ipr1 gene mediates innate immunity to tuberculosis. Nature. 2005;434:767–772. doi: 10.1038/nature03419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amit I, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samaniego LA, Neiderhiser L, DeLuca NA. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 47.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 48.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 49.Pichlmair A, et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487:486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- 50.Cristea IM, et al. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol. 2010;84:7803–7814. doi: 10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.