Abstract
In spite of intense interest in how altered epigenetic processes including DNA methylation may contribute to psychiatric and neurodevelopmental disorders, there is a limited understanding of how methylation processes change during early postnatal brain development. The present study used in situ hybridization to assess mRNA expression for the three major DNA methyltranserases (DNMTs) – DNMT1, DNMT3a and DNMT3b – in the developing rat brain at seven developmental timepoints: postnatal days (P) 1, 4, 7, 10, 14, 21, and 75. We also assessed 5-methylcytosine levels (an indicator of global DNA methylation) in selected brain regions during the first three postnatal weeks. DNMT1, DNMT3a and DNMT3b mRNAs are widely expressed throughout the adult and postnatal developing rat brain. Overall, DNMT mRNA levels reached their highest point in the first week of life and gradually decreased over the first three postnatal weeks within the hippocampus, amygdala, striatum, cingulate and lateral septum. Global DNA methylation levels did not follow this developmental pattern; methylation levels gradually increased over the first three postnatal weeks in the hippocampus, and remained stable in the developing amygdala and prefrontal cortex. Our results contribute to a growing understanding of how DNA methylation markers unfold in the developing brain, and highlight how these developmental processes may differ within distinct brain regions.
Keywords: DNA methyltransferase, DNA methylation, In situ hybridization, Neurodevelopment, Hippocampus, Amygdala
1. Introduction
Epigenetic research in neuroscience has exploded in recent years, with intense efforts aimed to determine whether epigenetic dysfunction plays a role in the neurobiology of psychiatric disease. Indeed, a growing number of studies have revealed epigenetic abnormalities in schizophrenia, depression (Akbarian, 2010; Burghardt et al., 2012; Carrard et al., 2011; Sun et al., 2013; Tsankova et al., 2007), autism, and comorbid developmental disorders like Fragile X and Rett Syndrome (Grafodatskaya et al., 2010; Miyake et al., 2012). However, there is much to learn regarding how epigenetic mechanisms dictate and influence brain function, either in health or disease.
DNA methylation – one of the most prominent and well-studied epigenetic processes – regulates the quantity, location, and timing of gene expression throughout life (Fagiolini et al., 2009; Mill and Petronis, 2007; Nelson and Monteggia, 2011). DNA methylation levels undergo drastic developmental changes (Cantone and Fisher, 2013; Okano et al., 1999; Smith and Meissner, 2013). These changes are required for normal development, and dysregulation can lead to imprinting disorders such as Beckwith–Wiedemann and Prader–Willi/Angelman syndromes or cancer (Paulsen and Ferguson-Smith, 2001). DNA methylation – the covalent addition of a methyl group to the 5′ position of cytosine residues – was traditionally thought to exclusively block gene transcription; however, recent studies suggest that it can both increase and decrease gene transcription depending upon the gene (Chen and Riggs, 2011). The process is catalyzed by DNA methyltransferases (DNMTs), a family of enzymes that includes DNMT1, DNMT3a, and DNMT3b.
There is a limited understanding of how enzymes involved in methylation change in the normal postnatal developing brain. Many previous studies focused on epigenetic modifications during embryonic development (Fan et al., 2001;Hirabayashi et al., 2013; Liang et al., 2011; Monk et al., 1987;Okano et al., 1999; Paulsen and Ferguson-Smith, 2001) or adult learning and memory processes (Feng et al., 2007; Liu et al., 2009; Lubin et al., 2008; Sweatt and Miller, 2007). Studies in developing human cortical tissue revealed dramatic shifts in cortical gene expression patterns between fetal and early postnatal life (Colantuoni et al., 2011). Not surprisingly, DNA methylation in the human (Numata et al., 2012; Siegmund et al., 2007) and mouse (Lister et al., 2013) neocortex also dynamically shifts throughout the lifespan. Epigenetic processes likely provide key regulatory mechanisms to guide these and other neurodevelopmental processes, and perturbation of epigenetic mechanisms at pivotal developmental timepoints likely evoke long-term neural and behavioral consequences. Studies in the developing mouse brain highlight dynamic changes in DNMT expression in both the embryonic and postnatal brain (Feng et al., 2005), and have found that DNMTs are differentially expressed within different cell types, with particular enrichment in GABAergic neurons (Kadriu et al., 2012).
Because there is still much to learn regarding dynamic DNA methylation changes that occur in early postnatal brain development, the present study assessed developmental expression of DNA methylation markers in developing rat brain. We assessed mRNA expression of three major DNMT enzymes (DNMT1, −3a, and −3b) and global DNA methylation (5-methylcytosine) levels in multiple brain regions across several developmental timepoints. We hypothesized that there would be a specific developmental patterning of DNA methylation markers that would vary over time, and perhaps vary across brain areas.
2. Results
2.1. Dnmt1, DNMT3a and DNMT3b mRNA expression in the adult rat brain
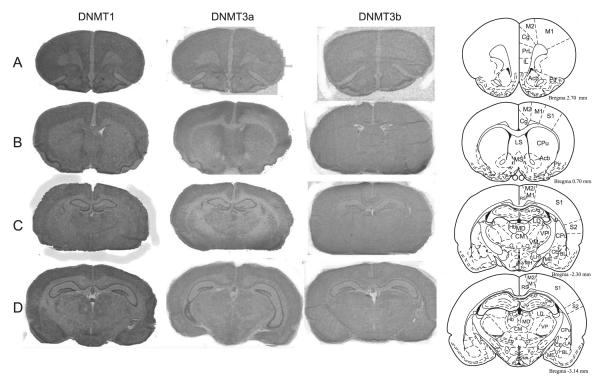
We first examined expression of DNMT1, DNMT3a, and DNMT3b mRNA in the adult rat brain (Fig. 1). DNMT1, −3a and −3b mRNAs were widely distributed throughout the adult brain, showing differential regional distribution (Fig. 1A–E; abbreviations as defined in Table 1). DNMT1 mRNA was highly expressed in the hippocampus (CA1–3 and dentate gyrus (DG) relative to other brain regions; Fig. 1C–E), as well as ventromedial hypothalamus (VMH), and habenula (Hb) (Fig. 1E). It was also present throughout the adult rat forebrain, including the cingulate (Cg), lateral septum (LS), nucleus accumbens (Acb), caudate putamen (Cpu), and several nuclei of the amygdala: the central (CE), medial (ME), lateral (LA) and basolateral (BL) nuclei (Fig. 1A–E). DNMT3a mRNA was also highly expressed in the hippocampus (CA1–3 and DG; Fig. 1C–E), moderately expressed in several regions of the forebrain, and expressed at very low levels in the amygdala (Fig. 1A–E) relative to other regions examined. Due to the extremely low levels apparent in the amygdala, we were unable to reliably identify the four amygdalar nuclei of interest, and therefore were unable to complete quantification there. DNMT3b was expressed at very low levels in the hippocampus (CA1–3 and DG; Fig. 1C–E) and hypothalamus relative to other regions and was unable to be detected in other brain regions including the amygdala and forebrain regions.
Fig. 1.
DNMT1, DNMT3a and DNMT3b mRNA expression in the adult rat brain. Columns show autoradiograms from representative tissue sections through the adult rat brain processed by in situ hybridization with antisense probes against rat DNMT1 (left), DNMT3a (middle) and DNMT3b (right) mRNA. X-ray films were exposed for 8 days for DNMT1, 3 days for DNMT3a, and 17 days for DNMT3b. Images show examples of coronal sections through the adult Sprague–Dawley rat brain, depicting regions such as the hippocampus, amygdala, caudate putamen, and hypothalamus. The diagrams in the far right column (adapted from Paxinos and Watson, 1997) indicate anatomical landmarks for each section. Abbreviations can be found in Table 1.
Table 1.
Abbreviations.
| A | anterior nucleus of thalamus |
| ac | anterior commisure |
| Acb | nucleus accumbens |
| AHA | anterior hypothalamic area |
| Arc | arcuate nucleus |
| BLA | basolateral amygdala |
| BM | basomedial nucleus of amygdala |
| BS | brainstem |
| BST | bed nucleus of the stria terminalis |
| CA1 | hippocampus, Cornu Ammonis 1 field of Ammon’s horn |
| CA2 | hippocampus, Cornu Ammonis 2 field of Ammon’s horn |
| CA3 | hippocampus, Cornu Ammonis 3 field of Ammon’s horn |
| Ce | central amygdaloid nucleus |
| Cg | cingulate |
| ChPlx | choriod plexus |
| CM | centromedial nucleus of thalamus |
| Co | cortical amygdaloid nucleus |
| Cpu | caudate putamen |
| DG | dentate gyrus |
| DLG | dorsal lateral geniculate nucleus |
| DMH | dorsomedial hypothalamus |
| DR | dorsal raphe |
| Dtt | dorsal tenia tecta |
| Ent | entorhinal cortex |
| GP | globus pallidus |
| Hb | habenula |
| HPC | hippocampus |
| Hyp | hypothalamus |
| ICjM | islands of Calleja, major island |
| IG | indusium griseum |
| LA | lateroanterior hypothalamic nucleus |
| La | lateral amygdaloid nucleus |
| LD | lateral dorsal nucleus of thalamus |
| LH | lateral hypothalamus |
| LS | lateral septum |
| M1 | primary motor cortex |
| M2 | secondary motor cortex |
| MB | midbrain |
| MD | mediodorsal nucleus of thalamus |
| Me | medial amygdaloid nucleus |
| MG | medial geniculate nucleus |
| MnPo | median preoptic nucleus |
| MPA | medial preoptic area |
| MR | median raphe |
| MS | medial septum |
| PAG | periaquaductral gray |
| Pin | pineal gland |
| Pir | piriform cortex |
| Pn | pontine nuclei |
| Po | posterior nucleus of thalamus |
| PrL | prelimbic cortex |
| PV | paraventricular nucleus of thalamus |
| PVN | paraventricular nucleus of hypothalamus |
| PVP | paraventricular nucleus of thalamus, posterior part |
| Re | reuniens nucleus of thalamus |
| Rh | rhomboid nucleus of thalamus |
| Rt | reticular nucleus of thalamus |
| S | subiculum |
| SC | superior colliculus |
| SCN | suprachiasmatic nucleus |
| sm | stria medullaris of the thalamus |
| SN | substantia nigra |
| SON | supraoptic nucleus |
| TegN | tegmental nucleus |
| VM | ventromedial nucleus of thalamus |
| VMH | ventromedial hypothalamus |
| VP | ventral posterior nucleus of thalamus |
| ZI | zona incerta |
2.2. Developmental expression of DNMT1, DNMT3a and DNMT3b mRNA
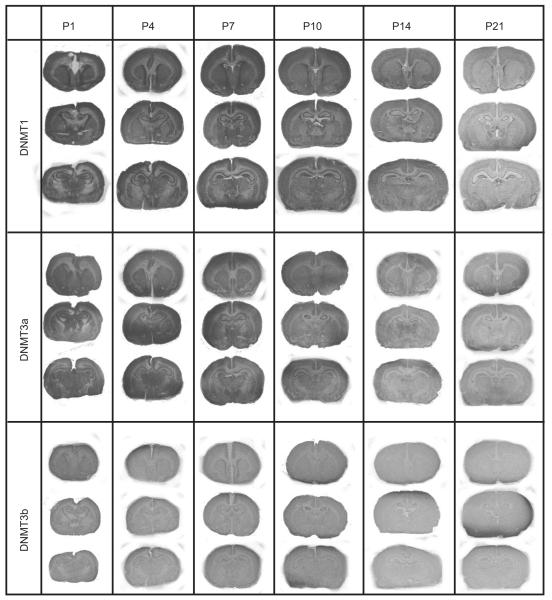
We also examined the pattern of DNMT1, DNMT3a, and DNMT3b mRNA expression in the developing rat brain (P1, P4, P7, P10, P14, and P21). Visual inspection of the autoradio-graphic films exposed to in situ hybridization-processed slides indicates a decrement of DNMT1 mRNA levels over time with relatively higher levels at P1 compared to P21 (top row, Fig. 2). This same pattern can be seen with DNMT3a (middle row, Fig. 2), but is not apparent with DNMT3b (bottom row, Fig. 2). We selected several representative brain areas – the hippocampus, amygdala, cingulate, caudate putamen, nucleus accumbens, and lateral septum – for quantitative analysis to determine developmental patterns of DNMT1, DNMT3a, and DNMT3b mRNA expression in the developing and adult rat brain (Fig. 3).
Fig. 2.
DNMT1, DNMT3a and DNMT3b mRNA expression in the developing rat brain. Columns show autoradiograms from representative tissue sections through the developmental rat brain at postnatal day (P)1, P4, P7, P10, P14, and P21 that were processed by in situ hybridization with antisense probe against rat DNMT1 (top column), DNMT3a (middle column) and DNMT3b (bottom column) mRNA. X-ray films were exposed for 8 days for DNMT1, 3 days for DNMT3a, and 17 days for DNMT3b.
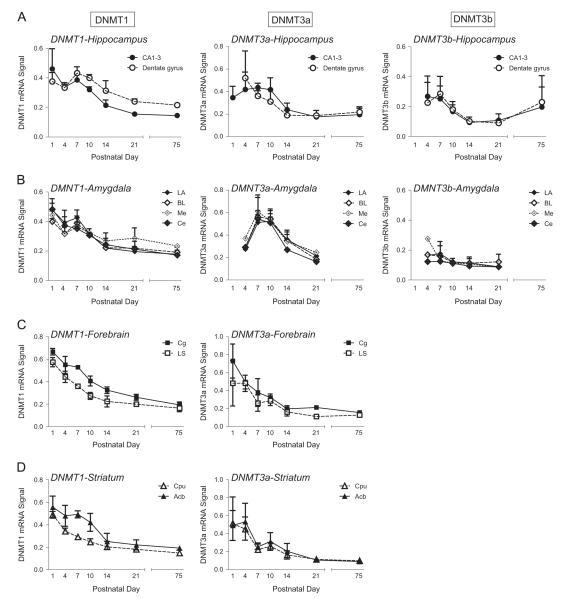
Fig. 3.
Developmental patterns of DNMT1, DNMT3a and DNMT3b mRNA expression in the early rodent postnatal period and adulthood. DNA methyltransferase (DNMT)1, DNMT3a, and DNMT3b mRNA expression levels were quantified in several regions of rat brains collected at 6 developmental timepoints – postnatal day (P)1, P4, P7, P10, P14, P21, and P75 (adulthood). (A) Developmental expression pattern of DNMT1, DNMT3a, and DNTM3b in subregions of the hippocampus (CA1–3 and dentate gyrus). (B) Developmental expression pattern of DNMT1, DNMT3a, and DNMT3b in subnuclei of the amygdala (lateral, basolateral, medial and central nucleus). (C) Developmental expression pattern of DNMT1 and DNMT3a in several forebrain regions, including cingulate (Cg) and lateral septum (LS). Due to low abundance, DNMT3b was unable to be quantified in these regions. (D) Developmental expression pattern of DNMT1 and DNMT3a in the striatum (caudate putamen (Cpu) and nucleus accumbens (Acb)). Due to low abundance, DNMT3b was unable to be quantified in these regions.
Fig. 3 shows relative DNMT1 (left column), DNMT3a (middle column) and DNMT3b (right column) mRNA expression by specific brain region in the developing rat brain. DNMT1 mRNA expression was quantified in 4 subregions of the hippocampus – CA1, CA2, CA3, and dentate gyrus (Fig. 3A). The two-way ANOVA showed a main effect of subregion (F1,72=11.469, p<0.001), with approximately 13% higher DNMT1 levels in the dentate gyrus compared to CA regions. There was also a main effect of developmental age (F5,72=41.898, p<0.0001), since DNMT1 levels declined with age (e.g., 66% decrease from P1 to P21 in CA regions and 36% reduction in the dentate gyrus). Post hoc comparisons showed that DNMT1 mRNA levels were higher at P1, P4, P7 and P10 compared to P14, P21 and P75 (p<0.05 for all comparisons). Finally, there was a subregion × age interaction (F5,72=5.465, p<0.0001); post hoc analysis showed that DNMT1 levels were specifically higher in the dentate gyrus versus CA regions at P10, P14, P21, and P75.
DNMT1 transcript expression was also quantified in the developing amygdala (Fig. 3B). The two-way ANOVA in the amygdala showed a main effect of age (F3,36=35.715, p<0.001), where again, DNMT1 levels generally declined with age (e.g., 49% decrease from P1 to P21 across the nuclei examined). Post hoc comparisons showed that DNMT1 mRNA levels were higher at P1, P4, and P7 compared to P14, P21 and P75 (p<0.05 for all comparisons). There was no effect of nucleus and no age × nucleus interaction on DNMT1 expression in the amygdala.
We also examined developmental expression of DNMT1 in two forebrain regions – the cingulate and lateral septum. Here we found a main effect of region (F5,18=62.709, p<0.0001) with 24% higher DNMT1 levels in the cingulate versus septum. There was also a main effect of age (F5,18=73.353, p<0.0001) as DNMT levels declined with increasing age (e.g., 61% decrease from P1 to P21 in the cingulate and 65% reduction in the septum). Post hoc comparisons showed that DNMT1 mRNA levels were higher at P1, P4, and P7 compared to P10, P14, P21 and P75 (p<0.05 for all comparisons). There was no region × age interaction (Fig. 3C).
Lastly, DNMT1 mRNA expression was quantified in the developing dorsal (caudate putamen) and ventral (nucleus accumbens) striatum (Fig. 3D). The two-way ANOVA showed a main effect of age (F5,19=24.462, p<0.0001) with declining DNMT1 levels with increasing age (e.g., 60% decrease from P1 to P21 in the caudate putamen and 63% reduction in the nucleus accumbens). Post hoc comparisons showed that DNMT1 mRNA levels were higher at P1, P4, and P7 compared to P14, P21 and P75 (p<0.05 for all comparisons). There was also a main effect of region (F5,19=19.391, p<0.0001) with generally higher DNMT1 levels in the accumbens versus caudate; there was no age × region interaction.
DNMT3a mRNA expression was quantified in 4 subregions of the hippocampus – CA1, CA2, CA3, and dentate gyrus (Fig. 3A). The two-way ANOVA showed a main effect developmental age (F5,66=14.089, p<0.0001) with steadily decreasing levels with increasing age (e.g., 49% decrease from P1 to P21 in the CA regions and 64% reduction in the dentate gyrus). Post hoc comparisons showed that DNMT3a mRNA levels were higher at P1, P4, and P7 compared to P14, P21 and P75 (p<0.05 for all comparisons). There was no effect of region, and no age × region interaction. DNMT3a transcript expression was also quantified in 4 nuclei of the amygdala (lateral, basolateral, medial, and central nuclei; Fig. 3B). The two-way ANOVA showed a main effect of age (F3,24=32.203, p<0.001), with DNMT3a levels decreasing with age (e.g., 34% decrease from P1 to P21 across the nuclei examined). Post hoc comparisons showed that DNMT3a mRNA levels were higher at P7 and P10 compared to P14 and P21 (p<0.05 for all comparisons). There was no effect of nucleus, and no age × region interaction. We compared DNMT3a levels in the developing cingulate and lateral septum. Here we found a main effect of age (F5,16=8.894, p<0.0001), with DNMT3a levels decreasing with age (e.g., 71% decrease from P1 to P21 in the cingulate and 77% reduction in the septum). Post hoc comparisons showed that DNMT3a mRNA levels were higher at P1, P4, and P7 compared to P14, P21 and P75 (p<0.05 for all comparisons). There was no main effect of region and no region × age interaction (Fig. 3C). Lastly DNMT3a mRNA expression was quantified in the developing dorsal (caudate putamen) and ventral (nucleus accumbens) striatum (Fig. 3D). The two-way ANOVA showed a main effect of age (F5,16=5.205, p<0.005), with DNMT3a levels decreasing with age (e.g., 78% decrease from P1 to P21 in both the caudate putamen and nucleus accumbens). Post hoc comparisons showed that DNMT3a mRNA levels were higher at P1 and P4 compared to P14, P21 and P75 (p<0.05 for all comparisons). There was no main effect of region and no age × region interaction.
DNMT3b mRNA expression was quantified in 4 subregions of the hippocampus – CA1, CA2, CA3, and dentate gyrus (Fig. 3A). The two-way ANOVA showed a main effect of age (F5,60=13.931, p<0.0001), with DNMT3b levels decreasing from early to later postnatal ages (e.g., 60% decrease from P1 to P21 in both the CA regions and dentate gyrus). Post hoc comparisons showed that DNMT3b levels were higher at P4 and P7 compared to P10, P14, and P21 (p<0.05). There was no effect of region, and no age × region interaction. DNMT3b expression was also quantified in four nuclei of the amygdala (detailed above; Fig. 3B). There was no effect of age or nucleus, and no age × nucleus interaction. Due to very low abundance, DNMT3b mRNA was not quantified in the fore-brain regions (cingulate and lateral septum) or striatum at any developmental age.
2.3. Global DNA methylation patterns in the developing rat brain
We next assessed global DNA methylation levels in select brain regions of interest by measuring amounts of 5-methylcytosine in our samples. Tissue punches from the hippocampus, amygdala and prefrontal cortex were collected from rat brains at 6 developmental ages: P1, P4, P7, P10, P14 and P21 (Fig. 4). The two-way ANOVA showed a main effect of age (F5,108=2.883, p<0.018) and a main effect of region (F2,108=15.260, p<0.001) and no age × region interaction. Global DNA methylation levels increased with age, although this effect was carried by changes in the developing hippocampus with minimal developmental changes found in the amygdala and prefrontal cortex. Global DNA methylation levels in the hippocampus were relatively high at P1, significantly dropped by about 25% from P4–14 (p<0.05 for P1 versus P4, P7, P10 and P14), and then rebounded to its highest level at P21 (p<0.05 P21 versus all other ages). For the comparison between brain regions, post hoc test showed that there was higher global DNA methylation in the hippocampus compared to the amygdala (p<0.0001) and pre-frontal cortex (p<0.0001). There was no significant interaction between age and brain region.
Fig. 4.
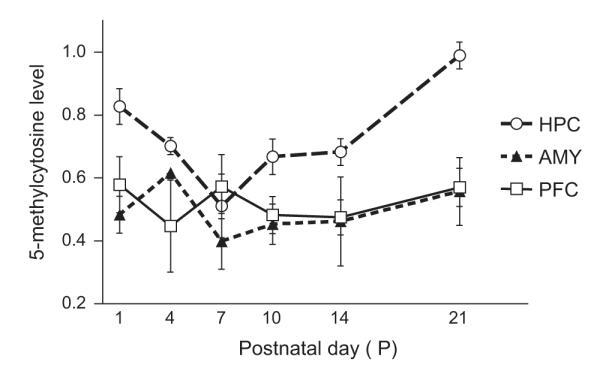
Patterns of global DNA methylation in select brain regions of the developing rat brain. Global DNA methylation levels (approximated by measuring levels of 5-methylcytosine) were measured at several postnatal time periods – postnatal day (P)1, P4, P7, P10, P14 and P21 in tissue punches from the hippocampus, amygdala, and prefrontal cortex. Global DNA methylation levels appeared to increase, although this effect is most apparent in the developing hippocampus. Also, when comparing global DNA methylation levels across the three brain regions, levels were significantly higher in hippocampus compared to amygdala and prefrontal cortex.
3. Discussion
The first three postnatal weeks represent a critical developmental period for rodent neural systems, evident in radical changes in behavior, neuroendocrine response, synaptic connectivity, and global neural gene expression (Eilam and Golani, 1988; Sapolsky and Meaney, 1986; Stead et al., 2006b). Dynamic gene expression and methylation changes occur in the developing brain across species (Feng et al., 2005; Han et al., 2009; Kang et al., 2011; Liang et al., 2011; Lister et al., 2013; Numata et al., 2012; Stead et al., 2006a), which likely drive numerous downstream effects on neurogenesis, synaptogenesis, neural circuit formation, and behavior. The present study examined how patterns of DNMT mRNAs unfold in the postnatal developing rat brain. We show that transcripts of DNMT1, DNMT3a and DNMT3b, three chief enzymes that catalyze DNA methylation, are widely expressed throughout the adult and postnatal developing rat brain. We found that DNMT1, DNMT3a, and DNMT3b mRNA levels reached their highest point in the first week of life and gradually decreased over the first three postnatal weeks within the hippocampus, amygdala, striatum, cingulate and lateral septum. Global DNA methylation levels did not follow this developmental pattern; methylation levels gradually increased over the first three postnatal weeks in the hippocampus, and remained stable in the developing amygdala and prefrontal cortex.
3.1. DNMT mRNA expression in the adult and developing brain
A handful of previous studies examined DNMT expression in the postnatal developing brain (Brown et al., 2008; Feng et al., 2005; Robertson et al., 1999). A study in normal human brain tissue showed higher DNMT1 levels compared to DNMT3a and DNMT3b in fetal tissue, although the study did not distinguish specific brain regions (Robertson et al., 1999). A study in rats reported relative DNMT mRNA levels in the adult rat hippocampus, showing that DNMT1 levels were highest in CA1 and dentate gyrus compared to CA2/3. DNMT3a levels were also highest in the DG while DNMT3b mRNA was highest in CA1 relative to other hippocampal subregions (Brown et al., 2008). Another study in the mouse cortex showed that DNMT3a mRNA levels were significantly higher than DNMT3b levels (Feng et al., 2005). This study also found that DNMT3b mRNA was particularly abundant during early neurogenesis, with levels dramatically dropping with increasing age. DNMT3a expression was high during the first two postnatal weeks, but its levels dropped precipitously by adulthood (Feng et al., 2005). The Allen Brain Atlas (http://www.brain-map.org/) also depicts DNMT-1, DNMT3a, and DNMT3b mRNA expression in the adult as well as developing mouse brain. Three of their chosen developmental time-points coincide with ones used in our study: P4, P14, and P56 (adulthood). Although they do not quantify gene expression within specific brain regions, heat-maps are provided to illustrate general abundance across ages. Based on the heat-maps, DNMT1 appears to be most abundant followed by DNMT3a and DNMT3b. It is interesting to note that DNMT3a mRNA levels in the developing mouse brain appear to transiently increase from P4 to P14, then drop back to a lower level by P28. This resembles our findings in the developing amygdala where DNMT3a levels appeared to increase around P7–10 relative to timepoints before and after that period (Fig. 3b; middle panel). Unfortunately due to the variability in the data and relatively low sample sizes, we were not able to detect statistically significant differences between the earliest time points (i.e. postnatal day 4 versus postnatal days 7 and 10). It would be useful to follow-up this finding with future experiments to (1) expand samples sizes and/or try another technique such as RT-PCR to confirm the expression pattern of DNMT3a mRNA, and (2) explore the functional implications of such patterning.
Overall, our current results are consistent with previous work demonstrating that all three DNMTs are most highly expressed in early life and levels drop significantly by adulthood in most brain regions. The current study examines the expression of full-length transcripts for DNMT1, DNMT3a, and DNMT3b, although each enzyme has alternative splice variants (Deng and Szyf, 1998; Golding and Westhusin, 2003;Liu et al., 2003). For example, rat DNMT1 has 7 alternate splice variants that are differentially expressed in various tissues, although the biological function of these variants is still unclear (Deng and Szyf, 1998). It would be useful if future studies examined developmental expression of DNMT splice variants that have important biological significance. Further, it should be noted that there is an additional DNMT that was not analyzed here – DNMT3L. DNMT3L is a DNMT that lacks methyltransferase activity due to absence of amino acid residues necessary for this function (Guenatri et al., 2013;Hata et al., 2002; Hu et al., 2008; Ooi et al., 2007; Suetake et al., 2004; Suetake et al., 2006). Future work could examine DNMT3L developmental expression and compare its distribution with that of DNMT3a.
3.2. Discord between patterns of DNMT mRNA expression and global DNA methylation levels
The discrepancy between developmental patterns of DNMT1/3a/3b mRNA expression and global DNA methylation was somewhat surprising. Overall DNMT mRNA expression decreased with age in all brain regions examined while global DNA methylation levels increased in the hippocampus and remained stable in the amygdala and prefrontal cortex over time. Few studies have directly examined the relationship between DNMT mRNA expression and global methylation levels (Numata et al., 2012), although it has been assumed that DNMT mRNA and protein levels correlate with DNA methylation levels (Brown et al., 2008). Several possible factors may account for the apparent disconnect between DNMT mRNA expression and global methylation levels reported here. For example, while DNMTs methylate DNA, other epigenetic processes such as histone deacetylases stabilize and maintain methylation levels; therefore, DNMT mRNA levels may fluctuate, but these other factors could maintain methylation levels (Fuks, 2005). It is also important to consider that a vast amount of methylation in the genome occurs within repetitive DNA elements that are widespread and typically heavily methylated (Yang et al., 2004). Thus, developmental fluctuations in methylation levels (as seen in the present study) may not necessarily occur within genomic regions associated with regulating gene expression. Interestingly, methylation has been shown to occur both at cytosine–guanine (CG) sites as well non-CG sites (e.g., cytosine–adenine, cytosine–cytosine, or cytosine–thymine), particularly in brain tissue (Stadler et al., 2011; Varley et al., 2013; Xie et al., 2012). A recent paper revealed that non-CG DNA methylation accumulates during early postnatal brain development (Lister et al., 2013). Neurons were particularly enriched in methylated non-CGs (relative to glia), and methylated non-CG levels accumulated most rapidly during early stages of synaptogenesis (postnatal days 14–28 in mouse) (Lister et al., 2013). It is possible that our observation of increased global DNA methylation in the developing hippocampus stems from a similar accumulation of methylated non-CG levels.
3.3. Summary
There is a limited understanding of how DNA methylation processes change during the course of early postnatal brain development. Characterizing developmental expression patterns of DNA methylation-related genes (e.g., the DNMTs) and downstream methylation markers (e.g., global DNA methylation, hydroxymethylation levels, gene-specific methylation) can help shed light on the important roles they play during neurodevelopment. Our results contribute to a growing understanding of how DNA methylation markers unfold in the developing brain, and highlight how these developmental processes may differ within distinct brain regions.
4. Experimental procedures
4.1. Animals and tissue preparation
Timed-pregnant Sprague–Dawley female rats (n=6, Charles River, Wilmington, MA) were delivered to our animal facility at gestational day 15. Animals were housed in standard laboratory cages with free access to food (Harlan 2014) and water, and were maintained on a 12:12 light:dark cycle, with lights on at 6:00 am. At birth (postnatal day (P)0), litters were culled to 6 male/6 female pups to control for litter size and sex composition, which can impact maternal behavior (Agnish and Keller, 1997). Maternal care can powerfully influence offspring’s physiology, neurodevelopment, and behavior (Levine et al., 1957; Meaney and Szyf, 2005;Sanchez et al., 2001; Weaver et al., 2004), so we balanced litters by size and sex, even though only males were studied. Male offspring from several developmental timepoints (P1, P4, P7, P10, P14, P21 and P75; n=9–13/timepoint) were sacrificed by rapid decapitation. Brains were removed, snap frozen, and stored at −80 °C. We took only 1–2 male offspring from a given litter at a given timepoint to ensure that multiple litters were represented in all samples. Procedures were conducted in accordance with the National Institutes of Health (NIH) guidelines on laboratory animal use and care, using a protocol approved by the University of Alabama-Birmingham Committee on Use and Care of Animals.
For the in situ hybridization study, we prepared n=3 brains from each per timepoint with each brain coming from a separate litter. Our previous work as well as other similar studies describing anatomical distribution of mRNAs in developing and adult brain indicated that an n=3 would be sufficient to determine the developmental expression patterns of the DNMTs (Beneyto and Meador-Woodruff, 2004;Broide et al., 2007; Clinton and Meador-Woodruff, 2002). Brains were cryostat sectioned at 12 μm and thaw-mounted on Fisherbrand Superfrost Plus Microscope slides (Fisher Scientific, http://www.fishersci.com/). Tissue was processed by in situ hybridization to assess DNMT1, DNMT3a and DNMT3b mRNA expression at 240 μm intervals throughout the brain (described below).
The remaining brains (n=6–10 per timepoint) were used for the Global DNA Methylation Assays to measure 5-methylcytosine – an indicator of methylated DNA (procedure described below). Preliminary studies indicated that an n=7–10 worked best for these assays based on variability in the data. Brains tissue punches were collected in a manner similar to Datson and colleagues (Datson et al., 1999). Briefly, brains were sectioned in a cryostat between −10 and −12 °C, with alternating sections of 20 μm and 300 μm collected. The 20 μm sections were stained with cresyl violet and compared to developing and adult rat brain atlases (Paxinos and Watson, 1986; Paxinos et al., 1994b) to identify appropriate anatomical regions in the 300 μm sections. The prefrontal cortex, hippocampus, and amygdala were removed from thick sections using a 0.5 mm tissue punch (Harris Micro-Punch, Ted Pella, Redding, CA), stored at −20 °C, and later homogenized. Total DNA was extracted (Yu et al., 2013) for subsequent global DNA methylation analysis (described below).
4.2. In situ hybridization
To generate subclones for riboprobe synthesis, we used PCR to amplify unique regions of rat DNMT1 (NCBI GenBank accession #D64060 – a 490 nucleotide fragment spanning nucleotides 183–673), DNMT3a (NCBI GenBank accession #NM001003958 – a 657 nucleotide fragment spanning nucleotides 1428–2084), and DNMT3b (NCBI GenBank accession #NM001003959 – a 573 nucleotide fragment spanning nucleotides 3423–3995) from a rat brain cDNA brain library (Edge-Biosystems, Gaithersburg, MD). Amplified cDNA segments were extracted (QIAquick Gel Extraction Kit, Qiagen, Valencia, CA), subcloned into a Bluescript vector, and confirmed by nucleotide sequencing.
The antisense strand DNMT1/DNMT3a/DNMT3b riboprobes were labeled in separate reaction mixtures and purified as previously described (Simmons et al., 2012). The hybridizations were run in three separate batches: (1) all slides representing all developmental ages were processed in parallel for DNMT1; (2) all slides representing all ages were run for DNMT3a; and finally (3) all slides representing all ages were run for DNMT3b.
Tissue sections from rat brains at all ages (P1, 4, 7, 10, 14, 21, and adult) for a given experiment (DNMT1, DNMT3a, or DNMT3b) were fixed and hybridized with riboprobes as previously described (Simmons et al., 2012). After this processing, slides were dried and apposed to Kodak XAR film (Eastman Kodak, Rochester, NY). For DNMT1, slides were exposed to film for 8 days (hippocampus and forebrain analysis) and 7 weeks (amygdala analysis). For DNMT3a, slides were exposed to film for 3 days (hippocampus and forebrain analysis) and 7 days (amygdala analysis). For DNMT3b, slides were exposed to film for 17 days (hippocampus and forebrain analysis) and 8 weeks (amygdala analysis). These film development times were chosen because they produced images within the linear portion of the development curve for each probes for the brain regions of interest across developmental ages. After the appropriate exposure period for each probe, autoradiograms were developed and digitized using a ScanMaker 1000XL Pro flatbed scanner (Microtek, Carson, CA) with LaserSoft Imaging software (AG, Kiel, Germany) at 1,600 dpi.
4.3. Global DNA methylation quantification
DNA was extracted in a manner similar to Bettscheider and colleagues (Yu et al., 2013) using a modified DNA column based extraction; DNA quantity and quality were then assessed using Thermo Scientific NanoDrop 2000 (seeking a 260/280 value around 1.8). For global DNA methylation quantification, the Epigentek Methylflash Methylated DNA Quantification Kit (Colorimetric) was used according to manufacture instructions. In this assay, DNA is bound to strip wells; the methylated fraction is detected using capture and detection antibodies and then quantified by measuring absorbance via microplate reader (Molecular Devices M3, Sunnyvale, CA). The Epigentek Methylflash Methylated DNA Quantification Kit utilizes optimized antibodies specific to 5-methylcytosine with no cross reactivity to unmethylated cytosine and negligible cross-reactivity to hydroxymethylcytosine.
4.4. Data analysis
In situ hybridization results were visualized on autoradiograhphic film. Macroscopic images were compared to plates and diagrams from atlases of the adult and developing rat brain (Paxinos et al., 1994a; Paxinos, 2004) for identification of structures. Digitized images were analyzed using ImageJ Analysis Software for PC from NIH. Optical density measurements relative to background (signal taken from white matter on each tissue section) were obtained for regions of interest in each animal at each timepoint. Specific signal, defined as 3.5 × the standard deviation of individual pixel signal values above mean background signal, was converted to optical density and multiplied by the area of signal to produce integrated optical density (IOD). Each brain region was sampled across 5–8 tissue sections collected in 240 μm increments; measurements for a given region were averaged across sections to give a single value per rat and then averaged across the 3 animals to provide data depicted in Fig. 3. We assessed the developmental pattern of DNMT1, DNMT3a, and DNMT3b mRNA expression in 8 brain regions: hippocampus, caudate putamen, lateral septum, nucleus accumbens, cingulate cortex, habenula, hypothalamus and amygdala. We used two-way ANOVAs to compare mRNA levels between certain brain areas (e.g. CA regions of the hippocampus versus dentate gyrus) across age (the early developmental timepoints as well as adulthood – P75). When necessary we used Fisher’s PLSD post hoc tests. For all tests, α=0.05. For all autoradiograms and photomicrographs, original images were exported to Adobe Photoshop for minor adjustments of brightness and contrast in final processing of images presented in this report.
For the methylated DNA quantification kit, DNA methylation levels were calculated using the following formula: (Sample OD-Blank OD)/(Slope of Standard Curve × 2). Samples were run in triplicate and averaged together after correction to generate one data point per sample. We assessed the developmental pattern of the DNA methylation levels by two-way ANOVA with age and region as between subject factors. When necessary we used Fisher’s PLSD post hoc tests. For all tests, α=0.05.
Acknowledgments
This study was supported by NIMH R00 MH085859-03 (SMC). We are grateful to Sharon Burke, Jennifer Fitzpatrick, and the laboratory of Dr. Stanley J. Watson at the University of Michigan for generously providing the DNMT1, DNMT3a, and DNMT3b riboprobes used for the in situ hybridization experiments. We would also like to thank Jasmine Howard (University of Alabama Birmingham) for excellent technical assistance.
REFERENCES
- Agnish ND, Keller KA. The rationale for culling of rodent litters. Fundam. Appl. Toxicol.: Off. J. Soc. Toxicol. 1997;38:2–6. doi: 10.1006/faat.1997.2318. [DOI] [PubMed] [Google Scholar]
- Akbarian S. Epigenetics of schizophrenia. Curr. Top. Behav. Neurosci. 2010;4:611–628. doi: 10.1007/7854_2010_38. [DOI] [PubMed] [Google Scholar]
- Beneyto M, Meador-Woodruff JH. Expression of transcripts encoding AMPA receptor subunits and associated postsynaptic proteins in the macaque brain. J. Comp. Neurol. 2004;468:530–554. doi: 10.1002/cne.10981. [DOI] [PubMed] [Google Scholar]
- Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1–11 in the rat brain. J. Mol. Neurosci.: MN. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- Brown SE, Weaver IC, Meaney MJ, Szyf M. Regional-specific global cytosine methylation and DNA methyltransferase expression in the adult rat hippocampus. Neurosci. Lett. 2008;440:49–53. doi: 10.1016/j.neulet.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Burghardt KJ, Pilsner JR, Bly MJ, Ellingrod VL. DNA methylation in schizophrenia subjects: gender and MTHFR 677C/T genotype differences. Epigenomics. 2012;4:261–268. doi: 10.2217/epi.12.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nat. Struct. Mol. Biol. 2013;20:282–289. doi: 10.1038/nsmb.2489. [DOI] [PubMed] [Google Scholar]
- Carrard A, Salzmann A, Malafosse A, Karege F. Increased DNA methylation status of the serotonin receptor 5HTR1A gene promoter in schizophrenia and bipolar disorder. J. Affective Disord. 2011;132:450–453. doi: 10.1016/j.jad.2011.03.018. [DOI] [PubMed] [Google Scholar]
- Chen ZX, Riggs AD. DNA methylation and demethylation in mammals. J. Biol. Chem. 2011;286:18347–18353. doi: 10.1074/jbc.R110.205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton SM, Meador-Woodruff JH. Nucleus-specific expression of NMDA receptor-associated postsynaptic density proteins in primate thalamus. Thalamus Relat. Syst. 2002;1:303–316. [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datson NA, van der Perk-de Jong J, van den Berg MP, de Kloet ER, Vreugdenhil E. MicroSAGE: a modified procedure for serial analysis of gene expression in limited amounts of tissue. Nucleic Acids Res. 1999;27:1300–1307. doi: 10.1093/nar/27.5.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Szyf M. Multiple isoforms of DNA methyltransferase are encoded by the vertebrate cytosine DNA methyltransferase gene. J. Biol. Chem. 1998;273:22869–22872. doi: 10.1074/jbc.273.36.22869. [DOI] [PubMed] [Google Scholar]
- Eilam D, Golani I. The ontogeny of exploratory behavior in the house rat (Rattus rattus): the mobility gradient. Dev. Psychobiol. 1988;21:679–710. doi: 10.1002/dev.420210707. [DOI] [PubMed] [Google Scholar]
- Fagiolini M, Jensen CL, Champagne FA. Epigenetic influences on brain development and plasticity. Curr. Opinion Neurobiol. 2009;19:207–212. doi: 10.1016/j.conb.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, Biniszkiewicz D, Bates B, Lee PP, Kuhn R, Trumpp A, Poon C, Wilson CB, Jaenisch R. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J. Neurosci.: Off. J. Soc. Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr. Res. 2007;61:58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opinion Genet. Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Golding MC, Westhusin ME. Analysis of DNA (cytosine 5) methyltransferase mRNA sequence and expression in bovine preimplantation embryos, fetal and adult tissues. Gene Expression Patterns: GEP. 2003;3:551–558. doi: 10.1016/s1567-133x(03)00121-2. [DOI] [PubMed] [Google Scholar]
- Grafodatskaya D, Chung B, Szatmari P, Weksberg R. Autism spectrum disorders and epigenetics. J. Am. Acad. Child Adolescent Psychiatry. 2010;49:794–809. doi: 10.1016/j.jaac.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Guenatri M, Duffie R, Iranzo J, Fauque P, Bourc’his D. Plasticity in Dnmt3L-dependent and -independent modes of de novo methylation in the developing mouse embryo. Development. 2013;140:562–572. doi: 10.1242/dev.089268. [DOI] [PubMed] [Google Scholar]
- Han X, Wu X, Chung WY, Li T, Nekrutenko A, Altman NS, Chen G, Ma H. Transcriptome of embryonic and neonatal mouse cortex by high-throughput RNA sequencing. Proc. Natl Acad. Sci. USA. 2009;106:12741–12746. doi: 10.1073/pnas.0902417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- Hirabayashi K, Shiota K, Yagi S. DNA methylation profile dynamics of tissue-dependent and differentially methylated regions during mouse brain development. BMC Genomics. 2013;14:82. doi: 10.1186/1471-2164-14-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YG, Hirasawa R, Hu JL, Hata K, Li CL, Jin Y, Chen T, Li E, Rigolet M, Viegas-Pequignot E, Sasaki H, Xu GL. Regulation of DNA methylation activity through Dnmt3L promoter methylation by Dnmt3 enzymes in embryonic development. Hum. Mol. Genet. 2008;17:2654–2664. doi: 10.1093/hmg/ddn165. [DOI] [PubMed] [Google Scholar]
- Kadriu B, Guidotti A, Chen Y, Grayson DR. DNA methyltransferases1 (DNMT1) and 3a (DNMT3a) colocalize with GAD67-positive neurons in the GAD67-GFP mouse brain. J. Comp. Neurol. 2012;520:1951–1964. doi: 10.1002/cne.23020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, Guennel T, Shin Y, Johnson MB, Krsnik Z, Mayer S, Fertuzinhos S, Umlauf S, Lisgo SN, Vortmeyer A, Weinberger DR, Mane S, Hyde TM, Huttner A, Reimers M, Kleinman JE, Sestan N. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine S, Alpert M, Lewis GW. Infantile experience and the maturation of the pituitary adrenal axis. Science. 1957;126:1347. doi: 10.1126/science.126.3287.1347. [DOI] [PubMed] [Google Scholar]
- Liang P, Song F, Ghosh S, Morien E, Qin M, Mahmood S, Fujiwara K, Igarashi J, Nagase H, Held WA. Genome-wide survey reveals dynamic widespread tissue-specific changes in DNA methylation during development. BMC Genomics. 2011;12:231. doi: 10.1186/1471-2164-12-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, Behrens MM, Ecker JR. Global epigenomic reconfiguration during mammalian brain development. Science. 2013 doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Wang YF, Cantemir C, Muller MT. Endogenous assays of DNA methyltransferases: evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol. Cell. Biol. 2003;23:2709–2719. doi: 10.1128/MCB.23.8.2709-2719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, van Groen T, Kadish I, Tollefsbol TO. DNA methylation impacts on learning and memory in aging. Neurobiol. Aging. 2009;30:549–560. doi: 10.1016/j.neurobiolaging.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M. Maternal care as a model for experience-dependent chromatin plasticity? Trends Neurosci. 2005;28:456–463. doi: 10.1016/j.tins.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Mill J, Petronis A. Molecular studies of major depressive disorder: the epigenetic perspective. Mol. Psychiatry. 2007;12:799–814. doi: 10.1038/sj.mp.4001992. [DOI] [PubMed] [Google Scholar]
- Miyake K, Hirasawa T, Koide T, Kubota T. Epigenetics in autism and other neurodevelopmental diseases. Adv. Exp. Med. Biol. 2012;724:91–98. doi: 10.1007/978-1-4614-0653-2_7. [DOI] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Nelson ED, Monteggia LM. Epigenetics in the mature mammalian brain: effects on behavior and synaptic transmission. Neurobiol. Learn. Mem. 2011;96:53–60. doi: 10.1016/j.nlm.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, Colantuoni C, Weinberger DR, Kleinman JE, Lipska BK. DNA methylation signatures in development and aging of the human prefrontal cortex. Am. J. Hum. Genet. 2012;91:765. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen M, Ferguson-Smith AC. DNA methylation in genomic imprinting, development, and disease. J. Pathol. 2001;195:97–110. doi: 10.1002/path.890. [DOI] [PubMed] [Google Scholar]
- Paxinos G. The Rat Nervous System. Elsevier Academic Press; Amsterdam; Boston: 2004. [Google Scholar]
- Paxinos G, Ashwell KWS, Tork I. Atlas of the Developing Rat Nervous System. Academic Press; New York: 1994a. [Google Scholar]
- Paxinos G, Ashwell KWS, Törk I. Atlas of the developing rat nervous system. Academic Press; San Diego: 1994b. [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. Acacemic Press Inc.; San Diego, CA: 1997. [Google Scholar]
- Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. Academic Press Inc; San Diego, CA: 1986. [Google Scholar]
- Sanchez MM, Ladd CO, Plotsky PM. Early adverse experience as a developmental risk factor for later psychopathology: evidence from rodent and primate models. Dev. Psychopathol. 2001;13:419–449. doi: 10.1017/s0954579401003029. [DOI] [PubMed] [Google Scholar]
- Sapolsky RM, Meaney MJ. Maturation of the adrenocortical stress response: neuroendocrine control mechanisms and the stress hyporesponsive period. Brain Res. 1986;396:64–76. doi: 10.1016/s0006-8993(86)80190-1. [DOI] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons RK, Howard JL, Simpson DN, Akil H, Clinton SM. DNA methylation in the developing hippocampus and amygdala of anxiety-prone versus risk-taking rats. Dev. Neurosci. 2012;34:58–67. doi: 10.1159/000336641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schubeler D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stead JD, Neal C, Meng F, Wang Y, Evans S, Vazquez DM, Watson SJ, Akil H. Transcriptional profiling of the developing rat brain reveals that the most dramatic regional differentiation in gene expression occurs postpartum. J. Neurosci. 2006a;26:345–353. doi: 10.1523/JNEUROSCI.2755-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead JD, Neal C, Meng F, Wang Y, Evans S, Vazquez DM, Watson SJ, Akil H. Transcriptional profiling of the developing rat brain reveals that the most dramatic regional differentiation in gene expression occurs postpartum. J. Neurosci. 2006b;26:345–353. doi: 10.1523/JNEUROSCI.2755-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- Suetake I, Morimoto Y, Fuchikami T, Abe K, Tajima S. Stimulation effect of Dnmt3L on the DNA methylation activity of Dnmt3a2. J. Biochem. 2006;140:553–559. doi: 10.1093/jb/mvj185. [DOI] [PubMed] [Google Scholar]
- Sun H, Kennedy PJ, Nestler EJ. Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology: Off. Publ. Am. Coll. Neuropsychopharmacology. 2013;38:124–137. doi: 10.1038/npp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD, Miller CA. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, Absher DM, Wold BJ, Myers RM. Dynamic DNA Methylation Across Diverse Human Cell Lines and Tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Mo Y, Ebenezer D, Bhattacharyya S, Liu H, Sundaravel S, Wonkatal S, Cartier J, Caces B, Artz A, Nischal S, Bhagat T, Bathon K, Maqbool S, Gligich O, Suzuki M, Steidl U, Godley L, Skoultchi A, Greally J, Wickrema A, Verma A. High resolution methylome analysis reveals widespread functional hypomethylation during adult human erythropoiesis. J. Biol. Chem. 2013 doi: 10.1074/jbc.M112.423756. [DOI] [PMC free article] [PubMed] [Google Scholar]