Abstract
Background
Many wounds are difficult to heal because of the large, complex community of microbes present within the wound.
The Problem
Classical laboratory culture methods do not provide an accurate picture of the microbial interactions or representation of microorganisms within a wound. There is an inherent bias in diagnosis based upon classical culture stemming from the ability of certain organisms to thrive in culture while others are underrepresented or fail to be identified in culture altogether. Chronic wounds also contain polymicrobial infections existing as a cooperative community that is resistant to antibiotic therapy.
Basic/Clinical Science Advances
New methods in molecular diagnostic medicine allow the identification of nearly all organisms present in a wound irrespective of the ability of these organisms to be grown in culture. Advances in DNA analyses allow absolute identification of microorganisms from very small clinical specimens. These new methods also provide a quantitative representation of all microorganisms contributing to these polymicrobial infections.
Clinical Care Relevance
Technological advances in laboratory diagnostics can significantly shorten the time required to heal chronic wounds. Identification of the genetic signatures of organisms present within a wound allows clinicians to identify and treat the primary organisms responsible for nonhealing wounds.
Conclusion
Advanced genetic technologies targeting the specific needs of wound care patients are now accessible to all wound care clinicians.

Owatha L. Tatum
Background
Patients harboring chronic wounds often present with a number of impairments that interfere with the healing trajectory of their chronic wound. Even when these factors are taken into consideration and controlled, it still may not be possible to heal these wounds. It is now understood that the large mass of bacterial material, whether considered bioburden or biofilm or colonizing infection, present within the wound is a primary and universal cause of recalcitrance to timely healing in chronic wounds. For many years, the standard of care for identification and diagnosis of the microorganisms present in a chronic wound has been classical microbiological culture. This diagnostic method can accurately identify bacteria present at the wound surface. However, in the vast majority of cases, it is not able to provide information about the organisms present in the total bioburden of the wound. The reasons for this stem from the nature of culture techniques. First, chronic wounds are not simple infections containing one microorganism. They are polymicrobial infections composed of sometimes dozens of bacterial species and yeasts.1–6 These populations of microorganisms exist as a cooperative community. It has been well established that these communities, referred to as biofilms or bioburden, cease to exhibit the response to antimicrobial treatments that would be expected in simple infections. Culture methods often misrepresent the contribution of individual bacterial species present in a wound's bioburden because of the bias created by the culture process—many bacteria thrive in wound environments but fail to propagate in culture. A dependence upon culture results may therefore lead a clinician into a treatment decision that is ineffective.6
Target Articles.
1. Wolcott RD, Cox SB, and Dowd SE: Healing and healing rates of chronic wounds in the age of molecular pathogen diagnostics. J Wound Care 2010; 19: 272.
2. Wolcott RD, Rhoads DD, Bennett ME, Wolcott BM, Gogokhia L, Costerton JW, et al.: Chronic wounds and the medical biofilm paradigm. J Wound Care 2010; 19: 45.
3. Wolcott RD, Gontcharova V, Sun Y, and Dowd SE: Evaluation of the bacterial diversity among and within individual venous leg ulcers using bacterial tag-encoded FLX and titanium amplicon pyrosequencing and metagenomic approaches. BMC Microbiol 2009; 9: 226.
Clinical Problem Addressed
Whether the patient has a chronic infection characterized by multiple species of slow-growing bacteria as is characteristic of a biofilm infection or a simple acute infection (i.e., cellulitis) containing one rapidly dividing bacterial species, the primary goal is the same: effectively treat the wound through accurate identification of the pathogens involved.2,7
Until now, identifying microorganisms has meant routine laboratory culture. However, there are important limitations to this technique. Most striking is cultivation bias. Interestingly, fewer than 2% of all known bacterial species can be routinely cultured in the clinical microbiology laboratory. Further, only a subset of these microorganisms will grow within the 24-h period prescribed for most cultures.8,9 A second limitation is that yeast including Candida species are rarely identified in culture.8,10–15 Because yeasts can be important components of a biofilm community, it is important to recognize their contribution to a chronic wound. Classical cultures are also poor at quantification. Swab cultures do not actually allow quantification of the microorganisms (i.e., it is impossible to know whether there were 10 organisms or 10 million organisms present in a given wound). Because the vast majority of microorganisms cannot be routinely cultured in the laboratory, culture also provides a very low level of confidence in identification. Although biochemical identification of bacteria and yeast is 80%–90% accurate, it can only account for the small percentage of organisms we can culture.
The 2% of bacteria that are easily cultured in a planktonic growth state fail to rapidly grow in the laboratory; so they can be thought of as viable, but not cultivable. Examples are anaerobic bacteria, which are traditionally difficult to propagate in the laboratory without specialized collection methods, growth media, and environmental control, yet anaerobes are a primary and until now unreported component of most recalcitrant wounds.1,2,4,6,10,16–18 Finally, some species are known to efficiently grow in laboratory conditions and outcompete other species. Thus, such diagnostic methods confer a selection bias of one species over another. In all, it is clear how dependence upon laboratory culture may easily misguide treatment choices.
Relevant Basic Science Context
The polymerase chain reaction (PCR) is a molecular technique utilized to target and “amplify” a single or few copies of a piece of DNA, generating billions or more copies of a specific DNA sequence. The amplification process thereby allows detection and analysis of the DNA fragments in the laboratory. The method relies on repeated cycles of heating and cooling for DNA melting and then enzymatic replication of the DNA. Microorganisms are identified by amplifying their unique DNA. This method requires only a minute amount of specimen material. It also identifies organisms directly, without requiring propagation in a microbiology laboratory. This technology is extremely sensitive and specific and can be done in hours. DNA sequencing complements the PCR diagnostic test development process by identifying novel pathogens previously unknown to be involved in chronic wound development and persistence.
Experimental Model or Material: Advantages and Limitations
Molecular diagnostic methods have been used to accurately identify the composition of chronic wounds and guide treatment in a number of study models. Most recently, bacterial diversity in decubitus ulcers was evaluated from molecular diagnostic methods based upon the PCR and genetic sequencing processes. Other models have included surgical site infections, venous leg ulcers, and burn wound sepsis.1,2,4,6,10,16–18 These models represent the vast majority of wounds encountered in practice. They also represent a full span of clinical microbial diversity and complexity.
Discussion of Findings and Relevant Literature
Use of molecular diagnostic methods has revealed some surprising findings. Wounds cultured with standard techniques grew only bacteria such as Staphylococcus aureus. In these same clinical specimens, molecular pathogen diagnostics revealed that chronic wounds were in fact colonized by many other bacteria and fungi (yeast), including anaerobes and other difficult-to-culture organisms.1,2,4,6,10,16–18 Without molecular genetic pathogen analysis, these anaerobic organisms, fungi, and other microorganisms were undetectable. When specific antibiotics targeting these other bacteria were used, the patients responded to therapy.17
An array of bacterial species may be identified in any given specimen. The diversity of microorganisms identified in a clinical sample may make a clinician uncomfortable in formulating a treatment regimen, because it is difficult to know which need to be targeted for therapy and how to tailor a treatment plan using a reasonable number of antibiotics, providing the broadest coverage of the bacteria identified. Using molecular-based diagnostics, we have observed many previously uncharacterized bacteria such as Bacteroides occurring in a majority of surgical site infections that were reported as “culture negative.”4
The interpretation of information from PCR diagnostics can be used in both surveillance and immediate management of an infection. In one example, S. aureus may be present in a wound containing a large bioburden with 90% of the bacteria exhibiting methicillin resistance (as indicated by the presence of the mecA gene detected by molecular methods). This type of result suggests a high-risk wound requiring treatment with first-line methicillin-resistant S. aureus (MRSA) antibiotics. However, in the case of a wound containing less than 10% MRSA of the total population in a wound with a large bioburden, and if the wound is not threatening a limb, it may be more appropriate to treat the infection less aggressively with second-line therapeutics such as trimethoprim-sulfa. Going further, if MRSA is present in very low amounts (<3%) in a given wound, it may be most appropriate to conservatively manage the patient and use molecular methods only as a means of surveillance.3,19–22 In these cases, the utility of quantitative information is obvious. Microbiological cultures would likely not distinguish between the compositions of these three very different wounds. In addition, if MRSA were only present in the biofilm within a wound, it may have been viable but noncultivable and not detected at all.
Molecular genetics-based pathogen diagnostics as described here are based upon the identification of unique DNA sequence of each type of microorganism. Although it may seem futuristic, one company has it (www.pathogenius.com). This method detects the unique genetic signature of bacterial species and, in some cases, subspecies. Clinical DNA sequencing and PCR also have the ability to simultaneously identify known antibiotic resistance factors, thereby eliminating the need for additional sensitivity testing. Genetic databases are actively maintained with this information for clinical use.
Innovation
The use of PCR analyses, particularly quantitative PCR analyses, of chronic wound specimens has the power to revolutionize the field of wound care. The technology is many orders of magnitude more sensitive than culture and days faster and covers an almost universal range of pathogens that may be detected. DNA sequencing technologies also promise to reveal as-yet-undiscovered players in wound pathology and course of infection. Molecular diagnostic technologies will have a dramatic impact on all areas of clinical care over the coming years. These technologies are undergoing a rapid advancement akin to that of the semiconductor industry in the 1980s. It is not inconceivable that within 5 years, a clinician may be able to perform this type of molecular diagnostic analysis at the time the patient visits the office and receive a treatment tailored to the specific wound within an hour.
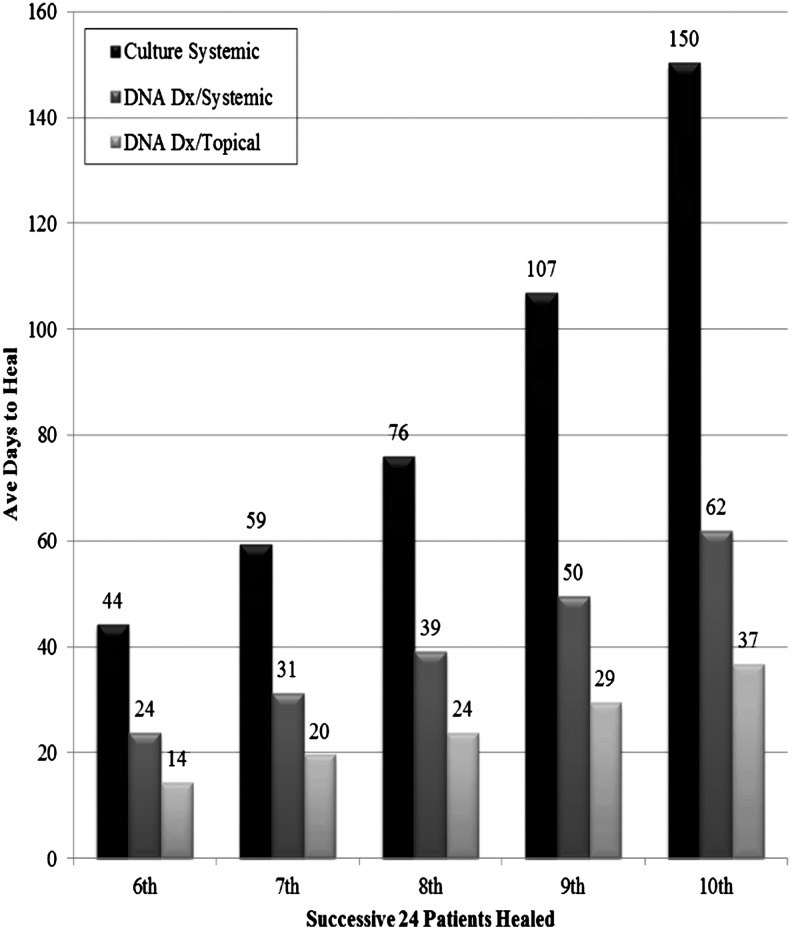
Summary Illustration
The figure illustrates the power of molecular or genetic-based pathogen diagnostics in the wound care clinic. In 2009, molecular genetics-based diagnosis of pathogens in wounds was instituted as standard practice in the clinic. The figure illustrates improvement in time to healing for several types of wounds. In 2007 (before the use of molecular diagnostics), 48.5% of wounds completely healed compared with 62.4% in 2009 and over 90% in 2010.17,23
Take-Home Message.
Basic science advances
• DNA sequencing and quantitative PCR now allow the identification and quantification of both known and previously unknown microbial species.
• Antibiotic resistance factors can be identified by their genetic signatures in clinical specimens.
Clinical science advances
• Complex polymicrobial communities can be quantified and their relative contribution to a nonhealing wound can be determined from molecular diagnostic methods.
• The contribution of nonculturable pathogens to persistence of a chronic wound can be assessed from genetic information.
• Microbial composition of wounds can be determined in hours as opposed to days using quantitative PCR.
Relevance to clinical care
• Treatments can be tailored to the specific needs of the patient with regard to appropriate antibiotic therapy.
• Healing time, expense, and suffering can all be significantly reduced with the use of molecular diagnostic methods in place of classical cultures.
Caution, Critical Remarks, and Recommendations
One concern that can arise when using highly sensitive molecular diagnostic methods is contamination of a specimen with other organisms not involved with the wound. In fact, this was one of the early concerns of the authors during development of these methods. For example, many wounds exhibited an abundance of 80% Corynebacterium spp. Conventionally, Corynebacterium in wounds has been thought to be a nonpathogen. However, this understanding was based on data from culture techniques. We have observed that Corynebacterium and many other surprising bacteria propagate in wounds and contribute to the severity of chronic infections. One concern among practitioners is that the data contained in molecular diagnostic laboratory reports are overwhelming and difficult to interpret. This is certainly true. It is common to see many organisms reported as present in one wound. Although diagnostic laboratories strive to provide information in a concise, easily digested format, the input of clinicians utilizing this information will continue to be important as technology continues to improve and yield even more information. Cost of PCR-based testing is often cited as a barrier to these tests. Although the individual tests can be expensive, they are comparable to classical microbiological culture (approximately $200 vs. $300) and provide clinicians with far more clinical information. In addition, most insurers are now accustomed to these types of testing technologies and they are also reimbursable by Medicare in most cases.
Future Development of Interest
Molecular diagnostic microbiology is experiencing an enormous amount of growth in the clinical laboratory. The power of genetic technologies and application of genetic data to clinical care have important and promising implications for the treatment of chronic wounds. Molecular methods are much more rapid, accurate, and comprehensive than culture methods. For these reasons, they promise to improve healing trajectory and reduce suffering while also reducing costs and long-term dependence on antibiotic therapy. As molecular methods evolve, they will likely become commonplace in the hospital laboratory and have the power to replace the culture-based methods in use for so many years.
Abbreviations and Acronyms
- MRSA
methicillin-resistant Staphylococcus aureus
- PCR
polymerase chain reaction
Acknowledgments and Funding Sources
The authors have not received funding for this work.
Author Disclosure and Ghostwriting
The authors have no disclosures relevant to this work. The content of this article was expressly written by the authors listed. No ghostwriters were used to write this article.
References
- 1.Gontcharova V, et al. A comparison of bacterial composition in diabetic ulcers and contralateral intact skin. Open Microbiol J. 2010;4:8. doi: 10.2174/1874285801004010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolcott RD. Gontcharova V. Sun Y. Dowd SE. Evaluation of the bacterial diversity among and within individual venous leg ulcers using bacterial tag-encoded FLX and titanium amplicon pyrosequencing and metagenomic approaches. BMC Microbiol. 2009;9:226. doi: 10.1186/1471-2180-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leake JL, et al. Identification of yeast in chronic wounds using new pathogen-detection technologies. J Wound Care. 2009;18:103. doi: 10.12968/jowc.2009.18.3.39810. [DOI] [PubMed] [Google Scholar]
- 4.Wolcott RD, et al. Bacterial diversity in surgical site infections: not just aerobic cocci any more. J Wound Care. 2009;18:317. doi: 10.12968/jowc.2009.18.8.43630. [DOI] [PubMed] [Google Scholar]
- 5.Dowd SE, et al. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS One. 2008;3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowd SE, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolcott RD. Rhoads DD. Bennett ME. Wolcott BM. Gogokhia L. Costerton JW, et al. Chronic wounds and the medical biofilm paradigm. J Wound Care. 2010;19:45. doi: 10.12968/jowc.2010.19.2.46966. [DOI] [PubMed] [Google Scholar]
- 8.Stephens P, et al. Anaerobic cocci populating the deep tissues of chronic wounds impair cellular wound healing responses in vitro. Br J Dermatol. 2003;148:456. doi: 10.1046/j.1365-2133.2003.05232.x. [DOI] [PubMed] [Google Scholar]
- 9.Thomsen TR, et al. The bacteriology of chronic venous leg ulcer examined by culture-independent molecular methods. Wound Repair Regen. 2010;18:38. doi: 10.1111/j.1524-475X.2009.00561.x. [DOI] [PubMed] [Google Scholar]
- 10.Dowd SE, et al. Research survey of fungi and yeast in polymicrobial infections in chronic wounds. J Wound Care. 2011;20:40. doi: 10.12968/jowc.2011.20.1.40. [DOI] [PubMed] [Google Scholar]
- 11.Bradshaw DJ, et al. Effect of oxygen, inoculum composition and flow rate on development of mixed-culture oral biofilms. Microbiology. 1996;142(Pt 3):623. doi: 10.1099/13500872-142-3-623. [DOI] [PubMed] [Google Scholar]
- 12.Rasmussen K. Lewandowski Z. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol Bioeng. 1998;59:302. doi: 10.1002/(sici)1097-0290(19980805)59:3<302::aid-bit6>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 13.Stott MB, et al. Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ Microbiol. 2008;10:2030. doi: 10.1111/j.1462-2920.2008.01621.x. [DOI] [PubMed] [Google Scholar]
- 14.Petti CA, et al. Utility of extended blood culture incubation for isolation of Haemophilus, Actinobacillus, Cardiobacterium, Eikenella, and Kingella organisms: a retrospective multicenter evaluation. J Clin Microbiol. 2006;44:257. doi: 10.1128/JCM.44.1.257-259.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forward KR. An evaluation of extended incubation time with blind subculture of blood cultures in patients with suspected endocarditis. Can J Infect Dis Med Microbiol. 2006;17:186. doi: 10.1155/2006/284019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith DM, et al. Evaluation of the bacterial diversity of Pressure ulcers using bTEFAP pyrosequencing. BMC Med Genomics. 2010;3:41. doi: 10.1186/1755-8794-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolcott RD. Cox SB. Dowd SE. Healing and healing rates of chronic wounds in the age of molecular pathogen diagnostics. J Wound Care. 2010;19:272. [PubMed] [Google Scholar]
- 18.Smith DM, et al. Evaluation of the bacterial diversity of pressure ulcers using bTEFAP pyrosequencing. BMC Med Genomics. 2010;3:41. doi: 10.1186/1755-8794-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephenson MF, et al. Molecular characterization of the polymicrobial flora in chronic rhinosinusitis. J Otolaryngol Head Neck Surg. 2010;39:182. [PubMed] [Google Scholar]
- 20.Wolcott RD. Kennedy JP. Dowd SE. Regular debridement is the main tool for maintaining a healthy wound bed in most chronic wounds. J Wound Care. 2009;18:54. doi: 10.12968/jowc.2009.18.2.38743. [DOI] [PubMed] [Google Scholar]
- 21.Wolcott RD. Rhoads DD. Dowd SE. Biofilms and chronic wound inflammation. J Wound Care. 2008;17:333. doi: 10.12968/jowc.2008.17.8.30796. [DOI] [PubMed] [Google Scholar]
- 22.Wolcott RD. Dowd SE. A rapid molecular method for characterising bacterial bioburden in chronic wounds. J Wound Care. 2008;17:513. doi: 10.12968/jowc.2008.17.12.31769. [DOI] [PubMed] [Google Scholar]
- 23.Dowd SE. Wolcott RD. Kennedy J. Jones C. Cox SB. Molecular diagnostics, personalised medicine in wound care: assessment of outcomes. J Wound Care. 2011;20:232. doi: 10.12968/jowc.2011.20.5.232. [DOI] [PubMed] [Google Scholar]