Abstract
Small interfering RNA (siRNA)-based therapeutics have been used in humans and offer distinct advantages over traditional therapies. However, previous investigations have shown that there are several technical obstacles that need to be overcome before routine clinical applications are used. Currently, we are launching a novel class of RNAi therapeutic agents (PnkRNA™, nkRNA) that show high resistance to degradation and are less immunogenic, less cytotoxic, and capable of efficient intracellular delivery. Here, we develop a novel platform to promote naked RNAi approaches administered through inhalation without sophisticated delivery technology in mice. Furthermore, a naked and unmodified novel RNAi agent, such as ribophorin II (RPN2-PnkRNA), which has been selected as a therapeutic target for lung cancer, resulted in efficient inhibition of tumor growth without any significant toxicity. Thus, this new technology using aerosol delivery could represent a safe, potentially RNAi-based strategy for clinical applications in lung cancer treatment without delivery vehicles.
RNA interference (RNAi) is a post-transcriptional gene-silencing mechanism. Small interfering RNA (siRNA) must be dissociated into their component single strands to act as a guide for the RNA-induced silencing complexes, which are the protein complexes that repress gene expression1. The development of siRNA technology has opened an avenue of opportunity to study gene function, as well as the possibility of novel forms of therapeutic intervention in several genetic diseases. In fact, siRNA-based therapy has enormous potential for the treatment of several diseases through either local or systemic administration of siRNAs that are being tested in experimental animal models or in clinical development2. Oncology is one of the medical fields that can benefit most from this powerful therapeutic strategy because this approach can modulate the expression of target genes involved in tumor initiation, growth, and metastasis3.
However, the clinical application of siRNAs has been impaired by problems related to their delivery, low biological stability, off-target gene silencing, and immunostimulatory effects4,5. Indeed, naked siRNAs are promptly degraded by nucleases in serum and extracellular fluids, and chemical modifications at specific positions or formulations with delivery vehicles have been shown to improve stability. However, these may attenuate the suppressive activity of siRNAs6. Furthermore, the cost of large-scale production is another obstacle to the clinical application of siRNAs7. For this reason, their translation to the clinical setting is dependent upon the development of an efficient delivery system that is able to improve the pharmacokinetic and biodistribution properties of siRNAs.
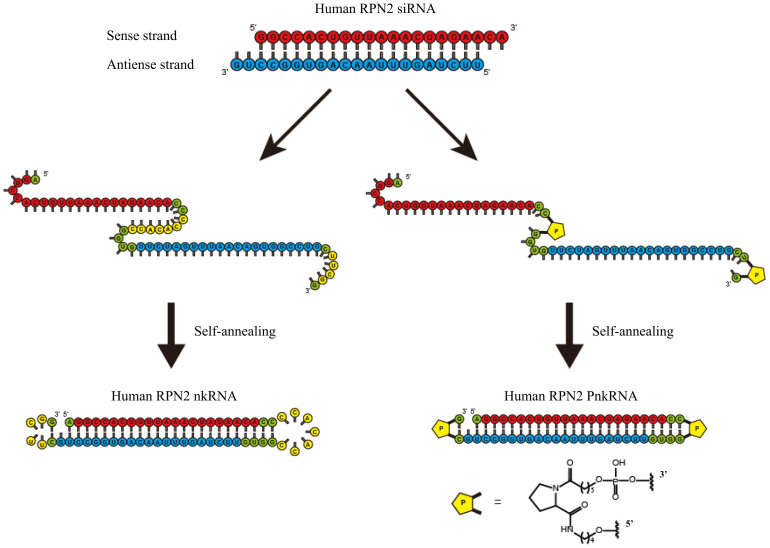
Recently, engineered designs, such as aptamer-siRNA chimeras and transferring-decorated nanoparticles, have continued to dramatically improve the precision of delivery for RNAi agents8. Advances in RNAi-based therapeutics may require new biochemical technologies to maximise drug potency while minimising off-target toxicity and immunogenicity. Meanwhile, we have already reported a novel class of RNAi therapeutic agents (PnkRNA, nkRNA) and evaluated their effectiveness9. We showed that PnkRNA and nkRNA directed against transforming growth factor (TGF)-β1 ameliorate outcomes in mouse models of acute lung injury and pulmonary fibrosis. This novel class of RNAi agents was synthesised on solid phase as single-stranded RNAs (ssRNAs) that self-anneal into a unique helical structure containing a central stem and two loops following synthesis (Fig. 1). The production of the novel RNAi agents is simple; because PnkRNA and nkRNA are synthesised as ssRNAs that spontaneously self-anneal, low-cost, large-scale production is possible. These novel RNAi agents have showed significant effectiveness in disease models and also superior resistance against nuclease degradation compared to canonical siRNAs. Additionally, by evaluating the induction of proinflammatory cytokines, our previous results suggest that none of the platforms were immunotoxic9. Thus, the novel RNAi therapeutic agents are safe and might be employed in clinical applications because they address several issues in siRNA-based therapy.
Figure 1. Structure of novel RNAi agents.
Both nkRNA and PnkRNA were prepared as single-stranded RNA oligomers that then self-anneal, as shown. Nucleotides in red indicate the sense strand of the target (RPN2); nucleotides in blue indicate the antisense strand; and nucleotides in green and yellow indicate the loop cassettes. P indicates a proline derivative.
Lung cancer is the leading cause of cancer-related death in the world. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancers. Approximately 70% of all newly diagnosed patients present with local advanced or metastatic disease and require systemic chemotherapy10,11. Although NSCLC patients with epidermal growth factor receptor (EGFR) mutations initially respond to EGFR tyrosine kinase inhibitors12, most patients experience a relapse within 1 year. Despite the development of novel molecular therapies13, the prognosis of lung cancer is still poor and shows a median survival time of approximately 18 months in the operable stages. Hence, novel and more effective approaches are needed for the treatment of advanced lung cancer. Lung diseases in general are attractive targets for siRNA-based therapeutics because of their lethality and prevalence. In addition, the lung is anatomically accessible to therapeutic agents via the intrapulmonary route. Accessibility is a key requirement for successful RNAi-based in vivo and clinical studies, and this characteristic offers several important benefits over systemic delivery, including the use of lower doses of siRNAs, the reduction of undesirable systemic side effects, and improved siRNA stability due to the lower nuclease activity in the airways compared to the serum14. The direct administration of siRNAs into the target organs is a promising strategy for overcoming the problems of intravenous administration15. On the basis of these promising findings, we explored the effectiveness of this novel class of RNAi therapeutic agents in lung cancer. In the current study, we prepared inhaled novel RNAi agents directed against luciferase and human-ribophorin II (RPN2) as candidate genes, and we compared their inhibitory activity to canonical siRNAs using in vitro assays and animal models of lung cancer. Furthermore, using this new technology, lung cancer xenograft studies showed that the aerosol delivery of a naked novel RNAi agent significantly inhibited tumor growth.
Results
In vitro stability of novel RNAi agents
The stability of the novel RNAi agents, PnkRNA and nkRNA, against degradation by ribonucleases was compared with that of siRNA. Each RNAi agent was incubated in the presence of ribonucleases, and the degree of degradation was followed overtime. siRNA was degraded after 5 min of incubation with ribonucleases, but both PnkRNA and nkRNA remained intact after 15 min of incubation (Supplementary Fig. 1). The novel RNAi platforms were more resistant to degradation than siRNA.
Distribution of fluorescent siRNA after inhalation
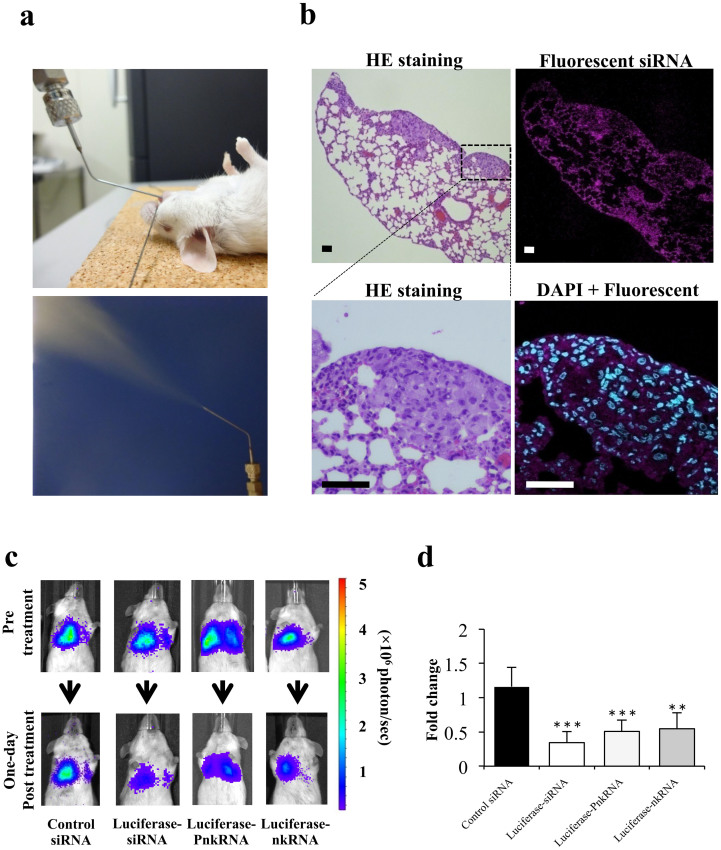
The inhaled administration of fluorescently labeled siRNA by MicroSprayer™ enabled the efficient intracellular distribution of lung cancer cells throughout the lung parenchyma. The efficient and homogeneous distribution of the RNAi agent to most parts of the lung tissue is a prerequisite for studying target-specific RNAi efficacy. The endotracheal application of suspended RNAi agents by a MicroSprayer™ can result in nanoparticle deposition in smaller airway and peripleural tumor cells (Fig. 2a,b).
Figure 2. Inhaled single administration study with novel RNAi platforms against luciferase gene in vivo.
(a) Intratracheal delivery route: RNAi therapeutic agents are sprayed directly from the mouth into the lungs using a MicroSprayer™ aeroliser. (b) Distribution of fluorescent siRNA in the lungs after inhalation. A sufficient pulmonary distribution of aerosolised siRNA was attained in mice by MicroSprayer™. In addition, intracellular fluorescent staining in lung cancer cells and bronchial epithelial cells was occasionally observed (magnified image, DAPI + Fluorescent siRNA). The scale bars indicate 50 μm. HE, Haematoxylin-eosin. (c) Monitoring luciferase inhibition in vivo with bioluminescent imaging. Representative images pre-treatment and on the first day post-treatment. (d) Normalised fold change (one day post-treatment/pre-treatment) of bioluminescence emitted from the whole bodies of mice. The data represent the means ± SD (n = 4). Statistical analysis was performed by the Bonferroni multiple-comparison test.***, P < 0.001 versus control siRNA group. **, P < 0.01 versus control siRNA group.
Monitoring luciferase inhibition of the inhaled novel RNAi agents' delivery system in vivo
Mice with an intravenous injection of A549-luc-C8 cells on day 0 were imaged twice a week up to day 28. In all mice, measurable lung tumors could be calipered within 2 weeks using this cell line. Four weeks after tumor injection, the observed patterns indicated lesions developing in the lungs of the mice. To estimate whether the aerosol delivery of novel RNAi agents had a valid gene-silencing effect on the lung tumors, the mice were treated with a luciferase siRNA, PnkRNA, and nkRNA, each of which was compared with the control siRNA. The activities of the siRNA and novel RNAi platforms are shown. On the next day, in mice receiving luciferase siRNA and novel RNAi agents, bioluminescence was inhibited by 50–60% in the whole body when compared with bioluminescence before treatment. On the other hand, the bioluminescent signals in the mice that were treated with the control siRNA had increased (Fig. 2c,d). In addition, we found that the RNAi effect of the novel platform would continue for at least five days (supplementary Fig. 2).
In vitro suppressive effect of novel RNAi agents for RPN2
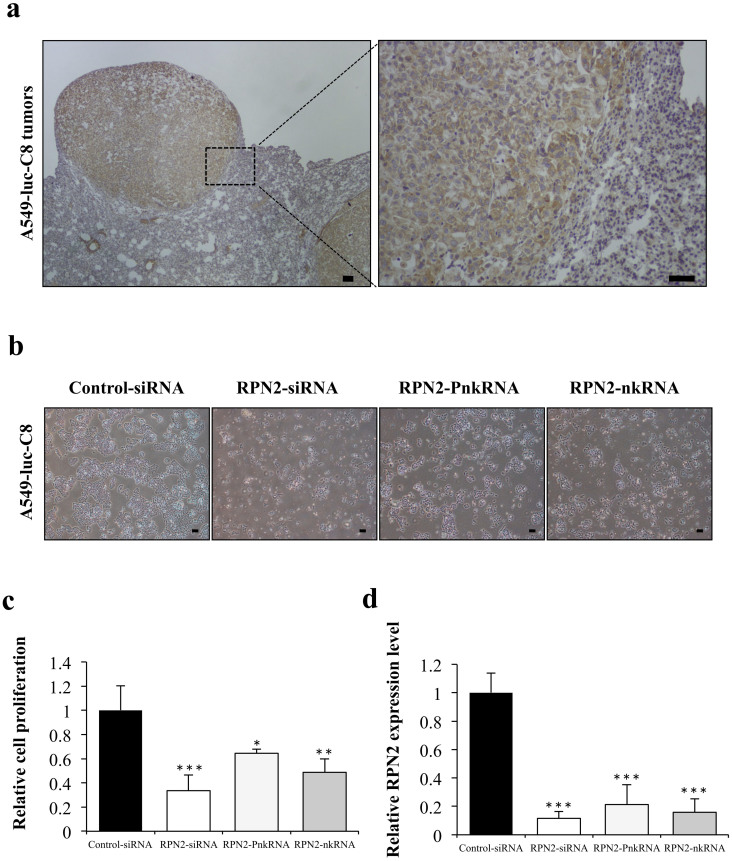
To screen for target genes showing the growth inhibition of A549-luc-C8 cells, RPN2 was selected as a target gene. A549-luc-C8 cells expressed RPN2 mRNA at high levels, as evaluated by real-time RT-PCR, and RPN2 protein expression on immunohistochemical staining was detected in the cytoplasm of cancer cells in the A549-luc-C8 xenograft model (Fig. 3a). To monitor cell growth and suppressive effects for RPN2, the RNAi cell transfection method was used. The inhibition of cell growth was observed on A549-luc-C8 cells treated with RPN2 siRNA, PnkRNA, nkRNA, or the control siRNA (Fig. 3b,c). The inhibition of the targeted mRNA levels was also shown (Fig. 3d). RPN2 PnkRNA and nkRNA, as well as siRNA, inhibited A549-luc-C8 cell proliferation and suppressed RPN2 expression. Furthermore, a rescue experiment indicated that exogenous RPN2 expression could rescue the functional changes induced by the specific RNAi knockdown (supplementary Fig. 3a,b). These results revealed that RPN2 may be the target of the inhibition of tumor growth of A549-luc-C8 cells.
Figure 3. Suppressive effect of novel RNAi agents for RPN2 in A549-luc-C8 cells.
(a) Immunohistochemical staining for RPN2 proteins in representative tumors of A549-luc-C8 xenograft lung cancer models. The scale bars indicate 100 μm. (b) Phase-contrast micrographs of A549-luc-C8 cells 96 h after transfection with RPN2 siRNA, RPN2 PnkRNA, RPN2 nkRNA or control siRNA using DharmaFECT 1 reagent. The scale bars indicate 10 μm. (c) Cell proliferation was measured 96 h after transfection with each of the RNAi therapeutic agents. Inhibition of cell growth was observed on A549-luc-C8 cells treated with RPN2 siRNA, PnkRNA, nkRNA, or the control siRNA. Statistical analysis was performed by the Bonferroni multiple-comparison test. The data represent the means ± SD (n = 3). ***, P < 0.001 versus control siRNA group. **, P < 0.01 versus control siRNA group. *, P < 0.05 versus control siRNA group. (d) The inhibition of the targeted mRNA levels is shown. Human RPN2 expression levels were normalised to beta-actin levels. Statistical analysis was performed by the Bonferroni multiple-comparison test. Data represent the means ± SD (n = 3). ***, P < 0.001 versus control siRNA group.
Inhibition of RPN2 expression by the inhaled novel RNAi agents' delivery system (single administration study, mice per group; n = 4)
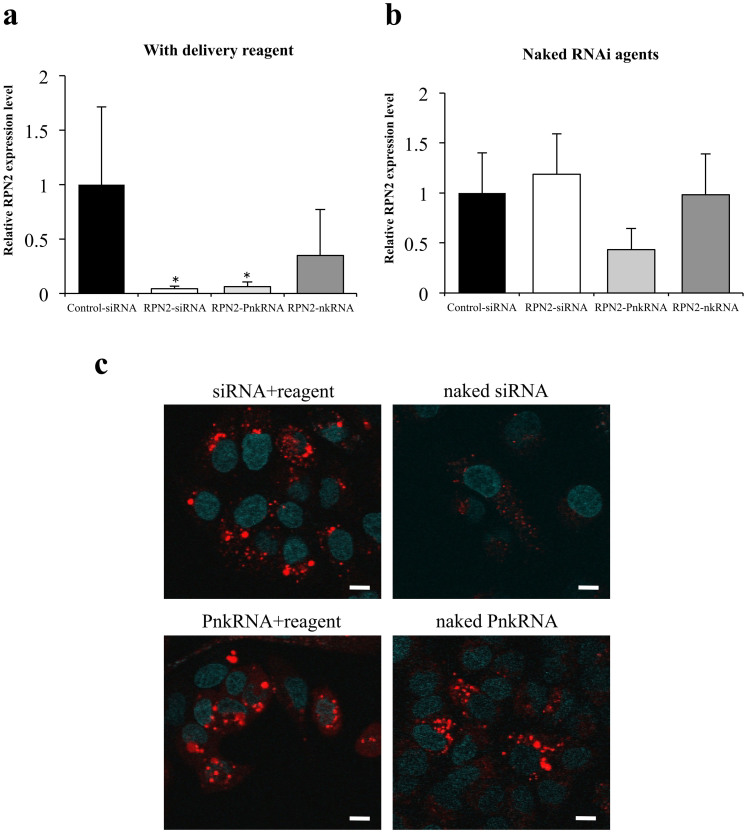
To investigate the inhibition of lung cancer by the inhaled novel RNAi agents' delivery system, RPN2 siRNA, PnkRNA and nkRNA were administered intratracheally into mice on day 28 of the intravenous injection of A549-luc-C8 cells. The treatment group received 15 μg RNAi agent by inhaled administration only on day 28. The total luminescence from all tumors was determined at different times post-treatment until 7 days post-treatment for each mouse. However, there was no suppression of luminescence in mice treated with RPN2 siRNA, PnkRNA, nkRNA, or control siRNA over the same observation period. Upon confirming the down-regulation of RPN2 expression, the xenograft tumor tissues were excised 24 h after the inhaled administration of RNAi agents, and quantitative real-time RT-PCR was performed. RPN2 mRNA was significantly down-regulated in the RPN2 siRNA- and PnkRNA-treated xenografts, compared with those treated with control siRNA (Fig. 4a). Moreover, we also tried to replicate the same inhaled administration study of RNAi agents without delivery vehicles. Remarkably, only naked RPN2-PnkRNA tended to suppress RPN2 mRNA expression, compared with the other three experimental groups (Fig. 4b). The endocytosis assay with labeled siRNAs and PnkRNAs using pHrodo™ Red succinimidyl ester showed the presence of the intracellular fluorescence signal in both A549-luc-C8 cells and PC14 cells transfected with not only RNAi therapeutic agents plus reagent but also naked PnkRNAs (Fig. 4c, Supplementary Fig. 4). This indicates that naked PnkRNAs were incorporated into the cell cytoplasm by endocytosis without delivery reagents. These results suggest that the repeated administration of naked RPN2-PnkRNA might be required for the inhibition of tumor growth. Therefore, we tested the hypothesis that a novel RNAi agent might have emerged as a powerful technology capable of suppressing the expression of target genes without delivery vehicles.
Figure 4. Single administration study with novel RNAi platforms against RPN2 gene in vivo and with an endocytosis assay in vitro.
The effects of transfection on the expression of RPN2-mRNA 24 h after the inhaled administration of RNAi therapeutic agents with (a) or without delivery reagents (b). Measurements of human RPN2 expression levels by real-time RT-PCR. Data represent the means ± SD (n = 4). *, P < 0.05 versus control siRNA group. (c) Endocytosis assay for naked RNAi therapeutic agents. Naked PnkRNAs were incorporated into the A549-luc-C8 cell cytoplasm through endocytosis in the absence of delivery reagents. The scale bars indicate 10 μm.
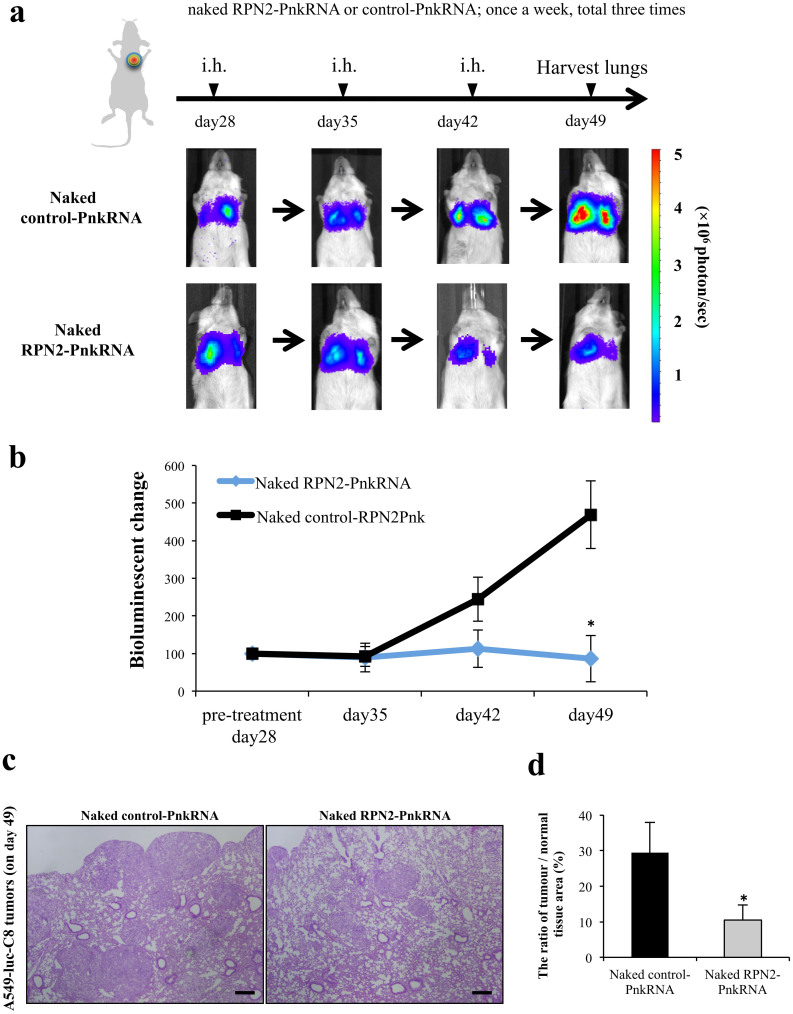
Analysis of the efficiency of naked RPN2-PnkRNA on the inhibition of lung tumor growth (repeated administration study, mice per group; n = 12)
Based on the observed PnkRNA pharmacodynamics (supplementary Fig. 2), we believe that the weekly administration schedule of PnkRNA-RPN2 is a reasonable RNAi-therapeutic strategy. To assess the efficiency of the naked novel RNAi (PnkRNA) agent on the inhibition of lung tumor growth, naked RPN2-PnkRNA was intratracheally administered on days 28, 35, and 42 post-inoculation. At the end of the experiment on day 49, mice with naked RPN2-PnkRNA showed inhibition of tumor growth, and there were significant differences between the naked RPN2-PnkRNA and the naked control-PnkRNA on day 49 (P < 0.05) (Fig. 5a,b). Histopathological analysis revealed that the A549-luc-C8 cell lung tumors were significantly inhibited by naked RPN2-PnkRNA (Fig. 5c, d). Furthermore, the lung specimens, including normal lung tissues, did not show any significant toxicity from the aerosol treatment (Fig. 5c). The significant effects did not involve weight loss and were associated with the specific down-regulation of mRNAs and protein at the molecular target (Supplementary Fig. 5a, b). In addition, the number of Ki-67 positive nuclei in naked RPN2-PnkRNA-treated cells was significantly decreased compared to the control group (Supplementary Fig. 5c). Therefore, the aerosol delivery of naked PnkRNA could be a unique and safe strategy for inhibiting lung cancer growth in vivo.
Figure 5. Repeat administration study with naked RPN2-PnkRNA in vivo.
(a) Analysis of the efficiency of naked RPN2-PnkRNA upon the inhibition of lung tumor growth after inhaled (i.h.) administration once a week for 3 weeks with bioluminescent imaging. Representative images of scid mice on days 28, 35, 42 and 49. Each experimental regimen consisted of twelve animals. (b) The bioluminescent change (days 35, 42 and 49/pre-treatment day 28) emitted from the whole bodies of the mice on day 28 was set to 100%. Statistical analysis was performed by the Bonferroni multiple-comparison test. The data represent the means ± SD (n = 12). *, P < 0.05 versus naked control-PnkRNA group. (c) Haematoxylin eosin-stained sections of each of the right lower lobes from the same mice that were imaged in Figure 5a, treated with naked control-PnkRNA or naked RPN2-PnkRNA on day 49. The scale bars indicate 100 μm. (d) The ratio of tumor/normal tissue area (%) of HE staining (on day 49). The data represent the means ± SD (n = 12). Statistical analysis was performed using the Bonferroni multiple-comparison test. *, P < 0.05 versus naked control-PnkRNA group.
Discussion
Our findings indicate that a novel class of RNAi therapeutic agents (PnkRNA, nkRNA) can be delivered to lung cancer by aerosols. Furthermore, we have demonstrated that a novel RNAi agent, RPN2, markedly suppressed the growth of A549-luc-C8 xenograft tumors in vivo. The novel RNAi agent was administered in naked, unmodified form by aerosol. To the best of our knowledge, our results present the first evidence that gene silencing by means of intrapulmonary delivery of naked, unmodified RNAi agents may have therapeutic potential in treating lung cancer. Despite the highly promising potential of RNAi for novel drug discovery, some obstacles still must be overcome before clinical applications5. The biggest hurdle remains a suitable delivery method. In this study, we were challenged to overcome this hurdle through a combination of unique RNAi platforms and a local delivery method.
A new class of RNAi agents, PnkRNA and nkRNA, have a unique helical structure containing a central stem and two loops. Their large-scale production at low cost is possible because they do not require an annealing step. Previously, we also reported that the intrapulmonary delivery of novel RNAi agents was not associated with the expression of interferon (IFN)-α or IFN-β in the mouse model of lung diseases, suggesting that they might provide a solution to safety concerns about the off-target effects of canonical siRNAs9. In this study, novel RNAi agents showed significant effectiveness in lung cancer xenograft models and proved to be more stable against nuclease degradation than canonical siRNAs. The in vitro assay with labeled RNAi therapeutic agents showed that naked PnkRNAs were incorporated into the cell cytoplasm by endocytosis without delivery vehicles; these results were reproduced in two independent lung cancer cell lines. Furthermore, we were successful at providing evidence for this technology by showing that a naked, unmodified PnkRNA significantly inhibited lung tumor growth without any serious toxicity. The naked RNAi therapeutic approach has high safety potential because no proinflammatory or toxicological response was induced by the delivery vehicles. In the specimens of xenograft mouse lungs, no necrosis, degeneration, anaplasia in pneumocytes, atelectasis or emphysema was detected after naked aerosol treatment (Fig. 5c). These data show that naked PnkRNA functions safely and efficiently in an aerosol delivery system. However, we believe that more dedicated studies with aerosol delivery are required to evaluate the possible untoward pulmonary toxicity in vivo. In general, naked RNAi agents can avoid many of the issues regarding side effects and toxicities that are related to the formulation of reagents and might be able to facilitate specific therapeutic molecular targeting, which is considered a promising strategy15,16. In addition, naked siRNAs can be administered to mice to down-regulate an endogenous or exogenous target without inducing an IFN response17. Taken together, strategies using naked novel RNAi agents can be altered to minimise the potential for off-target effects from RNAi therapeutic agents. As one significant limitation of this study, it is unknown whether these novel RNAi-therapeutics would be successful in a host with an intact immune system. We will test the efficacy of naked-PnkRNA delivery technology in immune-competent mice in a future study. Furthermore, we hope that the stability and effectiveness of this technology in humans will be evaluated in preclinical trials.
Dozens of RNAi-based therapeutics are currently undergoing preclinical and clinical trials, and these studies provide further opportunities for successful results2. Many of these studies are conducted through local administration to specific tissues, such as ocular, epidermal, pulmonary, colonic, and pancreatic tissues18. Thus, the direct-delivery approach can overcome barriers in clinical testing, and successful RNAi can offer high specificity to specific organs and fewer undesirable off-target effects compared to systemic administration19. The success of the delivery of RNAi-based therapeutics requires efficiency, convenience, and patient compliance with the delivery route. For these reasons, the aerosol delivery system of RNAi agents is a safe and powerful potential treatment for lung cancer; because the anatomical structure and location of the lungs make this simple, non-invasive approach possible and its high delivery efficiency reduces systemic side effects. Here, we provided evidence that the aerosol delivery of a naked, unmodified RNAi agent by the MicroSprayer™ technique results in a highly improved local distribution in the lung peripheries. Although further analysis is required, our novel technology delivered by aerosol can be altered to minimise the potential for off-target effects from RNAi therapeutic agents.
The potency and target specificity of the RNAi knockdown have generated considerable excitement as a new therapeutic modality for cancer therapy. There have already been significant improvements in RNAi-based therapeutics for the treatment of various cancers, including lung cancer20. Honma et al revealed that RPN2, which is part of an N-oligosaccharyltransferase complex, efficiently induced apoptosis in docetaxel-resistant human breast cancer cells21. Recently, RPN2 expression has also been shown to be a potential therapeutic target and promising prognostic marker for several types of cancer22,23. Remarkably, Zhu et al revealed that RPN2 is highly expressed in CD24+CD44+ stem-like pancreatic cancer cells22. We have also recently demonstrated that RPN2 is multifunctional, that it tightly regulates tumor survival, and that it is anti-apoptotic. RPN2 regulates cancer stem cell properties through the stabilization of mutant p53. Furthermore, RNAi-mediated knockdown in several types of cancer cells results in cell growth inhibition in vitro and in vivo24. These findings suggest that RPN2 could be a promising therapeutic target for several types of cancer. The data presented in this study demonstrate that the expression of RPN2 is involved in lung tumor growth in vitro and in lung cancer xenograft models. Furthermore, based on a rescue experiment, exogenous RPN2 expression can rescue the functional changes induced by specific RNAi knockdown. Therefore, our study indicated that RPN2 might serve as a potential target for gene therapy in lung cancer treatment. Although RPN2 RNAi agents efficiently inhibited the proliferation of A549-luc-C8 cells, further study is required to develop an RNAi-based therapy that induces cytocidal activity specific to the lung cancer cells. It would be worthwhile to investigate in a future study whether naked RPN2-PnkRNA treatment combined with docetaxel is a better therapeutic strategy against lung cancer.
A major limitation of this study is that it is entirely based on a single cell line, a single target. Non–small cell lung cancers (NSCLCs) harbor a single specific mutated oncogene like K-ras that is thought to be the primary genetic “driver” leading to lung cancer. In this experiment, we used mutant k-ras cell line and still do not know the RPN2 function is mutant k-ras-dependent phenotype or not. Therefore, we plan to utilize a naked RNAi-based therapeutic technology in other lung cancer cell lines and other gene targets to establish delivery efficacy in future studies.
In conclusion, we have developed a new technology with unique RNAi platforms delivered by aerosol. Our findings have provided new insights into the availability of naked and unmodified RNAi agents in RNAi therapeutic trials. These novel findings also have the potential to make an extremely valuable contribution to the development of lung cancer treatment as a reasonable class of RNAi-based therapy.
Methods
Reagents
The antibiotic solution (containing 10,000 U/mL penicillin and 10 mg/mL streptomycin), the trypsin-EDTA mixture (containing 0.05% trypsin and EDTA), RPMI-1640, FBS (foetal bovine serum) and pHrodo™ Red succinimidyl ester were obtained from Invitrogen (Carlsbad, CA). The pairs of each small interfering RNA (siRNA) and novel RNAi reagent, targeting luciferase mRNA, and human RPN2 mRNA (Supplementary Table 1, 2) were purchased from Bonac (Kurume, Japan). Allstars Negative Control siRNA was obtained from Qiagen (Hilden, Germany).
Preparation of novel RNAi agents
The preparation of proline diamide amidite and the novel RNAi agents has been previously described9. Novel RNAi agents are prepared as single-stranded RNA that self-anneals into a unique structure containing a double-stranded RNA with an unpaired site bound at the right and left ends by an oligonucleotide loop or by a non-nucleotide molecule (a proline derivative). Fig. 1 shows the schematic model of human RPN2, which was selected as a representative RNAi target.
RNA stability
To estimate the resistance to nucleases, 40 μl (2 μmol/l) of siRNA, nkRNA and PnkRNA directed against human RPN2 were incubated at 37°C with 1 μl of RNase Cocktail Enzyme Mix (Ambion, Foster City, CA) (RNase A 500 U/ml, RNase T1 20,000 U/ml). After the specified times, the ribonuclease reaction was stopped, and 2 μl of each sample was run on 3% agarose gel.
Cell line
A549-luc-C8 cells, a luciferase-expressing cell line derived from A549 human lung adenocarcinoma cells by stable transfection of the North American Firefly Luciferase gene expressed from the CMV promoter, were purchased from Xenogen. The human lung adenocarcinoma cell line PC14 was obtained from RIKEN BioResource Center (Tokyo, Japan). These cells were cultured in RPMI-1640 containing 10% heat-inactivated FBS and an antibiotic-antimycotic at 37°C in 5% CO2.
RNA extraction
Total RNA was extracted from cultured cells or mice lungs using QIAzol and miRNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. The purity and concentration of all RNA samples were quantified using NanoDrop ND-1000 (Thermo Scientific, San Jose, CA). For RPN2 mRNA analysis of mice lungs, the animals were sacrificed 24 h after the inhaled administration of each RNAi agent.
Quantitative Real-time PCR (qRT-PCR)
The reverse transcription reaction was performed with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) using a random hexamer primer. The synthesised cDNAs were quantified by SYBR Green I qRT-PCR. Quantitative real-time reverse transcription-PCR (qRT-PCR) analysis was conducted using primers for human RPN2 (forward:5′-CTTCCAGAGCCACTGTCCTC-3′; reverse: 5′-CCGGTTGTCACCTTCAACTT-3′). β-Actin (forward:5′-ATTGCCGACAGGATGCAGA-3′; reverse:5′-GAGTACTTGCGCTCAGGAGGA-3′) was used for normalisation. The relative amounts of RPN2 were measured using the 2(-Delta Delta C(T)) method. The reactions were performed with the ABI Prism 7300 Sequence Detection System (Applied Biosystems) at 95°C/10 min, followed by 40 cycles at 95°C/15 s and 60°C/30 s. All qRT-PCR reactions were performed in triplicate.
Transient transfection assays
A549-luc-C8 cells were plated on six-well plates at a density of 2 × 105 cells/well and grown overnight until 50–80% confluence was achieved to obtain maximum transfection efficiency. The cells were transfected with validated siRNA, the novel RNAi agents (PnkRNA, nkRNA) for RPN2, or Allstars Negative Control siRNA (Qiagen) at a final concentration of 25 nmol/l using the DharmaFECT 1 reagent (Thermo Scientific), according to the manufacturer's protocol. In the cDNA rescue experiment, an RPN2 Human cDNA ORF clone (Origene Technologies, Rockville, MD) or pEGFP-N1 (Clontech Laboratories, Mountain View, CA) was combined with each siRNA using DharmaFECT Duo (Thermo Scientific), according to the manufacturer's protocol. RPN2-siRNA targeting site is located 906 nt downstream of the ATG start codon of human RPN2 cDNA sequence on human RPN2 expression vector.
Cell Proliferation Assay (MTS assay)
A Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Kumamoto, Japan) was used in the cell proliferation assay. Five thousand cells per well were seeded in 96-well plates. The following day, the cells were replenished with fresh medium containing 25 nmol/l of each RNAi agent. After four days of culture, a plate was assayed by adding 10 μl of CCK-8 solution to each well, and the plate was further incubated for 4 h at 37°C. The absorbance at 450 nm was measured using Envision (PerkinElmer, Norwalk, CT).
RNAi therapeutic agent labelling with pHrodo™ Red succinimidyl ester and endocytosis assay
The fluorescence of the pHrodo™ Red dye increases as the pH decreases from neutral to acidic, making it an ideal tool to study endocytosis25. The amine-reactive forms of pHrodo™ Red succinimidyl ester were used for labelling the RNAi therapeutic agents. After being labeled according to the manufacturer's protocol, the oligonucleotides were purified by reverse-phase HPLC. A549-luc-C8 cells or PC14 cells were transfected with the labeled siRNAs or PnkRNAs. Each labeled oligonucleotide was transfected with or without DharmaFECT 1 reagent. Then, the plates were incubated at 37°C for 3 hours to allow endocytosis to run to completion. Hoechst 33342 was used as a DNA counter-stain (cyan). Microscopic analysis was performed with a FLUOVIEW FV10i confocal microscope.
In vivo imaging of RNAi therapeutic agents in mice with lung cancer
Animal experiments were performed in compliance with the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute. These studies were approved by the National Cancer Center Research Institute. Six- to seven-week-old male C.B-17/Icr-scid/scidJcl mice (CLEA Japan, Shizuoka, Japan) were used in the experiments. The animals were housed in a 12 h light/12 h dark cycle and provided with an autoclaved rodent diet and water ad libitum. The mice were injected intravenously with 2 × 106 A549-luc-C8 cells suspended in 0.25 ml of sterile Dulbecco's PBS via the tail vein (day 0). For in vivo imaging, the mice were administered 150 mg/kg D-luciferin (Promega, Madison, WI) by intraperitoneal injection. Ten minutes later, photons from the whole bodies of the animals were counted by measuring bioluminescence with an IVIS imaging system (Xenogen, Alameda, CA), according to the manufacturer's instructions. The data were analysed using LIVINGIMAGE 4.2 software (Xenogen). The development of lung cancer was monitored twice a week in vivo by bioluminescent imaging. Four weeks after tumor injection (day 28), the bioluminescence from the implanted cancer cells was measured, and the mice were divided into four treatment groups (single administration study, mice per group; n = 4) or two groups (repeated administration study, mice per group; n = 12) with equivalent levels of bioluminescence. In the single administration study, the individual mice were administered 15 μg of each RNAi agent with In vivo-jetPEI™ (Polyplus Transfection Inc, New York, NY) (resulting in a calculated 1:6 charge ratio of nucleic acid backbone phosphates to cationic lipid nitrogen atoms) in a volume of 25 μl on day 28 using an endotracheally inserted MicroSprayer™ aerosoliser (IA-1C; Penn-Century) and a high-pressure syringe (FMJ-250; Penn-Century, Philadelphia, PA)26. Data are from a representative experiment of three independent experiments. In the repeated administration study, the treatment—15 μg of each naked RNAi agent inhaled by using a MicroSprayer™ aerosoliser—was performed on days 28, 35, and 42 (once a week for 3 weeks, three treatments total). To control for mouse-to-mouse variability, the bioluminescence ratio for each mouse was normalised by dividing by each day of the post-treatment/pre-treatment (day28) ratio of luciferase intensity for that mouse. For in vivo knockdown analysis, animals were sacrificed 24 h after each RNAi application, and lungs were removed and processed for histology or knockdown determination by qRT-PCR (SYBR Green).
Lung histological findings
Lung tissues were fixed in 10% neutral buffered formalin, paraffin-processed, and sectioned at 5 μm. Formalin-fixed and paraffin-embedded slides were stained with haematoxylin and eosin (H&E) or used for immunohistochemical (IHC) staining. With regard to the histologic estimation of tumor burden, the freeware Image J (National Institutes of Health, Bethesda, Maryland, USA) was used. Upon the IHC staining, antigen retrieval was performed by autoclave in a 10 mmol/l sodium citrate buffer (pH 6.0), and the endogenous peroxidase activity was blocked with the Immuno Pure Peroxidase Suppressor (Pierce, Chester, UK). The slides were incubated with RPN2 (A-1, Santa Cruz Biotechnology, Santa Cruz, CA) or Ki-67 (M7240, Dako Cytomation, Copenhagen, Denmark) primary antibody at 4°C overnight. The next day, after washing, the samples were incubated with mouse peroxidase-conjugated anti-mouse IgG (ImmPRESS Reagent; Vector Labs, Burlingame, CA) for 1 h. The immunoreactions were visualised with diaminobenzidine, and the sections were counterstained with haematoxylin.
Immunofluorescence staining
To estimate the inhaled distribution of RNAi agents, 15 μg of Allstars NegativesiRNA Alexia Fluor 647 (Qiagen) with In vivo-jetPEI™ was aerosolised by means of a MicroSprayer™, and the lungs were harvested 6 h after application, processed for paraffin sectioning, and analysed by confocal microscopy. DAPI staining was carried out immediately before imaging. Imaging for the cyan (DAPI) and magenta (fluorescently labeled siRNA) channels was performed in sequential mode using the appropriate excitation and emission settings. Microscopic analysis was performed with a FLUOVIEW FV10i confocal microscope (OLYMPUS, Tokyo, Japan).
Statistical analysis
All experiments were repeated at least three times, and the results are expressed as the means ± SE. The statistical analyses were conducted using the Bonferroni multiple-comparison test. These analyses were performed with the Expert StatView analysis software (version 4; SAS Institute, Cary, NC). P < 0.05 was considered to be statistically significant.
Author Contributions
T. Ochiya and K.K. conceived the idea and coordinated the project. Y.F. performed a significant amount of the experimental work. T. Ochiya, Y.F., F.T., T.M. and T. Ohgi wrote the manuscript and prepared the figures and tables. In vivo experiments were carried out by Y.F. and F.T.
Supplementary Material
Supplemental Information
Acknowledgments
This work was supported in part by a grant-in-aid for the Third-Term Comprehensive 10-Year Strategy for Cancer Control of Japan; Project for Development of Innovative Research on Cancer Therapeutics (P-Direct); Scientific Research on Priority Areas Cancer, Scientific Research on Innovative Areas (“functional machinery for non-coding RNAs”) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology; the National Cancer Center Research and Development Fund (23-A-2, 23-A-7, 23-C-6,); the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NiBio), the Project for Development of Innovative Research on Cancer Therapeutics; and the Japan Society for the Promotion of Science (JSPS) through the “Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program)” initiated by the Council for Science and Technology Policy (CSTP). We thank Ayako Inoue and Maki Abe for her excellent technical assistance.
References
- Matranga C., Tomari Y., Shin C., Bartel D. P. & Zamore P. D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123, 607–620 (2005). [DOI] [PubMed] [Google Scholar]
- Davidson B. L. & McCray P. B. Jr Current prospects for RNA interference-based therapies. Nat Rev Genet 12, 329–340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita F. & Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci 97, 689–696 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H. & Rossi J. J. Strategies for silencing human disease using RNA interference. Nat Rev Genet 8, 173–184 (2007). [DOI] [PubMed] [Google Scholar]
- Pecot C. V., Calin G. A., Coleman R. L., Lopez-Berestein G. & Sood A. K. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer 11, 59–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernolovskaya E. L. & Zenkova M. A. Chemical modification of siRNA. Curr Opin Mol Ther 12, 158–167 (2010). [PubMed] [Google Scholar]
- Lares M. R., Rossi J. J. & Ouellet D. L. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol 28, 570–579 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Bobbin M. L., Burnett J. C. & Rossi J. J. Current progress of RNA aptamer-based therapeutics. Front Genet 3, 234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki T. et al. Efficacy of a novel class of RNA interference therapeutic agents. PLoS One 7, e42655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina J. R., Yang P., Cassivi S. D., Schild S. E. & Adjei A. A. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 83, 584–594 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam S. S., Owonikoko T. K. & Khuri F. R. Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin 61, 91–112 (2011). [DOI] [PubMed] [Google Scholar]
- Herbst R. S., Heymach J. V. & Lippman S. M. Lung cancer. N Engl J Med 359, 1367–1380 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W. & Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol 12, 175–180 (2011). [DOI] [PubMed] [Google Scholar]
- Agu R. U., Ugwoke M. I., Armand M., Kinget R. & Verbeke N. The lung as a route for systemic delivery of therapeutic proteins and peptides. Respir Res 2, 198–209 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J. K., Liang W. & Chan H. K. Pulmonary delivery of therapeutic siRNA. Adv Drug Deliv Rev 64, 1–15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A., Gallou-Kabani C., Mathieu J. R. & Cabon F. Systemic delivery and quantification of unformulated interfering RNAs in vivo. Curr Top Med Chem 9, 1117–1129 (2009). [DOI] [PubMed] [Google Scholar]
- Heidel J. D., Hu S., Liu X. F., Triche T. J. & Davis M. E. Lack of interferon response in animals to naked siRNAs. Nat Biotechnol 22, 1579–1582 (2004). [DOI] [PubMed] [Google Scholar]
- Burnett J. C. & Rossi J. J. RNA-based therapeutics: current progress and future prospects. Chem Biol 19, 60–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M., Hornung V. & Hartmann G. siRNA and isRNA: two edges of one sword. Mol Ther 14, 463–470 (2006). [DOI] [PubMed] [Google Scholar]
- Petrocca F. & Lieberman J. Promise and challenge of RNA interference-based therapy for cancer. J Clin Oncol 29, 747–754 (2011). [DOI] [PubMed] [Google Scholar]
- Honma K. et al. RPN2 gene confers docetaxel resistance in breast cancer. Nat Med 14, 939–948 (2008). [DOI] [PubMed] [Google Scholar]
- Zhu J., He J., Liu Y., Simeone D. M. & Lubman D. M. Identification of glycoprotein markers for pancreatic cancer CD24+CD44+ stem-like cells using nano-LC-MS/MS and tissue microarray. J Proteome Res 11, 2272–2281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurashige J. et al. RPN2 expression predicts response to docetaxel in oesophageal squamous cell carcinoma. Br J Cancer 107, 1233–1238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R. et al. Ribophorin II regulates breast tumor initiation and metastasis through the functional suppression of GSK3β. Sci. Rep. 3, 2474; 10.1038/srep02474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. & Burgess K. Fluorescent indicators for intracellular pH. Chem Rev 110, 2709–2728 (2010). [DOI] [PubMed] [Google Scholar]
- Bivas-Benita M., Zwier R., Junginger H. E. & Borchard G. Non-invasive pulmonary aerosol delivery in mice by the endotracheal route. Eur J Pharm Biopharm 61, 214–218 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information