Abstract
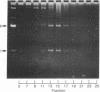
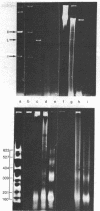
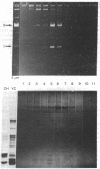
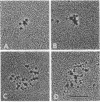
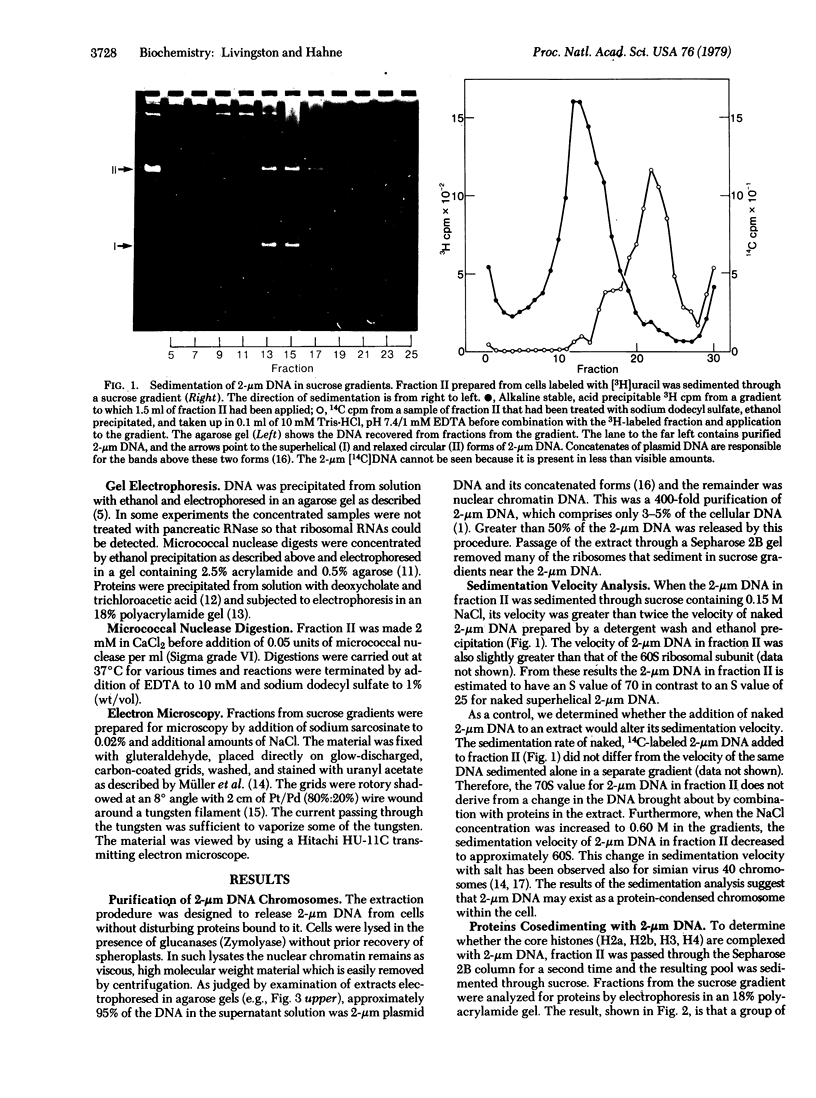
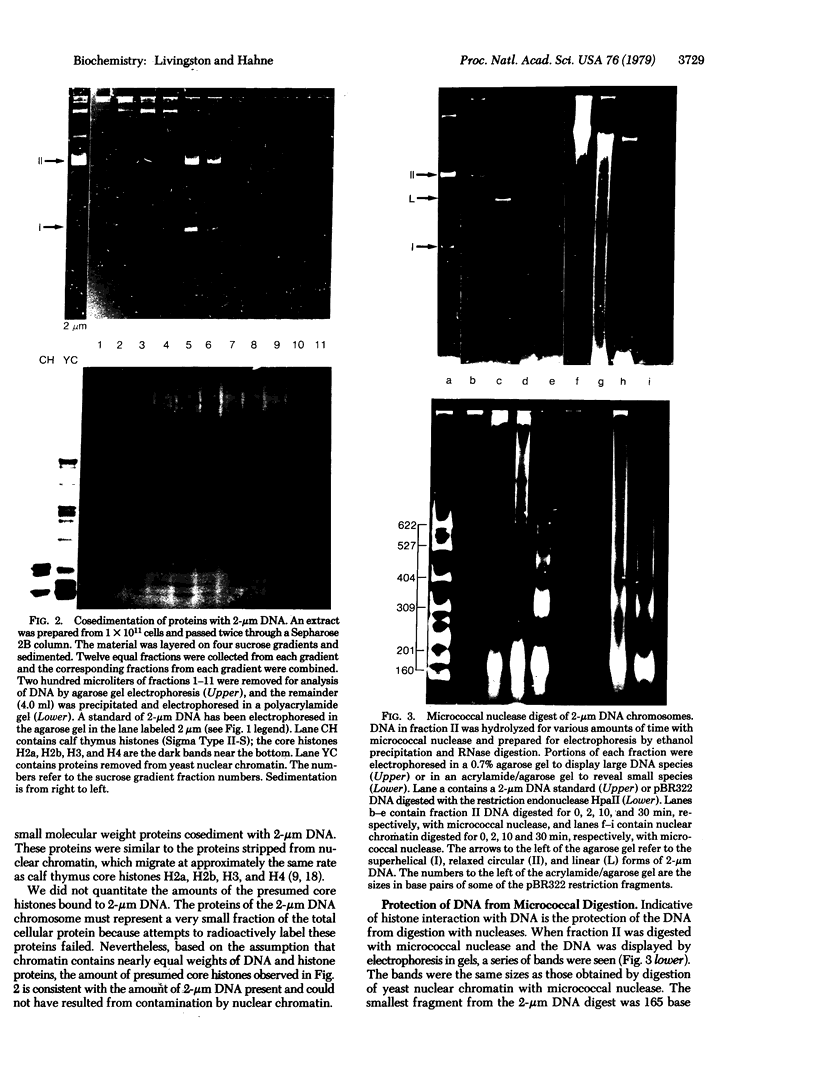
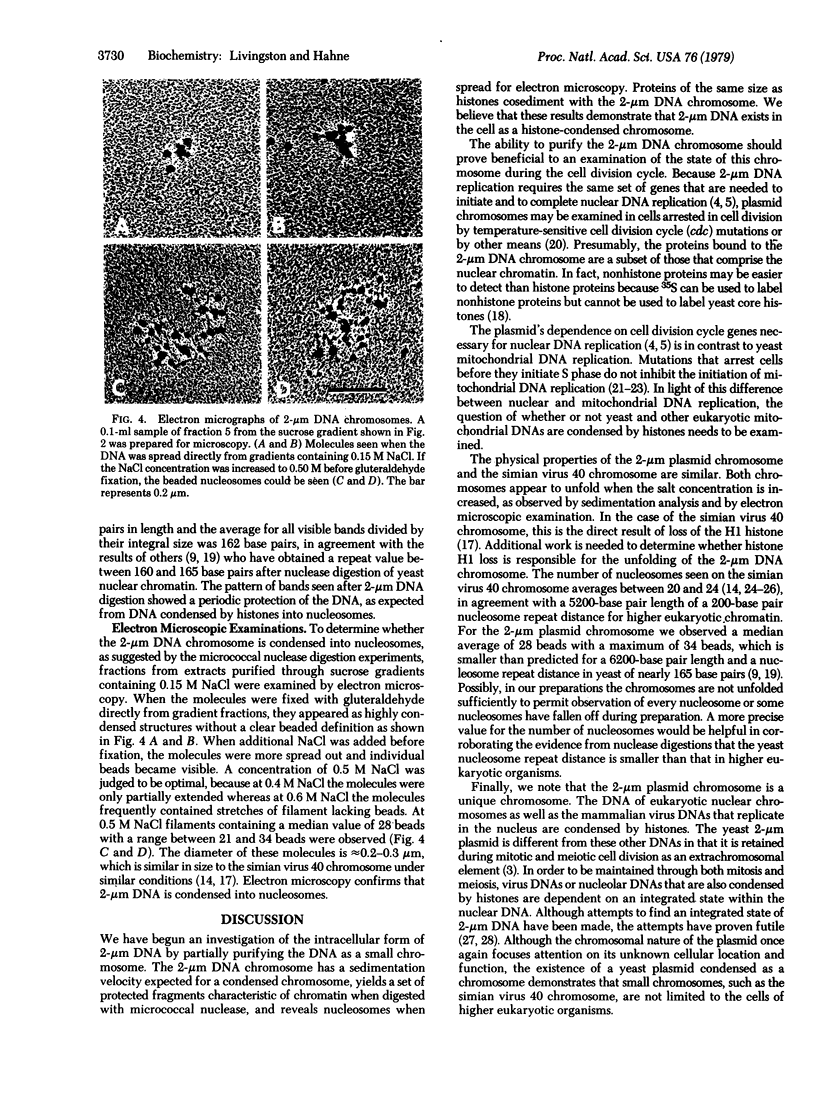
We have initiated an investigation of the proteins bound in vivo to the 2-micrometer DNA plasmid found in the yeast Saccharomyces cerevisiae by examining its intracellular form. To isolate 2-micrometer DNA without disturbing proteins bound to the plasmid, an extract was prepared from a strain lacking mitochondrial DNA and the nuclear chromatin was removed from the extract by centrifugation. When the DNA in this extract was sedimented through a sucrose gradient containing 0.15 M NaCl, plasmid DNA had a sedimentation coefficient of approximately 70. This S value is greater than the S value of 25 for naked, superhelical 2-micrometer DNA. Cosedimenting with the DNA were proteins of the same size as the histone proteins associated with yeast nuclear chromatin. Digestion of the plasmid DNA with micrococcal nuclease and electrophoresis of the resulting DNA fragments revealed that segments of discrete sizes are protected from degradation. Examination of the plasmid DNA protein complex by electron microscopy showed nucleosome structures. We conclude that 2-micron DNA exists as a condensed chromosome body within the cell.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adolphs K. W., Cheng S. M., Paulson J. R., Laemmli U. K. Isolation of a protein scaffold from mitotic HeLa cell chromosomes. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4937–4941. doi: 10.1073/pnas.74.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Oudet P., Germond J. E., Chambon P. Subunit structure of simian-virus-40 minichromosome. Eur J Biochem. 1976 Nov 15;70(2):543–553. doi: 10.1111/j.1432-1033.1976.tb11046.x. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Philippsen P., Davis R. W. Analysis of chromosomal integration and deletions of yeast plasmids. Nucleic Acids Res. 1977;4(5):1429–1448. doi: 10.1093/nar/4.5.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G., Griffith J. Salt and divalent cations affect the flexible nature of the natural beaded chromatin structure. Nucleic Acids Res. 1977 Jun;4(6):1837–1851. doi: 10.1093/nar/4.6.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D. Isolation of circular DNA from a mitochondrial fraction from yeast. Proc Natl Acad Sci U S A. 1972 Feb;69(2):388–392. doi: 10.1073/pnas.69.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D., Miklos G. L. Localization and quantification of circular DNA in yeast. Eur J Biochem. 1974 Jan 16;41(2):359–365. doi: 10.1111/j.1432-1033.1974.tb03278.x. [DOI] [PubMed] [Google Scholar]
- Cottrell S., Rabinowitz M., Getz G. S. Mitochondrial deoxyribonucleic acid synthesis in a temperature-sensitive mutant of deoxyribonucleic acid replication of Saccharomyces cerevisiae. Biochemistry. 1973 Oct 23;12(22):4374–4378. doi: 10.1021/bi00746a012. [DOI] [PubMed] [Google Scholar]
- Cremisi C., Pignatti P. F., Croissant O., Yaniv M. Chromatin-like structures in polyoma virus and simian virus 10 lytic cycle. J Virol. 1975 Jan;17(1):204–211. doi: 10.1128/jvi.17.1.204-211.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryer D. R., Goldthwaite C. D., Zinker S., Lam K. B., Storm E., Hirschberg R., Blamire J., Finkelstein D. B., Marmur J. Studies on nuclear and mitochondrial DNA of Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1974;38:17–29. doi: 10.1101/sqb.1974.038.01.005. [DOI] [PubMed] [Google Scholar]
- Franco L., Johns E. W., Navlet J. M. Histones from baker's yeast. Isolation and fractionation. Eur J Biochem. 1974 Jun 1;45(1):83–89. doi: 10.1111/j.1432-1033.1974.tb03532.x. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Saccharomyces cerevisiae cell cycle. Bacteriol Rev. 1974 Jun;38(2):164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston D. M. Inheritance of the 2 micrometer m DNA plasmid from Saccharomyces. Genetics. 1977 May;86(1):73–84. doi: 10.1093/genetics/86.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston D. M., Kupfer D. M. Control of Saccharomyces cerevisiae 2microN DNA replication by cell division cycle genes that control nuclear DNA replication. J Mol Biol. 1977 Oct 25;116(2):249–260. doi: 10.1016/0022-2836(77)90215-7. [DOI] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Yeast chromatin subunit structure. Science. 1975 Apr 11;188(4184):165–166. doi: 10.1126/science.1090006. [DOI] [PubMed] [Google Scholar]
- Mardian J. K., Isenberg I. Yeast inner histones and the evolutionary conservation of histone-histone interactions. Biochemistry. 1978 Sep 5;17(18):3825–3833. doi: 10.1021/bi00611a023. [DOI] [PubMed] [Google Scholar]
- Müller U., Zentgraf H., Eicken I., Keller W. Higher order structure of simian virus 40 chromatin. Science. 1978 Aug 4;201(4354):406–415. doi: 10.1126/science.208155. [DOI] [PubMed] [Google Scholar]
- Newlon C. S., Fangman W. L. Mitochondrial DNA synthesis in cell cycle mutants of Saccharomyces cerevisiae. Cell. 1975 Aug;5(4):423–428. doi: 10.1016/0092-8674(75)90061-6. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Molecular weight estimation and separation of ribonucleic acid by electrophoresis in agarose-acrylamide composite gels. Biochemistry. 1968 Feb;7(2):668–674. doi: 10.1021/bi00842a023. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Williamson D. H. Replicating circular DNA molecules in yeast. Cell. 1975 Mar;4(3):249–253. doi: 10.1016/0092-8674(75)90172-5. [DOI] [PubMed] [Google Scholar]
- Royer H. D., Hollenberg C. P. Saccharomyces cerevisiae 2-mum DNA. An analysis of the monomer and its multimers by electron microscopy. Mol Gen Genet. 1977 Feb 15;150(3):271–284. doi: 10.1007/BF00268126. [DOI] [PubMed] [Google Scholar]
- Scheer U., Trendelenburg M. F., Krohne G., Franke W. W. Lengths and patterns of transcriptional units in the amplified nucleoli of oocytes of Xenopus laevis. Chromosoma. 1977 Mar 16;60(2):147–167. doi: 10.1007/BF00288462. [DOI] [PubMed] [Google Scholar]
- Tabak H. F. Absence of 2 micrometer DNA sequences in Saccharomyces cerevisiae Y 379-5D. FEBS Lett. 1977 Dec 1;84(1):67–70. doi: 10.1016/0014-5793(77)81058-2. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Furber V. Yeast chromatin structure. FEBS Lett. 1976 Jul 15;66(2):274–280. doi: 10.1016/0014-5793(76)80521-2. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintersberger U., Smith P., Letnansky K. Yeast chromatin. Preparation from isolated nuclei, histone composition and transcription capacity. Eur J Biochem. 1973 Feb 15;33(1):123–130. doi: 10.1111/j.1432-1033.1973.tb02663.x. [DOI] [PubMed] [Google Scholar]