Abstract
When vertebrate cells exit mitosis various cellular structures are re-organized to build functional interphase cells1. This depends on Cdk1 (cyclin dependent kinase 1) inactivation and subsequent dephosphorylation of its substrates2–4. Members of the protein phosphatase 1 and 2A (PP1 and PP2A) families can dephosphorylate Cdk1 substrates in biochemical extracts during mitotic exit5,6, but how this relates to postmitotic reassembly of interphase structures in intact cells is not known. Here, we use a live-cell imaging assay and RNAi knockdown to screen a genome-wide library of protein phosphatases for mitotic exit functions in human cells. We identify a trimeric PP2A–B55α complex as a key factor in mitotic spindle breakdown and postmitotic reassembly of the nuclear envelope, Golgi apparatus and decondensed chromatin. Using a chemically induced mitotic exit assay, we find that PP2A–B55α functions downstream of Cdk1 inactivation. PP2A–B55α isolated from mitotic cells had reduced phosphatase activity towards the Cdk1 substrate, histone H1, and was hyper-phosphorylated on all subunits. Mitotic PP2A complexes co-purified with the nuclear transport factor importin-β1, and RNAi depletion of importin-β1 delayed mitotic exit synergistically with PP2A–B55α. This demonstrates that PP2A–B55α and importin-β1 cooperate in the regulation of postmitotic assembly mechanisms in human cells.
In the budding yeast, Saccharomyces cervisiae, Cdk1 substrate dephosphorylation and mitotic exit depend on the Cdc14 phosphatase7, but this function does not seem to be conserved in Cdc14 homologues of other species2,3,8–11. Studies in cycling Xenopus laevis embryonic extracts suggest that phosphatases of both the PP1 (ref. 5) and PP2A (ref. 6) families can contribute to Cdk1 substrate dephosphorylation during vertebrate mitotic exit, whereas Ca2+-triggered mitotic exit in cytostatic-factor-arrested egg extracts depends on calcineurin12,13. Early genetic studies in Drosophila melanogaster14,15 and Aspergillus nidulans16 reported defects in late mitosis of PP1 and PP2A mutants. However, the assays used in these studies were not specific for mitotic exit because they scored prometaphase arrest or anaphase chromosome bridges, which can result from defects in early mitosis.
Intracellular targeting of Ser/Thr phosphatase complexes to specific substrates is mediated by a diverse range of regulatory and targeting subunits that associate with a small group of catalytic subunits3,4,17. It is possible that mitotic exit in intact cells requires phosphatases that have not been detected by previous assays using in vitro extracts. In practice, the short duration of mitotic exit makes it difficult to assay this process, which explains why previous RNAi screening of cell division regulators18 did not annotate mitotic exit phenotypes.
To assay mitotic exit in live human cells, we measured the timing from anaphase onset until nuclear reformation. We generated a HeLa cell line stably expressing a chromatin marker (histone 2B fused to a red fluorescent protein; H2B–mCherry19) to visualize the metaphase–anaphase transition (Fig. 1a–c). To probe for postmitotic nuclear reassembly, we stably co-expressed a nuclear import substrate (importin-β-binding domain of importin-α fused to monomeric enhanced green fluorescent protein; IBB–eGFP20), which is cytoplasmic during mitosis and co-localizes with chromatin regions after reassembly of a functional nuclear envelope (Fig. 1a, b).
Figure 1.
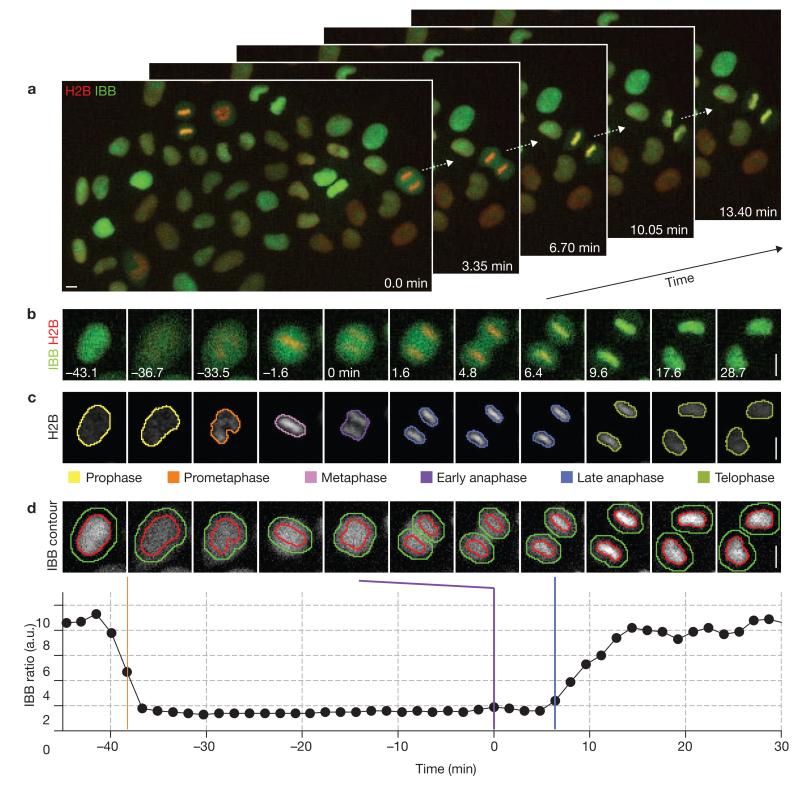
Live-cell imaging assay of mitotic exit timing. (a) Automated time-lapse microscopy imaging of a HeLa cell line stably expressing a chromatin marker (H2B–mCherry; red) and a nuclear import substrate (IBB–eGFP; green). The selected images show approximately 13% of a movie field-of-view. For full movie, see Supplementary Information, Movie S1. (b) Time-lapse microscopy of a single cell progressing through mitosis; onset of anaphase is marked as 0 min. (c) Automated detection of chromatin regions, tracking of cells over time, annotation of mitotic stages and classification of chromatin morphologies by supervised machine learning. For full movie, see Supplementary Information, Movie S2. (d) Automated detection of nuclear breakdown and reassembly. Chromatin regions (red outline) were defined by automated segmentation of the chromatin-associated H2B–mCherry fluorescence (as shown in c). Cytoplasmic regions (green outline indicates outer boundary, red outline is inner boundary) were derived by dilation of the chromatin regions. The ratio of mean IBB–eGFP fluorescence in chromatin versus cytoplasmic regions served to automatically determine nuclear envelope breakdown (orange bar) and reformation after anaphase (nuclear import of IBB, blue bar). Anaphase onset was defined by the classification of chromatin morphology (violet bar, see c). a.u., arbitrary units. Scale bars, 10 μm.
To annotate mitotic exit timing automatically in time-lapse microscopy movies, we used computational methods developed in-house (CellCognition21). Individual cells were detected and tracked over time, and the mitotic stage of each cell was assigned based on classification of chromatin morphology (Fig. 1c). Nuclear breakdown and reassembly was determined by recording changes in the ratio of mean IBB–eGFP fluorescence in chromatin regions versus surrounding cytoplasmic regions (Fig. 1d). Automated annotation of mitotic exit timing (4.70 ± 0.89 min; mean ± s.d.) closely matched manual annotation (4.88 ± 0.84 min; mean ± s.d.; n = 270 cells, Supplementary Information, Fig. S1).
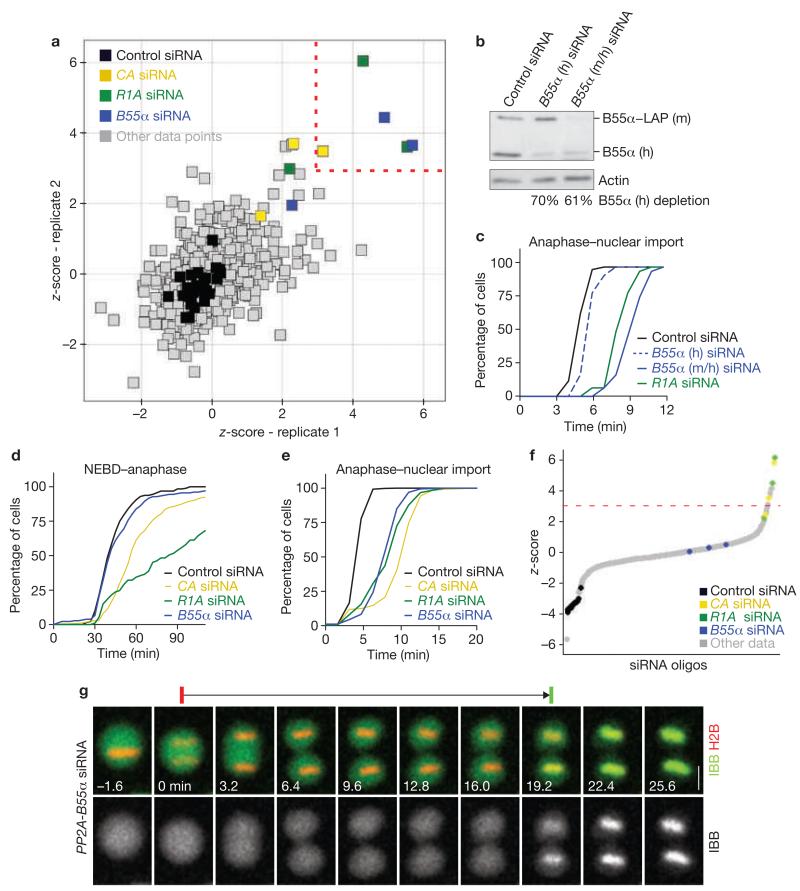
Cells were transfected with 675 different siRNAs targeting a genome-wide set of 225 annotated human protein phosphatases, including catalytic and associated regulatory and scaffolding subunits (three different siRNAs per gene, two experimental replicates; for a full list of siRNAs see Supplementary Information, Table 1). These cells were then imaged over approximately 24 h with time-intervals of approximately 3.8 min. On average, this yielded approximately 87 automatically annotated mitotic events per movie (total: 113,236 mitotic events). The mean mitotic exit timing was determined for all data sets that contained more than 10 mitotic events (n = 1,278 from 1,350 movies). Five siRNAs reproducibly scored as ‘hits’ above a significance cut-off at three standard deviations above the mean of all data points (z-score = 3; Fig. 2a). These five siRNAs targeted three distinct genes, encoding each of the three subunits of a heterotrimeric PP2A complex: PPP2CA (catalytic subunit α, subsequently labelled as CA), PPP2R1A (scaffold subunit α, subsequently labelled as R1A), and PPP2R2A (a regulatory B-type subunit, also termed PR55α or B55α and subsequently labelled as B55α). The remaining four siRNAs targeting the same PP2A genes also prolonged mitotic exit, albeit below the cut-off level (Fig. 2a).
Figure 2.
RNAi screen for mitotic exit regulators. (a) RNAi screen of a genome-wide library of annotated protein phosphatases using the mitotic exit assay shown in Figure 1. Individual data points correspond to the z-score based on the mean mitotic exit timing in individual movies, determined in two experimental replicates. Negative controls are black. siRNA oligos that resulted in phenotypes reproducibly scoring a z-score threshold of > 3 (dashed lines) were considered as hits (highlighted by colours as indicated in legend; siRNAs targeting the same genes as the hits but with z-scores below threshold are also highlighted). (b) RNAi depletion of B55α in a HeLa cell line stably expressing LAP-tagged mouse-B55α from a bacterial artificial chromosome (BAC; for localization see Supplementary Information, Fig. S6b). Cells were transfected with siRNA targeting either human B55α alone (B55α h), or both mouse- and human-B55α (B55α m/h). A quantitative western blot is shown, probed with an anti-B55α antibody that recognizes both mouse- and human-B55α. (c) Rescue of mitotic exit delay phenotype by exogenous B55α. HeLa cells expressing mouse-B55α–LAP were RNAi-depleted for human (h) or mouse/human (m/h) B55α (as validated in b), or for the scaffolding subunit of PP2A (R1A), and the timing of mitotic exit was then assessed. (d) Cumulative histograms for early mitotic progression. Nuclear envelope breakdown until anaphase onset was timed in control and RNAi-depleted HeLa cells stably expressing H2B–mCherry and IBB–eGFP (as in Figure 1; n ≥ 64, under all conditions). (e) Cumulative histograms of mitotic exit. Anaphase onset until nuclear import of IBB–eGFP was measured for the same cells shown in d. (f) Synthetic depletion RNAi screen for mitotic exit phosphatases. Assay, sample preparation and siRNA library was identical to the screen shown in a, except that siRNA targeting B55α was co-transfected in each experimental condition. Negative controls (black) were transfected by only non-targeting siRNA. The plot shows the ranked z-scores of a single replicate, calculated as in a. Dashed line indicates z-score threshold. (g) Time-lapse microscopy images of a cell depleted of all three PP2A–B55α subunits progressing from anaphase through mitotic exit (as indicated by red and green bars above the images). Full movie shown in Supplementary Information, Movie S4. For negative control, see Supplementary Information, Movie S3. Scale bar, 10 μm. Uncropped image of blot is shown in Supplementary Information, Fig. S9a.
To validate the hits, we tested an extended set of six siRNAs per gene, which all significantly prolonged mitotic exit (P < 0.001; Supplementary Information, Fig. S2a–c). The mRNA depletion levels correlated with phenotypic penetrance, indicating specificity of the phenotype (Supplementary Information, Fig. S2a–f). RNAi depletion was also efficient at the protein level (Supplementary Information, Fig. S2g). Next, we depleted endogenous B55α in a HeLa cell line stably expressing eGFP-tagged mouse B55α. Transfection of siRNA targeting a non-conserved sequence on the human B55α mRNA efficiently depleted endogenous B55α, but not the exogenous mouse B55α (Fig. 2b), and these cells showed normal mitotic exit timing (Fig. 2c). In contrast, transfection of siRNA targeting both human and mouse B55α mRNA, or R1A, efficiently delayed mitotic exit (Fig. 2c). This provides validation that on-target depletion of B55α mRNA causes the observed mitotic exit phenotype.
The PP2A protein phosphatase family is involved in many cellular processes, including earlier mitotic stages3. It is generally accepted that the regulatory B-type subunit confers substrate specificity and thereby regulates diverse functions of PP2A3,17. Depletion of CA or R1A significantly prolonged mitotic progression from nuclear envelope breakdown until anaphase (P < 0.001; Fig. 2d; same cells shown in Fig. 2e), as expected from the known functions of PP2A complexes in spindle assembly and chromosome cohesion (which involve other B-type subunits3). In contrast, early mitotic progression was unaffected in B55α-depleted cells. These data indicate that PP2A function is required at all stages of mitosis, whereas the B55α subunit is rate-limiting only for post-anaphase progression.
Depletion of PP2A–B55α subunits delayed, but did not arrest, mitotic exit (Fig. 2e). This may be explained by incomplete RNAi depletion or the involvement of other unknown factors with redundant function. To investigate if additional phosphatases become limiting in the absence of B55α, we screened the phosphatase-targeting siRNA library in a background of synthetic B55α RNAi depletion. Increased mitotic exit delay occurred upon co-depletion of R1A or CA subunits with B55α (2–3 oligos scoring above a z-score threshold of 3; Fig. 2f). However, none of the other 222 phosphatases showed a consistent additive increase in mitotic exit delay (five other siRNA oligos delayed slightly above the z-score threshold, but could not be confirmed by different siRNAs targeting the same genes). Co-transfection of siRNAs targeting all three PP2A–B55α subunits (labelled as PP2A–B55α siRNA throughout the manuscript) caused the most pronounced prolongation of mitotic exit (14.76 ± 6.50 min, n = 205 versus 4.86 ± 1.07 min in the control, n = 251; mean ± s.d.; Fig. 2g and Supplementary Information, Fig. S2g). The additive effect of combinatorial PP2A–B55α subunit depletions suggests that residual levels of this phosphatase may account for a slow and gradual mitotic exit. The fact that no other RNAi conditions further delayed mitotic exit in combination with B55α depletion underlines the particular importance of PP2A–B55α for mitotic exit.
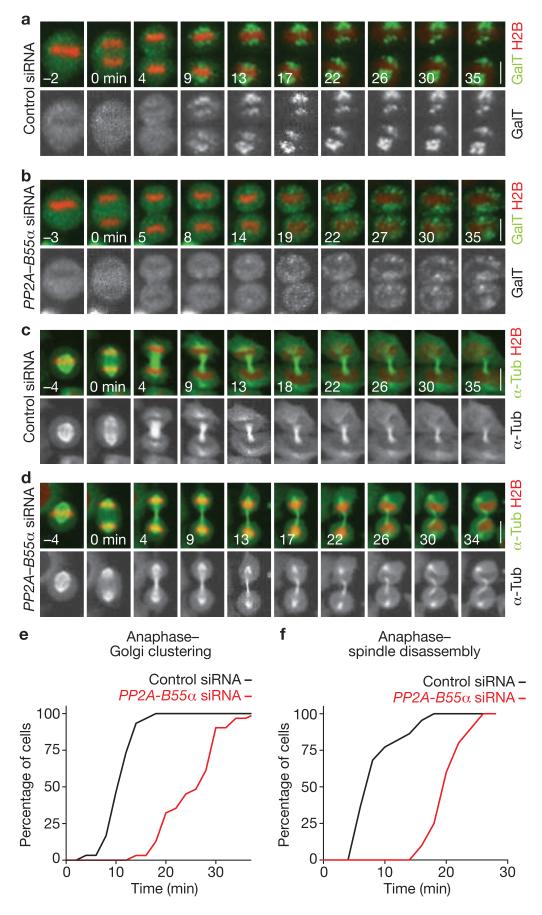
To address if PP2A–B55α controls postmitotic reassembly of cellular structures other than the nucleus, we generated a HeLa cell line stably expressing a fluorescent Golgi marker (galactosyl transferase, GalT–eGFP22) and H2B–mCherry (Fig. 3a). Time-lapse confocal microscopy showed that depletion of the PP2A–B55α complex significantly delayed postmitotic clustering of Golgi fragments (P < 0.001; 25.5 ± 5.86 min, n = 31 versus 11.17 ± 2.73 min in the control, n = 30; mean ± s.d.; Fig. 3a, b, e and see Supplementary Information, Fig. S3b, c for single depletion of B55α). In another mitotic exit assay, we imaged a HeLa cell line stably expressing H2B–mCherry and eGFP–α-tubulin19 (Fig. 3c). PP2A–B55α-depleted cells showed significantly delayed disassembly of spindle-pole-associated microtubules (P < 0.001; Fig. 3c, d, f). Postmitotic chromosome decondensation was also significantly delayed in PP2A–B55α-depleted cells (P < 0.001; 18.29 ± 2.29 min, n = 98 versus 8.94 ± 5.89 min in the control, n = 158; mean ± s.d.; see also Supplementary Information Fig. S3d). These data show that PP2A–B55α contributes to postmitotic reassembly of various interphase cell structures.
Figure 3.
PP2A–B55α controls postmitotic Golgi assembly, spindle breakdown and chromatin decondensation. (a) Images from a confocal microscopy time-lapse movie of a control cell expressing H2B–mCherry and the Golgi marker, GalT–eGFP (for full movie, see Supplementary Information, Movie S5). Golgi reassembly was scored based on clustering of the fluorescence into two distinct patches per cell (t = 9 min). (b) Golgi reassembly in a PP2A–B55α-depleted cell (for full movie, see Supplementary Information, Movie S6). (c) Confocal microscopy time-lapse images of mitotic spindle disassembly and chromosome decondensation in a control cell (for full movie, see Supplementary Information, Movie S7). Spindle disassembly was scored based on the first apparent detachment of spindle-pole-associated microtubules from chromatin masses (t = 9 min). (d) Mitotic spindle disassembly in a PP2A–B55α-depleted cell (for full movie, see Supplementary Information, Movie S8). (e, f) Cumulative histograms of postmitotic Golgi clustering (e), or spindle disassembly (f) relative to anaphase onset (t = 0 min). Scale bars, 10 μm.
We next investigated how depletion of PP2A–B55α affects progression through interphase, using a monoclonal cell line expressing H2B–mCherry and a DNA replication marker (proliferating cell nuclear antigen) tagged with enhanced GFP (eGFP–PCNA23; Supplementary Information, Fig. S4a–d). PP2A–B55α-depleted cells had a prolonged G1 phase (10.49 ± 2.70 h, n = 31 versus 5.77 ± 1.11 h in the control, n = 39; mean ± s.d.; Supplementary Information, Fig. S4b), as expected after perturbation of mitotic exit. Conversely, PP2A–B55α-depleted cells had a significantly shorter G2 phase (P < 0.001; 2.91 ± 0.39 h, n = 42 versus 3.95 ± 0.78 h in the control, n = 37; mean ± s.d.; Supplementary Information, Fig. S4d). This is consistent with previous observations in Xenopus embryonic extracts6,24.
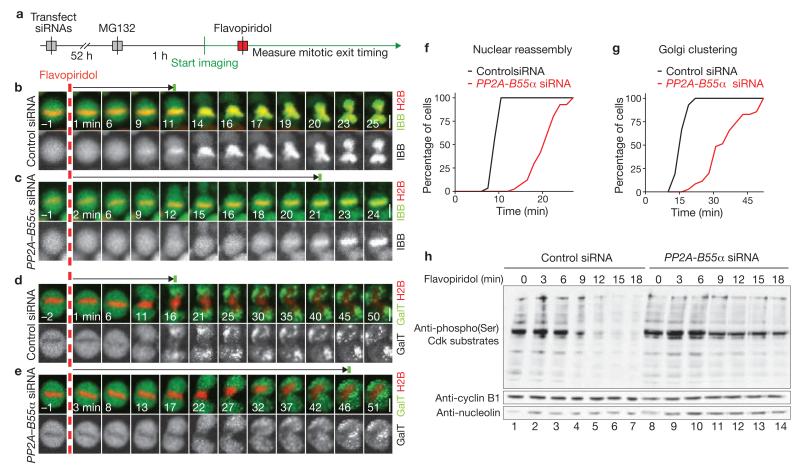
The mitotic exit phenotypes observed after PP2A–B55α depletion could be caused by a failure in Cdk1 inactivation, misregulated Cdk1 substrate dephosphorylation or both. To discriminate between these possibilities, we established a mitotic exit assay using chemical inactivation of Cdk1. Cells were first arrested in metaphase by the proteasome inhibitor MG132 and then forced to exit from mitosis by adding the Cdk inhibitor flavopiridol (still in the presence of MG132) to a final concentration of 20 μM (ref. 25; Fig. 4a). This treatment promoted changes that are indicative of mitotic exit, including nuclear reassembly (> 95% of all metaphase cells, n = 99), Golgi clustering (> 90%, n = 103), chromosome decondensation and re-attachment to the substratum (Fig. 4b, d). Only chromosome segregation did not occur, probably owing to the suppression of securin degradation by MG132. Higher concentrations of flavopiridol did not further accelerate nuclear reassembly (9.4 ± 1.1 min at 160 μM, n = 30 compared with 9.3 ± 1.3 min at 20 μM, n = 30; mean ± s.d.), indicating that Cdk1 inhibition was complete. PP2A–B55α depletion significantly delayed nuclear reassembly in this assay (P < 0.001; 20.67 ± 3.30 min, n = 41 versus 9.86 ± 0.95 min in the control, n = 29; mean ± s.d.; Fig. 4b, c, f and see Supplementary Information, Fig. S5a–c for single depletion of B55α). Golgi reformation was also delayed (36.01 ± 9.38 min, n = 35 versus 16.91 ± 2.27 min in the control, n = 42; mean ± s.d.; Fig. 4d, e, g). We conclude that a main function of PP2A–B55α in promoting mitotic exit must be downstream of Cdk1 inactivation.
Figure 4.
PP2A–B55α functions downstream of Cdk1 inactivation. (a) Experimental protocol for observation of mitotic exit induced by chemical inactivation of Cdks in absence of proteasome-mediated degradation. MG132 is the proteasome inhibitor and flavopiridol is the Cdk inhibitor. (b, c) Time-lapse microscopy images of cells expressing H2B–mCherry and IBB–eGFP. A control cell is shown in b (for full movie, see Supplementary Information, Movie S9) and a cell transfected with siRNA targeting PP2A–B55α is shown in c (for full movie, see Supplementary Information, Movie S10). Dashed red line indicates addition of flavopiridol, green bar indicates onset of IBB–eGFP nuclear import. (d, e) Golgi reassembly after chemically induced mitotic exit in cells expressing H2B–mCherry and the Golgi marker, GalT–eGFP. A control cell is shown in d and a cell transfected with siRNA targeting PP2A–B55α is shown in e. Dashed red line indicates addition of flavopiridol, green bar indicates onset of Golgi clustering. (f, g) Cumulative histograms of nuclear reassembly timing (f) and Golgi reassembly timing (g) based on the data shown in b–e. (h) Detection of Cdk1 substrate phosphorylation by an anti-phosphorylated-Ser antibody that specifically recognizes the Cdk target sequence K/R-pS-P-X-K/R (where X is any residue and pS is phosphorylated Ser) on a western blot. Samples were prepared after chemical induction of mitotic exit in synchronized cells in presence of proteasome inhibitor as in a. In control cells, Cdk substrates dephosphorylate rapidly (Control siRNA, lanes 1–7). Cells depleted for PP2A–B55α show delayed dephosphorylation (lanes 8–14). Scale bars, 10 μm. Uncropped image of blot is shown in Supplementary Information, Fig. S9b.
To test if PP2A–B55α depletion affected dephosphorylation of Cdk1 substrates, extracts were prepared from HeLa cells synchronized to different times after chemical induction of mitotic exit. Cdk1 substrate phosphorylation, detected on quantitative western blots by an anti-phosphorylated-Ser antibody that specifically recognizes the Cdk target sequence K/R-pS-P-X-K/R (where X is any residue and pS is phosphorylated Ser), dropped to approximately 23% within 18 min of flavopiridol addition in control cells, whereas it remained at approximately 66% in PP2A–B55α-depleted cells (Fig. 4h and see Supplementary Information, Fig. S5e for single depletion of B55α). This supports the conclusion that PP2A–B55α functions downstream of Cdk inactivation during mitotic exit and shows that PP2A–B55α is required for timely Cdk1 substrate dephosphorylation.
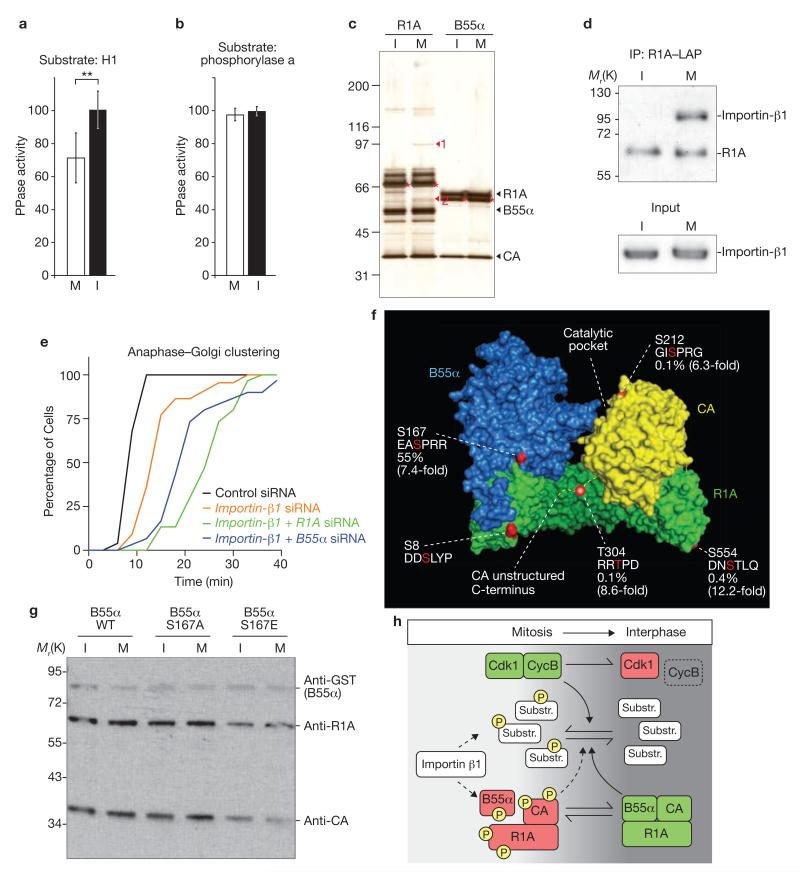
Previous studies in Xenopus embryonic extracts5,13 and HeLa cells26 have indicated that there is reduced phosphatase activity towards Cdk1 targets during early mitosis. Human PP2A–B55α isolated from mitotic cells had significantly reduced activity towards Cdk1-phosphorylated histone H1 (P < 0.001; 71 ± 15% versus 100 ± 11% in interphase; mean ± s.d., n = 15; Fig. 5a). However, we did not detect any cell cycle-dependent differences in the phosphatase activity towards another well-characterized model substrate, phosphorylase a (Fig. 5b). This suggests cell cycle-regulated changes of PP2A–B55α substrate specificity.
Figure 5.
Cell-cycle-dependent regulation of PP2A–B55α. (a, b) PP2A–B55α, isolated from nocodazol-treated mitotic (M) or unsynchronized interphase cells (I) by pulldown of GST–B55α, was assayed for (a) phosphatase activity towards Cdk1–cyclin B-phosphorylated histone H1 (n = 15, values indicate means ± s.d., asterisks denote P < 0.001; values normalized to interphase cells) and (b) activity towards its substrate, phosphorylase a (n = 3; values normalized to interphase cells. (c) Purification of PP2A complexes from interphase (I) or mitotic (M) HeLa cells stably expressing LAP-tagged R1A or B55α baits, resolved by SDS–PAGE and silver staining. Two mitosis-specific bands were identified by mass spectrometry: importin-β1 (1), and importin-α1 (2). Expected positions of the bands from endogenous PP2A subunits are indicated on the right and migration positions of mouse baits are marked with asterisks. (d) PP2A complexes were purified with R1A–LAP by immunoprecipitation and resolved on a western blot by probing with anti-R1A and anti-importin-β1 antibodies. (e) Importin-β1 function in mitotic exit. Cells expressing H2B–mCherry and GalT–eGFP were transfected with siRNAs as indicated. Timing from anaphase (t = 0 min) until Golgi reassembly was assayed as in Fig. 3 (n ≥ 30 for each condition). (f) Phosphorylation sites on PP2A–B55α were identified by mass spectrometry and are highlighted in red on the 3D structure of PP2A–B55α34 and in the associated primary sequences. The abundance of phosphorylated peptide in the mitotic sample was estimated by peak area quantification of the elution profiles and is indicated as percentage of total peptide based on elution profile peak area normalization. Mitotic phosphorylation increase (indicated in brackets) was estimated by comparing the normalized peak area quantifications of phosphorylated peptides in interphase with mitotic samples. (g) Phosphorylation of B55α Ser 167 affects PP2A complex assembly. GST-tagged wild-type-B55α or GST-tagged substitution mutants of B55α (a non-phosphorylatable S167A mutant or a phospho-mimicking S167E mutant) were isolated from unsynchronized (I) or mitotic (M) cells by GST-pulldown. PP2A subunits were detected on western blots by anti-GST, anti-R1A and anti-CA antibodies. (h) Model for mitotic exit control. Dephosphorylation of a broad range of mitotic Cdk1 substrates promotes reassembly of interphase cells during mitotic exit. A balance of kinase (Cdk1–cyclin B) and phosphatase (PP2A–B55α) activities determines the substrate dephosphorylation kinetics during mitotic exit. Green indicates activated state, red indicates lower activity and P indicates phosphorylation. Uncropped images of blot are shown in Supplementary Information, Fig. S9c.
All three PP2A–B55α subunits were expressed at similar levels in interphase and mitosis (Supplementary Information, Fig. S6a), indicating that mitotic PP2A–B55α is unlikely to be regulated at the protein level. We addressed potential changes in PP2A complex composition at different cell cycle stages by purification of LAP (localization and affinity purification)-tagged R1A or B55α, stably expressed from endogenous promoters27. R1A co-purified with several mitosis-specific binding partners (Fig. 5c), two of which were identified by mass spectrometry as the nuclear transport factors, importin-α1 and importin-β1. In addition to a function in nuclear transport during interphase, importin-β1 is part of a mitotic regulatory system involving the small GTPase Ran, which is known to control mitotic spindle and nuclear envelope assembly28. After validation of the mitosis-specific interaction between importin-β1 and R1A by western blotting (Fig. 5d), we investigated if importin-β1 contributes to mitotic exit progression. Because the specificity of the nuclear reassembly assay may be compromised after depletion of a nuclear import factor, we assayed mitotic exit by monitoring Golgi reformation. Importin-β1 depletion (Supplementary Information, Fig. S2h, i) significantly delayed postmitotic Golgi reassembly (P ≤ 0.001; 14.7 ± 5.0 min, n = 30 versus 10.1 ± 1.4 min in the control, n = 30; mean ± s.d.; Fig. 5e), which was further increased after co-transfection of siRNAs targeting R1A or B55α (25.9 ± 6.3 min, n = 30 and 22.5 ± 8.1 min, n = 25, respectively). Importin-β1 depletion also prolonged earlier mitotic stages (Supplementary Information, Fig. S7), as expected from its known function in spindle assembly. These data demonstrate that PP2A–B55α and importin-β1 jointly promote mitotic exit.
By mass spectrometry, we detected five phosphorylation sites on the PP2A complex purified from mitotic cells (Fig. 5f). The relative quantities of phosphorylated peptides were estimated by a semi-quantitative approach, using the extracted ion chromatogram for peak area quantification of the peptide elution profiles (Supplementary Information, Fig. S8). Phosphorylation of B55α at Ser 167 was estimated to be highly abundant at 55% (mean; n = 2 independent experiments), whereas the other sites were phosphorylated on < 1% of the eluted peptides (quantifications provided in Fig. 5f; the phosphorylation levels of S8 on R1A could not be determined because no unmodified peptides containing this site were detected). PP2A is known to auto-dephosphorylate29, therefore the absolute phosphorylation levels in cells may be higher. All four of the quantified phosphorylations were enriched more than fivefold on PP2A purified from mitotic cells (Fig. 5f; Supplementary Information, Fig. S8). To test if PP2A complex assembly is regulated by the phosphorylation of B55α at Ser 167, we isolated PP2A complexes from interphase and mitotic HeLa cells by expression of a GST (glutathione S-transferase)-tagged B55α phospho-mimicking S167E mutant. Indeed, the S167E mutant bound less efficiently to the CA and R1A subunits, compared with wild-type B55α or a non-phosphorylatable S167A mutant (Fig. 5g and Supplementary Information, Fig. S9h).
This study provides a comprehensive screen for mitotic exit phosphatases in human cells. B55α has been previously shown to increase PP2A activity towards Cdk1 phosphorylation sites30, consistent with the possibility that PP2A–B55α promotes mitotic exit by direct dephosphorylation of Cdk1 substrates. The in vitro phosphatase activity of mitotic PP2A–B55α towards Cdk1-phyosphorylated H1 was downregulated. However, it is not known which of the many putative PP2A substrates may be affected by this regulation, and therefore we cannot precisely estimate the extent to which PP2A regulation shapes the kinetics of Cdk1 substrate dephosphorylation.
Our data raise interesting possibilities for PP2A regulatory mechanisms (Fig. 5h). A B55α phospho-mimicking S167E mutation impaired the binding of R1A and CA subunits, consistent with the possibility that phosphorylation contributes to a cell cycle-dependent regulation of PP2A–B55α complex assembly. This may also involve mitotic hyper-phosphorylation of the CA subunit at Thr 304, as a previous study showed that a phospho-mimicking T304D mutation also suppresses assembly of B55α into PP2A complexes31. Importin-β1 may regulate PP2A by direct binding, by a nuclear-cytoplasmic targeting mechanism or as a molecular chaperone28. In this context, it is interesting to note that importin-β1 is structurally related to the R1A subunit and members of the B’ family of regulatory PP2A subunits32. Even though it is possible that importin-β1 functions in a mitotic exit pathway independently of PP2A, the physical and functional interaction observed here suggests a link between the importin-/Ran and Cdk1-phosphorylation regulatory systems.
In contrast to the Xenopus embryonic extract system6, depletion of B55δ in HeLa cells (down to 20% mRNA level; Supplementary Information, Fig. S2j) did not delay mitotic exit. This may reflect different relative expression levels of the B55-subfamily isoforms in the two systems or technical limitations of the depletion methods. A previous study proposed that PP1 dominates as a Cdk1-counteracting phosphatase in cycling Xenopus embryonic extracts5. The fact that we did not detect mitotic exit delays after RNAi depletion of any of the PP1 catalytic or regulatory subunits may be related to the cellular context of our phenotypic assays instead of homogenized extracts or to differences between embryonic and somatic mitosis. However, potential functional redundancy between different PP1 catalytic isoforms that were targeted only individually in our screen may have masked phenotypes; therefore we cannot rule out the possibility that PP1 also contributes to mitotic exit in human somatic cells.
In conclusion, our study reveals PP2A–B55α and importin-β1 as key regulators of cellular reassembly mechanisms during mitotic exit. Mitotic exit has been recently recognized as a target for improved, next-generation cancer therapeutics33. B55α and importin-β1 are therefore good targets for the development of mitotic exit-specific inhibitors.
METHODS
Cell lines and plasmids
The HeLa Kyoto cell line was obtained from S. Narumiya (Kyoto University, Japan) and cultured in Dulbecco’s modified eagle medium (DMEM; GIBCO) supplemented with 10% (v/v) foetal bovine serum (PAA Laboratories) and 1% (v/v) penicillin–streptomycin (Invitrogen). All live-cell imaging experiments were performed using monoclonal reporter cell lines that were generated as previously described35. For a complete list of plasmids and cell lines see Supplementary Information, Tables S3 and S4. Cells were grown either in 96-well microtitre plates (Greiner) or on LabTek chambered coverslips (Nunc) for live-cell microscopy. Live-cell imaging was performed in DMEM containing 10% (v/v) foetal calf serum and 1% (v/v) penicillin–streptomycin, but without phenol red and riboflavin to reduce autofluorescence of the medium35.
The bacterial artificial chromosomes (BACs), RP24-103C16, harbouring mouse PP2A–B55α (PPP2R2A), and RP24-255O20, harbouring mouse PPP2CA, were obtained from the BACPAC Resources Center (http://bacpac.chori.org). The LAP (eGFP–IRES (internal ribosome entry site)–neomycin) cassette was PCR amplified using primers that contain 50 nucleotides homologous to the carboxy terminus of each of the target genes. Recombineering and stable transfection of the modified BAC was performed as previously described27.
Live-cell imaging
Automated microscopy with reflection-based laser autofocus was performed on a Molecular Devices ImageXpress Micro screening microscope equipped with a ×10, 0.5 N.A. S Fluor dry objective (Nikon), controlled by Metamorph macros developed in-house21. Cells were maintained in a microscope stage incubator at 37 °C in a humidified atmosphere of 5% CO2 throughout the experiment. Illumination was adjusted so that the cell death rate was below 5% in untreated control cells. Confocal microscopy was performed on a customized Zeiss LSM 510 Axiovert microscope using a ×20, 0.8 N.A. Plan-Apochromat dry objective, a ×40, 1.3 N.A. oil DIC EC Plan-Neofluar objective, or a ×63, 1.4 N.A. oil Plan-Apochromat objective (Zeiss). The microscope was equipped with piezo focus drives (piezosystem jena), custom-designed filters (Chroma) and an EMBL incubation chamber (European Molecular Biology Laboratory), which provided a humidified atmosphere at 37 °C with 5% CO2.
For imaging chemically induced mitotic exit, cells were seeded in chambered LabTek coverslips overnight and then transfected with the indicated siRNAs. After transfection (52 h), the medium was replaced with imaging medium containing 30 μM MG132 (proteasome inhibitor; Sigma). The chambered coverslips were placed into the microscope stage incubator, which was maintained at 37 °C in a humidified atmosphere of 5% CO2, for 45 min, when imaging locations with metaphase-arrested cells were selected. These cells were imaged for 10–15 min, before mitotic exit was induced with 20 μM flavopiridol (Cdk inhibitor)25.
Image analysis and statistical analysis
Automated image analysis was performed by CellCognition software, which was developed in-house21 (http://www.cellcognition.org). Cell nuclei and mitotic chromosome masses were detected by local adaptive thresholding. Cytoplasmic regions were derived by region-growing the chromatin segmentation to a fixed size. Next, texture and shape features were calculated for each object and samples for mitotic classes were manually annotated for supervised classification. Support vector machine classification was performed by radial-based kernel and probability estimates. Cells were tracked over time using a constrained nearest-neighbour approach, with an algorithm that supported trajectory splitting and merging. Mitotic events were detected in the graph structure on the basis of the transition from prophase to prometaphase. Nuclear envelope reassembly was defined as an increase in the ratio of the mean nuclear versus cytoplasmic IBB–eGFP fluorescence > 1.5-fold above the ratio at the time of chromosome segregation. In the RNAi screen, mean mitotic exit timing was normalized per 96-well plate to compensate for slight differences in the temporal sampling rate. z-scores were calculated based on the mean and standard deviation of all data points. All statistical testing was by a two-tailed Student’s t-test.
RNAi
The human siRNA phosphatase library was based on version V3.0 from Qiagen and complemented with custom siRNAs targeting missing phosphatases. For a complete list of siRNA oligos see Supplementary Information, Tables S1 and S2. RNAi duplexes were transfected in liquid phase with either Oligofectamine (Invitrogen) or HiPerfect (Qiagen) according to the manufacturer’s instructions. Final siRNA concentrations were 50 nM for Oligofectamine or 10 nM for HiPerfect. Cells were reverse transfected in 96-well microtitre plates and incubated for approximately 40 h before imaging.
Quantitative real-time PCR
mRNA was extracted from cells 40 h after transfection of siRNAs using the TurboCapture 8 mRNA Kit (Qiagen) and cDNA was prepared using random hexamer primers (Microsynth) and Ready-To-Go You-Prime First-Strand beads (GE Healthcare). Real-time PCR was performed using the LightCycler 480 SYBR Green I Master system (Roche Diagnostics). Primers were designed using AutoPrime software (www.autoprime.de) or Clone Manager. Primer pairs for the indicated genes were: CA (5′-GGAGCTGGTTACACCTTTG-3′ and 5′-CCAGTTATATCCCTCCATCAC-3′), R1A (5′-CTTCAATGTGGCCAAGTCTC-3′ and 5′-TCTAGGATGGGCTTGACTTC-3′) B55α (5′-ATTCGGCTATGTGACATGAG-3′ and 5′-GACCTGTTACTGGGATCTTC-3′), B55δ (5′-CTGAAAGACGAAGATGGAAG-3′ and 5′-AATATTGGGACCCGTAGC-3′), importin-β1 (5′- CAGATACG AGGGTACGAGTG-3′ and 5′-TTTCATTGCTTCGATTGTG-3′) and GAPDH (5′-CGTGTCAGTGGTGGACCTGACC-3′ and 5′-CTGCTTCACCACCTTCTTGATGTCA-3′).
Protein blotting
Pelleted cells were washed with PBS and total protein lysates prepared in lysis buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 1% (v/v) Nonidet P-40, 10% (v/v) glycerol and 2 mM EDTA), supplemented with Mini-Complete protease inhibitor tablet (Roche), and 20 mM β-glycerophosphate. Protein concentrations were determined using a BCA kit (Pierce). The following primary antibodies were used: polyclonal rabbit-anti-PPP2CA (1:2,000, Cell Signaling), polyclonal rabbit-anti-PPP2R1A (1:2,000, Cell Signaling), monoclonal mouse-anti-PP2A–B55α (1:500, Santa Cruz), polyclonal rabbit-anti-phosphorylated-Ser Cdks substrate (1:1,000, Cell Signaling), monoclonal mouse-anti-cyclin B1 (1:5,000, Santa Cruz) and monoclonal mouse-anti-actin (1:50,000, Millipore).
For the chemically induced mitotic exit assay, nocodazole-arrested cells (100 ng ml−1 for 17 h; Sigma) were incubated for 30 min in 30 μM MG132 (Sigma), collected by shake-off, washed in PBS containing MG132 and resuspended in 800 μl of PBS (containing MG132). This suspension was divided into 100 μl aliquots in 1.5 ml centrifugation tubes at 37 °C. Mitotic exit was induced by adding flavopiridol to a final concentration of 20 μM (provided by the National Cancer Institute with permission by Sanofi-aventis). Cell aliquots were lysed at 3 min intervals over 18 min by adding 5× concentrated SDS (sodium dodecyl sulphate) cracking buffer to the respective aliquot, and boiling it for 5 min at 95 °C. Quantitative western blotting was performed using the Odyssey system (LICOR) or the FluorChem system (Alpha Innotec).
Immunoprecipitation and GST pulldowns
Subconfluent HeLa Kyoto cells expressing GST–B55α were untreated or arrested in mitosis with nocodazole (100 ng ml−1 for 16 h; M). Following collection by scraping (untreated cells) or shake off (cells arrested in mitosis), cells were washed in PBS, pooled and lysed in 1 ml NET buffer (50 mM Tris at pH 7.4, 150 mM NaCl, 1% (w/v) Nonidet P-40 and 15 mM EDTA), supplemented with Mini-Complete protease inhibitors (Roche) and, when appropriate, with phosphatase-inhibitor tablet (Roche). Equal amounts of the cleared lysates (as measured using the BCA kit; Pierce) were added to 25 μl of glutathione–Sepharose (GE Healthcare) and allowed to bind for 1 h at 4 °C. As previously shown31, this method allows isolation of catalytically competent PP2A–B55α trimers. After four washes in NENT-100 (ref. 31) and one wash in PP2A assay buffer (20 mM Tris at pH 7.4 and 5 mM DTT; dithiothreitol), PP2A assay buffer was added to the beads for phosphatase assays on two different substrates (histone H1 and phosphorylase a) for at least two times and with three replicates for each measurement (final volume 100–120 μl). A small aliquot was used for western blotting with anti-C and anti-A (monoclonals, 1:2,000 dilution, S. Dilworth) and anti-GST antibodies (1:10,000 dilution, Sigma) to check for equal input.
Phosphatase assays
Histone H1 (6 μg; Roche) was phosphorylated by Cdk1–cyclin B1 (Biaffin) or Cdk2–cyclin A36 in the presence of 32P-labelled ATP (GE Healthcare) to a level of 5–7 moles per mole of histone. 32P-radiolabelled histone H1 was precipitated in 25% (v/v) trichloroacetic acid (TCA) and washed; twice in 25% (v/v) TCA, once in a solution of acetone containing HCl (200:1) and once in acetone. This pellet was solubilized in PP2A assay buffer and used at a concentration of 0.3 μM. Following different incubation times with PP2A–B55α at 30 °C (2, 5, 10 or 20 min), release of inorganic 32P-phosphate was measured through extraction of a phosphate–molybdate complex and scintillation counting37. Under these conditions, phosphate release was < 10% of the phosphorylated substrate and thus remained in the linear range. Phosphorylase a phosphatase assays were performed as previously described38, by measuring the liberated TCA-soluble 32P-labelled phosphate following a 10- or 20-min incubation with PP2A–B55α. Each measurement of phosphatase activity was performed in triplicate. PP2A–B55α activities from mitotic cells were normalized to PP2A–B55α activities from interphase cells.
Affinity purification of protein complexes
HeLa cell pools expressing LAP-tagged mouse PP2A subunits from bacterial artificial chromosomes (BACs) were cultured in ten 25-cm square tissue culture trays. Cells were harvested from two culture conditions: prometaphase arrest induced by incubation in 0.1 μg ml−1 nocodazole for 18 h and during exponential growth, yielding cells typically > 90% in interphase. PP2A complexes were isolated from concentrated extracts of these cells using the two-step purification procedure previously described27, except that okadaic acid was included in the extract buffer and all subsequent solutions to inhibit auto-dephosphorylation. Purified protein complex (20%) was analysed by SDS–PAGE and silver staining. The remaining sample was processed as previously described39, then divided into two aliquots for parallel in-solution digestion by trypsin and subtilisin and analysis by liquid chromatography–mass spectrometry for protein identification and phosphorylation site mapping.
Trypsin digestion of SDS–PAGE gel slices
Selected silver-stained protein bands were excised from an SDS–PAGE gel, cut into smaller pieces and washed by incubating in a shaking incubator once with 200 μl of 50 mM triethyl ammonium bicarbonate (TEAB), then once with 100 μl acetonitrile (ACN) plus 100 μl of 50 mM TEAB. Each wash step was performed in a shaking incubator at room temperature. This two-step wash procedure was repeated, and then excess liquid removed. To shrink the gel pieces, 100 μl of ACN was added.
Proteins were reduced by incubating the gel pieces in 100 μl of 1 mg ml−1 DTT in 50 mM TEAB at 57 °C for 30 min, and removing the excess liquid. Proteins were alkylated by incubating the gel pieces with 100 μl of 5 mg ml−1 iodoacetamide in 50 mM TEAB at room temperature, in the dark, for 35 min. Gel pieces were subjected to the two-step wash and shrinking procedures, as previously described, and were then centrifuged in a vacuum concentrator (SpeedVac, Thermo Scientific) until dry (5–7 min).
Trypsin Gold (Promega) was first dissolved to a concentration of 100 ng μl−1 in 50 mM acetic acid, then diluted in 50 mM TEAB to a concentration of 12 ng μl−1. Gel pieces were incubated with 20 μl trypsin–TEAB solution at 4 °C for 5 min. Excess liquid was removed, and 20 μl of 50 mM TEAB was added, followed by incubation at 37 °C overnight. After centrifugation, the supernatant was transferred into a fresh tube and stored at 5 °C. Formic acid (20 μl of a 5% (v/v) solution) was added to the gel pieces, followed by sonication for 10 min in a cooled ultrasonic waterbath. The sample was spun and the supernatant transferred to the tube. This formic acid/sonication step was repeated once, and the supernatant pooled with the first, generating a sample of volume 60 μl, ready for analysis by mass spectrometry.
Nano-liquid chromatography–mass spectrometry
The nano-HPLC (high-performance liquid chromatography) system used in all experiments was an UltiMate 3000 Dual Gradient HPLC system (Dionex), equipped with a nanospray source (Proxeon), coupled to an LTQ FT mass spectrometer (Thermo Fisher Scientific) in the first analysis and coupled to an LTQ Velos Orbitrap mass spectrometer (Thermo Fisher Scientific) in the second analysis. The LTQ FT was operated in data-dependent mode using a full scan in the ICR (ion cyclotron resonance) cell followed by tandem mass spectrometry scans of the five most abundant ions in the linear ion trap. Tandem mass spectrometry spectra were acquired in the multistage activation mode, where subsequent activation was performed on fragment ions resulting from the neutral loss of −98, −49 or −32.6 m/z. Precursor ions selected for fragmentation were put on a dynamic exclusion list for 180 s and monoisotopic precursor selection was enabled. Phosphorylated peptides identified by database search and validated by manual inspection were put onto an inclusion list and replicate analyses were carried out on an LTQ Velos Orbitrap mass spectrometer.
Analysis of mass spectrometry data
For peptide identification, all tandem mass spectrometry spectra were analysed using Mascot 2.2.0 (Matrix Science) against a customized protein sequence database comprising the complete human sequences from Swiss-Prot, TrEMBL, PIR, GenBank, EMBL, DDBJ, RefSeq and Celera (hKBMS), plus the Swiss-Prot database entries corresponding to the mouse ‘bait’ proteins. The following search parameters were used: carbamidomethylation on cysteine was set as a fixed modification, and oxidation on methionine, and phosphorylation on serine, threonine and tyrosine were set as variable modifications. Monoisotopic masses were analysed using an unrestricted range of protein masses for tryptic peptides. The peptide mass tolerance was set to ± 5 p.p.m. and the fragment mass tolerance to ± 0.5. The maximal number of missed cleavages was set to two. Each tandem mass spectrometry spectrum corresponding to a predicted phosphorylated peptide was validated by visual inspection. A label-free quantification approach was used to determine abundance change from interphase to mitosis of the different peptides by peak area integration. For peak area quantification, we extracted the ion chromatograms of the peptides with a mass tolerance of 3 p.p.m. using the Qual Browser application from the Xcalibur software package. The mass traces of peak area integration for the phosphorylated peptides shown in Fig. 5f were extracted from the base peak chromatogram with ± 3p.p.m. mass tolerance, manually reviewed and are shown in Supplementary Information, Fig. S8.
Supplementary Material
Figure S1 Validation of automated image analysis. (a) Histograms of mitotic exit timing based on IBB-import after anaphase onset (t = 0) of the same dataset analysed manually (left, white bars), or automatically (right, black bars). (b) Automated annotation is highly reproducibly between movies (three independent movies shown).
Figure S2 Validation of RNAi efficiency. (a-c) Mitotic exit timing measured as in Figure 1, for six different siRNAs targeting CA (n ≥ 382 for each siRNA condition; mean±s.d) (a), R1A (n ≥ 175 for each siRNA condition) (b), or B55α (n ≥ 108 for each siRNA condition) (c). (d-f) Quantification of mRNA knockdown 40 h post transfection by real-time PCR for the same siRNAs as in (a-c), targeting CA (d), R1A (e), or B55α (f), normalized against GAPDH (n = 3 for each condition; mean±s.d). See Supplementary Information, Table 2 for siRNA sequences. (g) Protein depletion levels of CA, R1A, and B55α, detected by Western blotting in cells depleted for the indicated siRNAs 60 h post-transfection. Note that depletion of CA or R1A co-depletes other subunits of the PP2A-B55α complex, consistent with previous reports39, 40. (h) Quantification of Importin β1 mRNA knockdown 48 h post transfection. (i) Importin β1 protein levels, detected by Western blotting 64 h after siRNA transfection. (j) Quantification of B55δ mRNA knockdown 48 h post transfection.
Figure S3 Phenotypes of B55α single depletions. (a) Nuclear reassembly timing in a B55α-depleted cell, see also Supplementary Information, Movie S11. (b) Golgi reassembly timing in a B55α-depleted cell, see also Supplementary Information, Movie S12. (c-e) Cumulative histograms of postmitotic Golgi clustering (c), chromosome decondensation (d), and spindle disassembly (e) relative to anaphase onset (0 min). Scale bars: 10 μm.
Figure S4 Effect of PP2A-B55α-depletion on cell cycle progression. (a) Cell cycle staging by DNA replication factor pattern of EGFP-PCNA. (b, c, d) Cumulative histograms of G1-phase (b), S-phase (c), and G2-phase (d) duration. Scale bars: 10 μm.
Figure S5 B55α functions downstream of Cdk1-cyclin B inactivation. (a) Movie of a cell transfected with non-silencing control siRNA, following the protocol indicated in Figure 4a. Arrowhead indicates onset of nuclear import of IBB-EGFP. (b) Time-lapse images of a cell transfected with siRNA targeting B55α, see also Supplementary Information, Movie S13. (c, d) Cumulative histograms of nuclear envelope assembly (c) and Golgi clustering (d) timing based on the data shown in (a-b, and data not shown). (e) Chemical induction of mitotic exit in presence of proteasome inhibitor in nocodazole arrested mitotic cells results in rapid dephosphorylation of Cdk substrates (siControl, lanes 1-7). Cells depleted for B55α show delayed dephosphorylation (siB55α, lanes 8-14).Scale bars: 10 μm.
Figure S6 Expression levels and localization of PP2A-B55α. (a) PP2A-B55α subunits are expressed at constant levels during the cell cycle. Whole cell lysates were probed by Western Blotting using the antibodies against CA, R1A, and B55α. Left lane: 17 h nocodazole arrested cells (noc), right lane: unsynchronized interphase cells (log). Note that CA migrates as a doublet band (see also Supplementary Information, Figure S2g). (b) Localization of B55α. Confocal live images of cells expressing EGFP-mouse-B55α from endogenous promotor (see Fig. 2b). Scale bar: 10 μm.
Figure S7 Early mitotic progression in Importin β1-depleted cells. Timing from prometaphase until anaphase was measured for the same cells shown in Fig. 5e.
Figure S8 Quantification of phosphorylation levels by extracted ion chromatograms. Extracted ion chromatograms (XICs) of phosphopeptides identified (hash indicates the phosphorylated amino acid), and their unmodified counterparts, from purified PP2A complexes. Peak-area values for the phosphopeptides were used for quantification of the mitotic increase in phosphorylation abundance, whereas those of the unmodified peptides were used for normalization, as described in the legend for Fig. 5f. The panels show: 1. XIC featuring the phosphorylated peptide identified from the PP2A complex purified from interphase (log) cells. 2. XIC featuring the same phosphorylated peptide identified from the mitotic (noc) cell state. 3. XIC featuring the unmodified counterpart peptide from interphase. 4. XIC featuring the unmodified counterpart peptide from the mitotic (noc) cell state. (a) Quantitation of S554 phosphorylation on R1A. (b) S8 phosphorylation on R1A. (c) T304 phosphorylation on CA. (d) S212 phosphorylation on CA. (e) S167 phosphorylation on B55α.
Figure S9 Full scans of blots. (a) Full scan for B55α and Actin immunoblot of Figure 2b. (b) Full scan for phospho(Ser)Cdk substrates (upper) and Cyclin B1, Nucleolin, and Actin (lower) immunoblot of Figure 4f. (c) Full scan for Importin ß1 and R1A IP-blot (left) and Importin ß1 Input-blot (right) of Figure 5d. (d) Full scan for CA (upper left), R1A and B55α (lower left), and loading control blots (right panels) of Figure S2g. (e) Full scan for Importin ß1 and Actin immunoblot of Figure S2i. (f) Full scan for phospho(Ser)Cdk substrates (upper) and Cyclin B1, and Nucleolin (lower) immunoblot of Figure S5e. (g) Full scan for CA, R1A, and B55α (right) and Actin (left) immunoblot of Figure S6a. (Sliced bands are indicated with arrows). (h) Relevance of B55α-S167 phosphorylation for PP2A complex assembly. Independent replica of the experiment shown in Fig. 5g.
Supplementary Information Table 1. Library of siRNA oligos
Supplementary Information Table 2. SiRNA oligos targeting PP2A-B55α, B55δ and Importin ß1.
Supplementary Information Table 3. Plasmids generated for this study
For efficient generation of cell lines stably expressing fluorescently tagged marker proteins, the genes were subcloned into pIRES-puro2 and pIRES-neo3 vectors (Clontech) that allow expression of resistance genes and tagged proteins from a single transcript.
Supplementary Information Table 4. Monoclonal fluorescent reporter cell lines
Stable cell lines were generated as described in 5.
ACKNOWLEDGMENTS
The authors thank F. Uhlmann and B. Novak for critical comments on the manuscript. We thank S. Maar, the ETHZ Light Microscopy Centre (LMC), the ETHZ RNAi Screening Centre (RISC), M. Augsburg (The Max Planck Institute of Molecular Cell Biology and Genetics; MPI-CBG), M. Leuschner (MPI-CBG) and A. Ssykor (MPI-CBG) for technical assistance. We thank U. Kutay (ETHZ) for anti-importin-β1 and anti-nucleolin antibodies, J. Rohrer (University of Zurich) for providing GalT–eGFP plasmid, J. Ellenberg (EMBL, Heidelberg) for IBB–eGFP plasmid, M.C. Cardoso (Technical University, Darmstadt) for eGFP–PCNA plasmid and Sanofi Aventis and the National Cancer Institute for providing flavopiridol. This work was supported by Swiss National Science Foundation (SNF) research grant 3100A0-114120, SNF ProDoc grant PDFMP3_124904, a European Young Investigator (EURYI) award of the European Science Foundation to D.W.G., a MBL Summer Research Fellowship by the Evelyn and Melvin Spiegel Fund to D.W.G., a Roche Ph.D. fellowship to M.H.A.S. and a Mueller fellowship of the Molecular Life Sciences Ph.D. programme Zurich to M.H. M.H. and M.H.A.S. are fellows of the Zurich Ph.D. programme in Molecular Life Sciences. V.J. and J.G. were supported by grants of the ‘Geconcerteerde OnderzoeksActies’ of the Flemish government, the ‘Interuniversitary Attraction Poles’ of the Belgian Science Policy P6/28 and the ‘Fonds voor Wetenschappelijk Onderzoek-Vlaanderen’. A.I.L. is a Wellcome Trust Principal Research Fellow. A.A.H. acknowledges funding by the Max Planck Society, MitoCheck (the EU-FP6 integrated project), and a BMBF (Bundesministerium für Bildung and Forschung) grant, DiGtoP (01GS0859). Work in the groups of K.M. and J.M.P. was supported by MitoCheck (the EU-FP6 integrated project), Boehringer Ingelheim, the GEN-AU programme of the Austrian Federal Ministry of Science and Research (Austrian Proteomics Platform III), MeioSys within the Seventh Framework Programme of the European Commission and by Chromosome Dynamics, which is funded by the Austrian Science Foundation (FWF).
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Morgan DO. The cell cycle: principles of control. Oxford University Press; 2007. [Google Scholar]
- 2.Queralt E, Uhlmann F. Cdk-counteracting phosphatases unlock mitotic exit. Curr. Opin. Cell Biol. 2008;20:661–668. doi: 10.1016/j.ceb.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bollen M, Gerlich DW, Lesage B. Mitotic phosphatases: from entry guards to exit guides. Trends Cell Biol. 2009;19:531–541. doi: 10.1016/j.tcb.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Trinkle-Mulcahy L, Lamond AI. Mitotic phosphatases: no longer silent partners. Curr. Opin. Cell Biol. 2006;18:623–631. doi: 10.1016/j.ceb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Wu JQ, et al. PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat. Cell Biol. 2009;11:644–651. doi: 10.1038/ncb1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mochida S, Ikeo S, Gannon J, Hunt T. Regulated activity of PP2A–B55δ is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 2009;28:2777–2785. doi: 10.1038/emboj.2009.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- 8.Berdougo E, Nachury MV, Jackson PK, Jallepalli PV. The nucleolar phosphatase Cdc14B is dispensable for chromosome segregation and mitotic exit in human cells. Cell Cycle. 2008;7:1184–1190. doi: 10.4161/cc.7.9.5792. [DOI] [PubMed] [Google Scholar]
- 9.Gruneberg U, Glotzer M, Gartner A, Nigg EA. The CeCDC-14 phosphatase is required for cytokinesis in the Caenorhabditis elegans embryo. J. Cell Biol. 2002;158:901–914. doi: 10.1083/jcb.200202054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mailand N, et al. Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat. Cell. Biol. 2002;4:317–322. doi: 10.1038/ncb777. [DOI] [PubMed] [Google Scholar]
- 11.Mocciaro A, et al. Vertebrate cells genetically deficient for Cdc14A or Cdc14B retain DNA damage checkpoint proficiency but are impaired in DNA repair. J. Cell Biol. 2010;189:631–639. doi: 10.1083/jcb.200910057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishiyama T, Yoshizaki N, Kishimoto T, Ohsumi K. Transient activation of calcineurin is essential to initiate embryonic development in Xenopus laevis. Nature. 2007;449:341–345. doi: 10.1038/nature06136. [DOI] [PubMed] [Google Scholar]
- 13.Mochida S, Hunt T. Calcineurin is required to release Xenopus egg extracts from meiotic M phase. Nature. 2007;449:336–340. doi: 10.1038/nature06121. [DOI] [PubMed] [Google Scholar]
- 14.Mayer-Jaekel RE, et al. The 55 kd regulatory subunit of Drosophila protein phosphatase 2A is required for anaphase. Cell. 1993;72:621–633. doi: 10.1016/0092-8674(93)90080-a. [DOI] [PubMed] [Google Scholar]
- 15.Axton JM, Dombradi V, Cohen PT, Glover DM. One of the protein phosphatase 1 isoenzymes in Drosophila is essential for mitosis. Cell. 1990;63:33–46. doi: 10.1016/0092-8674(90)90286-n. [DOI] [PubMed] [Google Scholar]
- 16.Doonan JH, Morris NR. The bimG gene of Aspergillus nidulans, required for completion of anaphase, encodes a homolog of mammalian phosphoprotein phosphatase 1. Cell. 1989;57:987–996. doi: 10.1016/0092-8674(89)90337-1. [DOI] [PubMed] [Google Scholar]
- 17.Janssens V, Longin S, Goris J. PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail) Trends Biochem. Sci. 2008;33:113–121. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Neumann B, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steigemann P, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 20.Dultz E, et al. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Held M, et al. CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nat. Methods. doi: 10.1038/nmeth.1486. doi: 10.1038/nmeth.1486 (in the press) [DOI] [PubMed] [Google Scholar]
- 22.Schaub BE, Berger B, Berger EG, Rohrer J. Transition of galactosyltransferase 1 from trans-Golgi cisterna to the trans-Golgi network is signal mediated. Mol. Biol. Cell. 2006;17:5153–5162. doi: 10.1091/mbc.E06-08-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonhardt H, et al. Dynamics of DNA replication factories in living cells. J. Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee TH, Turck C, Kirschner MW. Inhibition of cdc2 activation by INH/PP2A. Mol. Biol. Cell. 1994;5:323–338. doi: 10.1091/mbc.5.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potapova TA, et al. The reversibility of mitotic exit in vertebrate cells. Nature. 2006;440:954–958. doi: 10.1038/nature04652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skoufias DA, Indorato RL, Lacroix F, Panopoulos A, Margolis RL. Mitosis persists in the absence of Cdk1 activity when proteolysis or protein phosphatase activity is suppressed. J. Cell Biol. 2007;179:671–685. doi: 10.1083/jcb.200704117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poser I, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harel A, Forbes DJ. Importin-β: conducting a much larger cellular symphony. Mol. Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 29.Guo H, Damuni Z. Autophosphorylation-activated protein kinase phosphorylates and inactivates protein phosphatase 2A. Proc. Natl Acad. Sci. USA. 1993;90:2500–2504. doi: 10.1073/pnas.90.6.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer-Jaekel RE, et al. Drosophila mutants in the 55 kDa regulatory subunit of protein phosphatase 2A show strongly reduced ability to dephosphorylate substrates of p34cdc2. J. Cell Sci. 1994;107:2609–2616. doi: 10.1242/jcs.107.9.2609. [DOI] [PubMed] [Google Scholar]
- 31.Longin S, et al. Selection of protein phosphatase 2A regulatory subunits is mediated by the C terminus of the catalytic Subunit. J. Biol. Chem. 2007;282:26971–26980. doi: 10.1074/jbc.M704059200. [DOI] [PubMed] [Google Scholar]
- 32.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Huang HC, Shi J, Orth JD, Mitchison TJ. Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell. 2009;16:347–358. doi: 10.1016/j.ccr.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Chen Y, Zhang P, Jeffrey PD, Shi Y. Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation. Mol. Cell. 2008;31:873–885. doi: 10.1016/j.molcel.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 35.Schmitz MH, Gerlich DW. Automated live microscopy to study mitotic gene function in fluorescent reporter cell lines. Methods Mol. Biol. 2009;545:113–134. doi: 10.1007/978-1-60327-993-2_7. [DOI] [PubMed] [Google Scholar]
- 36.Amniai L, et al. Alzheimer disease specific phosphoepitopes of Tau interfere with assembly of tubulin but not binding to microtubules. FASEB J. 2009;23:1146–1152. doi: 10.1096/fj.08-121590. [DOI] [PubMed] [Google Scholar]
- 37.Shacter-Noiman E, Chock PB. Properties of a Mr = 38, 000 phosphoprotein phosphatase. Modulation by divalent cations, ATP, and fluoride. J. Biol. Chem. 1983;258:4214–4219. [PubMed] [Google Scholar]
- 38.Waelkens E, Goris J, Merlevede W. Purification and properties of polycation-stimulated phosphorylase phosphatases from rabbit skeletal muscle. J. Biol. Chem. 1987;262:1049–1059. [PubMed] [Google Scholar]
- 39.Gregan J, et al. Tandem affinity purification of functional TAP-tagged proteins from human cells. Nat. Protoc. 2007;2:1145–1151. doi: 10.1038/nprot.2007.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Validation of automated image analysis. (a) Histograms of mitotic exit timing based on IBB-import after anaphase onset (t = 0) of the same dataset analysed manually (left, white bars), or automatically (right, black bars). (b) Automated annotation is highly reproducibly between movies (three independent movies shown).
Figure S2 Validation of RNAi efficiency. (a-c) Mitotic exit timing measured as in Figure 1, for six different siRNAs targeting CA (n ≥ 382 for each siRNA condition; mean±s.d) (a), R1A (n ≥ 175 for each siRNA condition) (b), or B55α (n ≥ 108 for each siRNA condition) (c). (d-f) Quantification of mRNA knockdown 40 h post transfection by real-time PCR for the same siRNAs as in (a-c), targeting CA (d), R1A (e), or B55α (f), normalized against GAPDH (n = 3 for each condition; mean±s.d). See Supplementary Information, Table 2 for siRNA sequences. (g) Protein depletion levels of CA, R1A, and B55α, detected by Western blotting in cells depleted for the indicated siRNAs 60 h post-transfection. Note that depletion of CA or R1A co-depletes other subunits of the PP2A-B55α complex, consistent with previous reports39, 40. (h) Quantification of Importin β1 mRNA knockdown 48 h post transfection. (i) Importin β1 protein levels, detected by Western blotting 64 h after siRNA transfection. (j) Quantification of B55δ mRNA knockdown 48 h post transfection.
Figure S3 Phenotypes of B55α single depletions. (a) Nuclear reassembly timing in a B55α-depleted cell, see also Supplementary Information, Movie S11. (b) Golgi reassembly timing in a B55α-depleted cell, see also Supplementary Information, Movie S12. (c-e) Cumulative histograms of postmitotic Golgi clustering (c), chromosome decondensation (d), and spindle disassembly (e) relative to anaphase onset (0 min). Scale bars: 10 μm.
Figure S4 Effect of PP2A-B55α-depletion on cell cycle progression. (a) Cell cycle staging by DNA replication factor pattern of EGFP-PCNA. (b, c, d) Cumulative histograms of G1-phase (b), S-phase (c), and G2-phase (d) duration. Scale bars: 10 μm.
Figure S5 B55α functions downstream of Cdk1-cyclin B inactivation. (a) Movie of a cell transfected with non-silencing control siRNA, following the protocol indicated in Figure 4a. Arrowhead indicates onset of nuclear import of IBB-EGFP. (b) Time-lapse images of a cell transfected with siRNA targeting B55α, see also Supplementary Information, Movie S13. (c, d) Cumulative histograms of nuclear envelope assembly (c) and Golgi clustering (d) timing based on the data shown in (a-b, and data not shown). (e) Chemical induction of mitotic exit in presence of proteasome inhibitor in nocodazole arrested mitotic cells results in rapid dephosphorylation of Cdk substrates (siControl, lanes 1-7). Cells depleted for B55α show delayed dephosphorylation (siB55α, lanes 8-14).Scale bars: 10 μm.
Figure S6 Expression levels and localization of PP2A-B55α. (a) PP2A-B55α subunits are expressed at constant levels during the cell cycle. Whole cell lysates were probed by Western Blotting using the antibodies against CA, R1A, and B55α. Left lane: 17 h nocodazole arrested cells (noc), right lane: unsynchronized interphase cells (log). Note that CA migrates as a doublet band (see also Supplementary Information, Figure S2g). (b) Localization of B55α. Confocal live images of cells expressing EGFP-mouse-B55α from endogenous promotor (see Fig. 2b). Scale bar: 10 μm.
Figure S7 Early mitotic progression in Importin β1-depleted cells. Timing from prometaphase until anaphase was measured for the same cells shown in Fig. 5e.
Figure S8 Quantification of phosphorylation levels by extracted ion chromatograms. Extracted ion chromatograms (XICs) of phosphopeptides identified (hash indicates the phosphorylated amino acid), and their unmodified counterparts, from purified PP2A complexes. Peak-area values for the phosphopeptides were used for quantification of the mitotic increase in phosphorylation abundance, whereas those of the unmodified peptides were used for normalization, as described in the legend for Fig. 5f. The panels show: 1. XIC featuring the phosphorylated peptide identified from the PP2A complex purified from interphase (log) cells. 2. XIC featuring the same phosphorylated peptide identified from the mitotic (noc) cell state. 3. XIC featuring the unmodified counterpart peptide from interphase. 4. XIC featuring the unmodified counterpart peptide from the mitotic (noc) cell state. (a) Quantitation of S554 phosphorylation on R1A. (b) S8 phosphorylation on R1A. (c) T304 phosphorylation on CA. (d) S212 phosphorylation on CA. (e) S167 phosphorylation on B55α.
Figure S9 Full scans of blots. (a) Full scan for B55α and Actin immunoblot of Figure 2b. (b) Full scan for phospho(Ser)Cdk substrates (upper) and Cyclin B1, Nucleolin, and Actin (lower) immunoblot of Figure 4f. (c) Full scan for Importin ß1 and R1A IP-blot (left) and Importin ß1 Input-blot (right) of Figure 5d. (d) Full scan for CA (upper left), R1A and B55α (lower left), and loading control blots (right panels) of Figure S2g. (e) Full scan for Importin ß1 and Actin immunoblot of Figure S2i. (f) Full scan for phospho(Ser)Cdk substrates (upper) and Cyclin B1, and Nucleolin (lower) immunoblot of Figure S5e. (g) Full scan for CA, R1A, and B55α (right) and Actin (left) immunoblot of Figure S6a. (Sliced bands are indicated with arrows). (h) Relevance of B55α-S167 phosphorylation for PP2A complex assembly. Independent replica of the experiment shown in Fig. 5g.
Supplementary Information Table 1. Library of siRNA oligos
Supplementary Information Table 2. SiRNA oligos targeting PP2A-B55α, B55δ and Importin ß1.
Supplementary Information Table 3. Plasmids generated for this study
For efficient generation of cell lines stably expressing fluorescently tagged marker proteins, the genes were subcloned into pIRES-puro2 and pIRES-neo3 vectors (Clontech) that allow expression of resistance genes and tagged proteins from a single transcript.
Supplementary Information Table 4. Monoclonal fluorescent reporter cell lines
Stable cell lines were generated as described in 5.