Abstract
hCINAP is an atypical nucleoplasmic enzyme, combining structural features of adenylate kinases and ATPases, which exhibits dual enzymatic activity. It interacts with the Cajal Body marker coilin and its level of expression and enzymatic activity influence Cajal Body numbers. Here we show that upon specific transcriptional inhibition of RNA pol.II, hCINAP segregates in perinuclear caps identified as Dark Nucleolar Caps (DNCs). These are distinct from perinucleolar caps where coilin and fibrillarin (both Cajal Body components) accumulate. In DNCs, hCINAP co-localizes with Paraspeckle Protein (PSP1) and also co-segregates with PSP1, and not coilin, in nuclear and nucleolar foci upon UV irradiation.
Keywords: Nuclear organization, Cajal Body, Coilin, Paraspeckle Protein 1, Paraspeckle
1. Introduction
Human Coilin Interacting Nuclear ATPase Protein (hCINAP) is a nuclear factor, originally identified as a protein interacting with the Cajal Body marker protein, p80 coilin [1]. hCINAP exhibits several unusual or unique properties. First, the hCINAP mRNA is an alternatively spliced transcript from the TAF9 locus, which also encodes the basal transcription factor TAFIID32, although the two proteins have no identity in their sequence due to differential usage of ATG starting codons and reading frames in the translation of the alternative transcripts [1]. Second, crystallographic analysis shows that while hCINAP has a structure typical for an adenylate kinase (AK), it also contains features characteristic of ATPase/GTPase proteins. Furthermore, it displays dual enzymatic activity of both an atypical AK, with unusually broad substrate specificity, and of an ATPase, an activity not reported for any other human adenylate kinase [2,3]. Intriguingly, His79, a B-motif amino acid residue, is crucial for the regulation of hCINAP’s dual enzyme selectivity, in response to intracellular substrate concentration [3].
At steady state hCINAP has a diffuse nucleoplasmic localization, excluding nucleoli, and although it does not concentrate in Cajal Bodies (CBs), its levels of expression and also its enzymatic activity influence CB organization. Specifically, overexpression of hCINAP decreases the average number of CBs per nucleus [1], while depletion of hCINAP causes defects in CB formation and redistribution of CB components [4]. Expression of an hCINAP mutant with His79 changed to Gly deregulates CB number, both increasing the average number of CBs per nucleus and also dramatically altering the frequency distribution of CBs, with numbers ranging from 0 to 30 per cell rather than 1–7 as seen when wild-type hCINAP is exogenously expressed [3]. Cajal Bodies are conserved nuclear organelles that serve as macromolecular assembly platforms (scaffolds), facilitating the maturation of splicing snRNPs and snoRNPs and other small nuclear RNPs involved in nuclear metabolic processes [5-8].
These findings have highlighted the putative importance of hCINAP in nucleotide homeostasis in the mammalian nucleus and in the assembly and/or stability of CBs. The challenge remains however to elucidate what is the mechanism through which hCIN-AP can impact on nuclear organization and eventually what is the biological function that is associated with hCINAP’s AK and ATPase activity in the nucleus, where movement or assembly of chromatin and nuclear organelles are ATP-dependent processes [9,10]. In this study we present new findings on hCINAP’s role in nuclear dynamics and organization.
2. Materials and methods
2.1. Cell culture and drug treatments
HeLa cells were cultured in GMax-DMEM (Invitrogen) supplemented with 10% fetal bovine serum and 100 U/ml penicillin-streptomycin at 37 °C with 5% CO2.
DRB (5,6-dichloro-β-d-ribofuranosylbenzimidazole) was added at a final concentration of 25 or 50 μg/ml and Actinomycin D at 0.04 μg/ml or 1 μg/ml. Cells were incubated for 3 h before sampling.
2.2. UV-Cirradiation
Semiconfluent cells were washed with PBS and the medium was collected and kept at 37 °C. The cells were irradiated in a UV Stratalinker 2400 oven at 254 nm with 30 J/m2. The saved medium was added back and cells were incubated for 6 h, prior to microscopic examination.
2.3. Generation of HeLaGFP-hCINAPstable cell lines
For the establishment of HeLaGFP-hCINAP, 5 μg of EGFP-hCINAP plasmid [circular (C) or linear (L)] was transfected into a 6 cm dish of HeLa cells using Lipofectamine 2000 (Invitrogen). After 18 h, cells were split at different dilutions (1:10–1:500) and medium containing 400 μg/ml G418 was added to select for cells that had stably incorporated the plasmid into their genomic DNA. After 14 days, visible colonies were picked, subcloned and expanded for biochemical and microscopic analyses.
2.4. Antibodies
Primary antibodies were: rabbit anti-hCINAP [1:1000 for Western blot (WB) and 1:800 for immunofluorescence (IF)] [1], mouse monoclonal anti-coilin 5P10 (1:50, IF) [11], rabbit anti-PSP1_48 (1:250, IF) [12], mouse monoclonal anti-fibrillarin AFB01 (1:200, IF, tebu-bio), mouse anti-GFP (1:1000, WB, Roche), mouse monoclonal anti-dynein (1:600, WB, Santa Cruz) and mouse monoclonal anti-α tubulin T5168 (1:6000, IF, Sigma). Secondary antibodies were: TRITC-conjugated goat anti rabbit IgGs (1:500, Jackson ImmunoResearch Laboratories), goat Cy5 anti mouse IgG (1:100, Jackson ImmunoResearch Laboratories), donkey Alexa Fluor 568 anti-rabbit IgG (1:600, Molecular Probes), donkey Alexa Fluor 555 anti-mouse IgG (1:1500, Molecular Probes), goat Alexa Fluor 350 anti-mouse IgG (H+L) (1:100, Molecular Probes), sheep HRP anti mouse IgG (1:6000, Amersham Pharmacia Biotech) and donkey HRP anti rabbit IgG (1:30 000, Santa Cruz). Nuclei were stained with Hoechst 33342 (0.5 μg/ml, Invitrogen).
2.5. Immunofluorescence microscopy
Immunofluorescence labeling was performed as previously described [3]. Deconvoluted fluorescent images were acquired with a Deltavision Restoration Microscope (Applied Precision) and a Micromax KAF1400 (Kodak) camera, and conventional fluorescent images with a C. Zeiss Axiovert 200M inverted fluorescence microscope equipped with an AxioCam MRm camera, using a ×63 1.3 or ×100 oil Plan-Apochromat objective lenses.
2.6. Reverse transcription-polymerase chain reaction (RT-PCR)
Poly A+ RNA from HeLa cultures was purified with the RNeasy Mini Kit (Qiagen) and cDNA was reverse transcribed from 2 μg of RNA using the Protoscript Kit (New England Biolabs). For the detection of endogenous hCINAP (excluding GFP-hCINAP) by semi-quantitative PCR, the upstream primer CINAP5UTR (gtagagcaaagggcacgtgagcgag) and the downstream CINAPRV (ccggaattcttaagtagctagccttataag) were used (product size 620 bp). For the detection of GFP-hCINAP (excluding endogenous hCINAP) the upstream primer GFPUP (ctcgtgaccaccctgacctac) was used in conjunction with primer CINAPRV (product size 1055 bp). The amplification of house keeping gene Pumilio 1 (PUM1) with primers PUM1UP (tgggaacaagagggcatctg) and PUM1RV (tgaggtgtgcaccatgaac) (product size 186 bp), was used as an internal reference reaction to normalize reaction conditions across samples. A mock RT reaction (2 μg of RNA, no reverse transcriptase) was used as negative control.
2.7. Cell cycle analysis by flow cytometry and analysis of mitotic progression by microscopy
To analyze cell cycle progression, cells in exponential growth were fixed with 70% ethanol for 2 h, stained for DNA content with propidium iodide, scored for their fluorescence on a FACSVantage SE and analyzed for their cell-cycle distribution using ModFit (Verity Software House).
The determination of the percentage of mitotic cells (mitotic index) and their assignment to mitotic subphases (n = 6000 cells) was performed by visual analysis using fluorescence microscopy of cells immunofluorescently labeled for α-tubulin and counterstained with Hoechst 33342. Statistical significance was assigned by two-way ANOVA analysis with Bonferroni post-test (GraphPad Prism).
3. Results and discussion
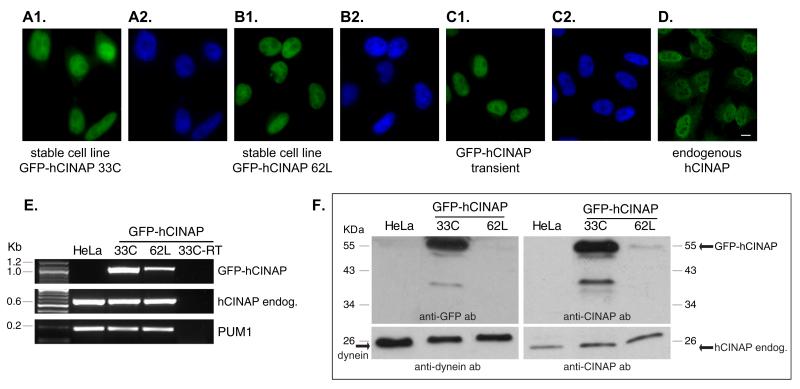
As a starting point, we constructed a HeLa cell line stably expressing GFP-hCINAP, following integration into the genome. Two stable clones, designated 33C and 62L, were pursued after purification with limiting dilution and expansion. Both stable clones displayed correct nuclear localization of GFP-hCINAP (Fig. 1A1, A2, B1, and B2, respectively), identical to GFP-hCINAP localization observed after transient transfection (Fig. 1C1 and C2) and also identical to endogenous hCINAP (Fig. 1D1 and D2). The level of expression of GFP-hCINAP was markedly greater in clone 33C compared with 62L, as assessed by fluorescence microscopy (note that exposure time in Fig. 1A1 is seven times lower than in B1) and confirmed (a) by RT-PCR, using oligonucleotide primers that would specifically amplify the GFP-hCINAP transcript (Fig. 1E, top panel) and (b) by Western immunoblotting, using anti-GFP tag antibodies (Fig. 1F, middle left panel) and anti-hCINAP antibodies (Fig. 1F, middle right panel). Expression levels of endogenous hCIN-AP transcript and endogenous protein in both clones were comparable to wild-type HeLa cells (Fig. 1E, middle panel; Fig. 1F, bottom right panel) while expression of GFP-hCINAP protein in clone 33C was much higher than that of endogenous hCINAP protein (note that the exposure time of right top immunoblot panel in Fig. 1F is half of that in the right bottom panel). We further characterized clones 33C and 62L, by evaluating their cell cycle profiles (Suppl. Fig. S1 and Suppl. Table 1), quantifying their mitotic indexes and mitotic phase distribution (Suppl. Fig. S2), and measuring the number of nuclei per cell (data not shown). There were minor differences in both clones as compared with wild-type cells, most notably a small increase in the percentage of aneuploid cells (Suppl. Fig. S1 and Suppl. Table 1) and small fluctuations in the distribution of cell cycle or mitotic subphases (Suppl. Fig. S2). However, both stable clones maintained a robust proliferation rate, normal morphology, stable expression and correct localization of hCINAP. Stable clone 33C was utilized for subsequent experiments.
Fig. 1.
Generation of stable cell lines. Expression of GFP-hCINAP in stable cell line 33C (A1 and A2), in stable cell line 62L (B1 and B2), or after transient transfection (C1 and C2), and endogenous hCINAP as detected by antibody staining (D1 and D2). Green panels represent GFP-hCINAP fluorescence or secondary antibody labeling (D1) and nuclei were labeled with Hoechst 33342 (blue panels). Scale bar 10 μm. (E) Detection by RT-PCR of the cDNA of GFP-hCINAP (top panel), endogenous hCINAP (middle panel), and equivalent reactions for house keeping gene PUM1 as internal control (bottom panel), in the stable cell lines 33C and 62L. (F) Detection of GFP-hCINAP protein and of endogenous hCINAP by Western immunoblot in the stable cell lines 33C and 62L by anti-GFP and anti-hCINAP antidodies, as indicated. Detection of dynein was used as internal loading control. Endog.: endogenous. All four blot panels derive from two identical SDS-PAGE gels that were run simultaneously but cut and probed with different antibodies. Exposure time for right bottom panel is 20 s and for all other panels 10 s.
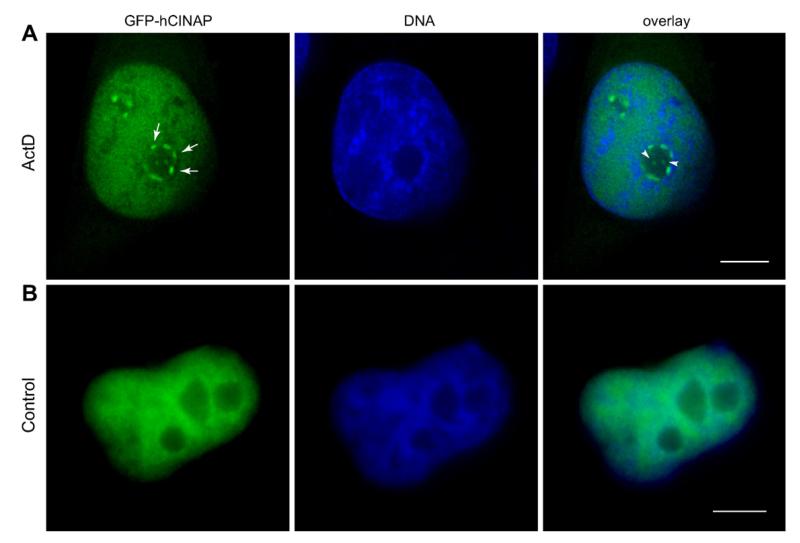
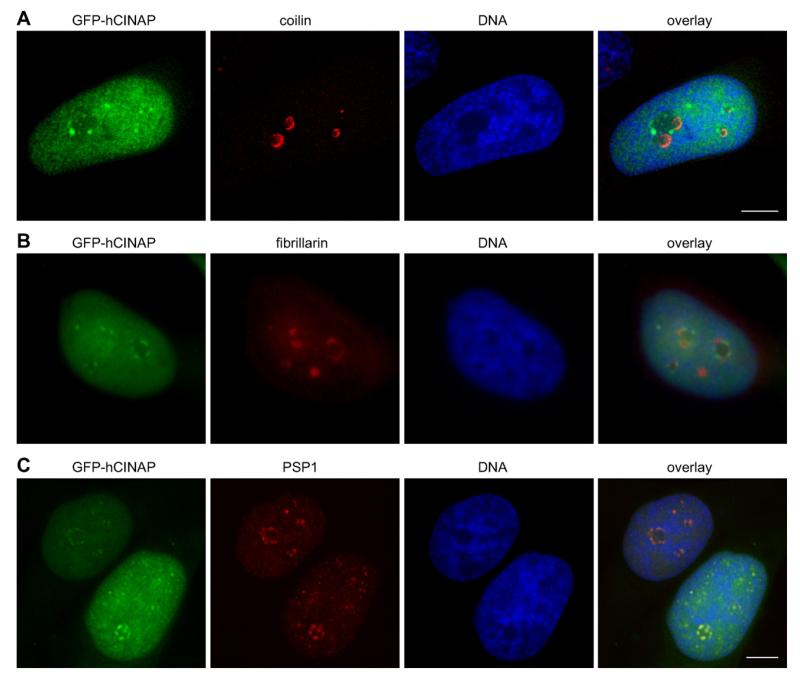
We subjected clone 33C cells to transcriptional arrest, using Actinomycin D at a concentration that causes inhibition of both RNA pol.II and pol.I (1 μg/ml), and observed a reproducible segregation of at least part of the nucleoplasmic GFP-hCINAP to perinucleolar caps and intranucleolar inclusions (Fig. 2, arrows and arrowheads, typically in >80% of treated cells). We confirmed that this phenotype was also detectable with endogenous hCINAP in wild-type HeLa cells (Suppl. Fig. S3). These perinucleolar caps were reminiscent of typical structures formed upon either physiological or experimentally-induced transcriptional arrest in mammalian cells [13,14]. In such conditions, the nucleolar, nucleoplasmic and nuclear body proteins and RNAs segregate and specifically relocalize into different nuclear subdomains, including distinct types of perinucleolar caps [10]. The most prominent of such caps are the so-called Dark Nucleolar Caps (DNCs) and the Light Nucleolar Caps (LNCs) [10]. Because of the known interaction of hCINAP with p80 coilin [1] and since coilin has been shown to segregate into LNCs during transcriptional arrest [15,16], we carried out double labeling for GFP-hCINAP and coilin to test whether the two proteins would be co-localized in the perinucleolar caps (Fig. 3A). In parallel, we also performed double labeling for GFP-hCINAP and fibrillarin (Fig. 3B), a nucleolar and Cajal Body component, also known to segregate to LNCs [10] and colocalizing with coilin when transcription is inhibited [17,10]. Surprisingly, in both sets of experiments, the lack of co-localization of GFP-hCINAP with coilin or fibrillarin (Fig. 3A and B, see also Suppl. Fig. 3 for endogenous hCINAP and Suppl. Fig. S4 for controls) indicated that (a) despite hCINAP’s interaction with coilin, the two proteins segregated differentially, and (b) the perinucleolar caps containing hCINAP were not LNCs.
Fig. 2.
GFP-hCINAP segregates in perinucleolar caps. (A) Transcriptional inhibition of RNA pol.II with Actinomycin D (1 μg/ml) causes the formation of GFP-hCINAP perinucleolar caps (arrows) and intranucleolar inclusions (arrowheads). GFP-hCINAP in green, nuclei in blue. (B) Equivalent control samples. Scale bar 5 μm.
Fig. 3.
GFP-hCINAP co-segregates with PSP1 in DNC, and not LNC, perinucleolar caps and intranucleolar inclusions upon transcriptional inhibition. HeLa 33C stable cell line was treated with Actinomycin D (1 μg/ml). Red panels: HeLa 33C stable cell line probed with antibodies coilin (A); fibrillarin (B); PSP1 (C); green panels: GFP-hCINAP fluorescence. Blue panels: nuclei labeled with Hoechst 33342. Equivalent control samples are shown in Suppl. Fig. S4. Scale bars 5 μm.
Paraspeckle Protein 1 (PPS1) is a marker protein of the nuclear organelle Paraspeckles and when RNA pol.II is inhibited it accumulates in perinucleolar caps [18] that have positively been identified as DNCs. Hence PPS1 can serve as a marker for DNCs following transcriptional inhibition [10]. Further analysis demonstrated that GFP-hCINAP and PPS1 co-localized in the same perinucleolar caps, therefore identifying these caps as DNCs (Fig. 3C).
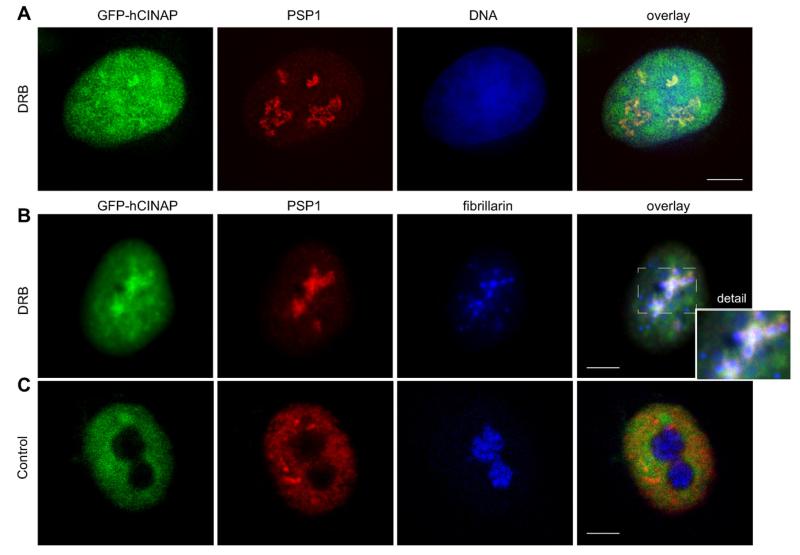
Because a high concentration of Actinomycin D causes a simultaneous inhibition of both RNA pol.I and pol.II, we sought to better characterize the co-segregation of hCINAP and PSP1 in DNCs. We utilized DRB, a widely used nucleoside analog that inhibits certain CTD kinases (carboxyterminal domain kinases) and affects positive and negative RNA pol.II elongation factors, thus acting as a specific inhibitor of RNA pol.II transcription. First, treatment of 33C cells with DRB, confirmed co-localization of GFP-hCINAP and PSP1 in DNCs, typically in >80% of treated cells (Fig. 4A for double GFP-hCINAP/PSP1 labeling). Including anti-fibrillarin as a nucleolar marker in triple labeling experiments, we also confirmed, as before with Actinomycin D, absence of co-localization of GFP-hCINAP/PSP1 with fibrillarin-positive fragments, resulting from the segregation of the nucleolus (Fig. 4B, enlarged detail in inset, and Fig. 4C for equivalent controls). Furthermore, when Actinomycin D was used at a low concentration (0.04 μg/ml), known to only inhibit RNA pol.I, we observed that GFP-hCINAP did not form perinucleolar caps or co-segregate with PSP1 (Suppl. Fig. S5).
Fig. 4.
Co-localization of GFP-hCINAP with PSP1 is specific to inhibition of RNA pol.II. (A) Double labeling of HeLa 33C stable cell line, treated with DRB. GFP-hCINAP fluorescence in green, PSP1 labeling in red and nuclei in blue. (B) Triple labeling of HeLa 33C, treated with DRB, showing GFP-hCINAP fluorescence (green), PSP1 (red) and fibrillarin (blue). The extracted area from the overlay image (dotted rectangle) is shown magnified in the inset and illustrates that fibrillarin-positive fragments of the nucleolus (blue) are in close proximity or in contact with but are distinct from co-segregated GFP-hCINAP (green) and PSP1 (red) caps. Note that segregation of the nucleolus, resulting from DRB treatment, has a less compact and more spread-out morphology, compared with the typical morphology resulting from Actinomycin treatment (as also documented in the literature [22-24]). (C) Equivalent examples as in B from control samples. Scale bar 5 μm.
These combined experiments therefore revealed that the re-distribution of hCINAP from its nucleoplasmic distribution to DNCs and its co-segregation with PSP1, recruited from the disassembled Paraspeckles, is specific to transcriptional arrest caused by RNA pol.II inhibition.
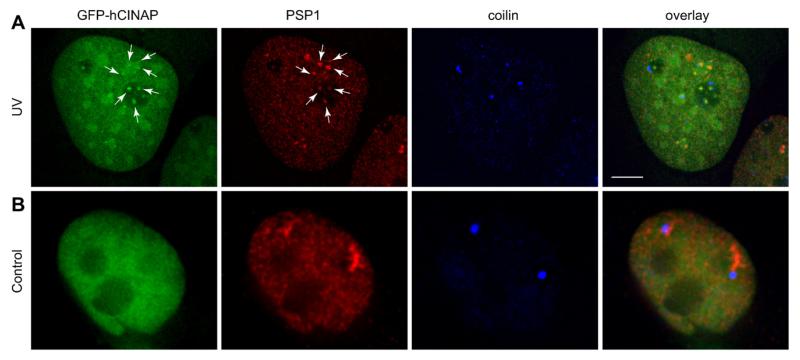
We next tested whether the co-segregation of the hCINAP and PSP1 proteins was a phenomenon that was only associated with transcriptional arrest, or if it also resulted from stress responses known to affect nuclear dynamics. UV-C irradiation, for instance, causes the disassembly of Cajal bodies to nuclear microfoci and differentially redistributes a subset of CB components [19]. Interestingly, when we subjected 33C cells to UV-C irradiation (254 nm at 30 J/m2) for 6 h, we found that GFP-hCINAP did not form perinucleolar caps but, instead, formed large rounded nuclear structures as well as small nuclear and intranucleolar foci. GFP-hCINAP and PSP1 were again co-localized in both nuclear and intranucleolar foci, which were, however, distinct from the coilin-containing microfoci resulting from UV-fragmentation of Cajal Bodies (Fig. 5A, compare with control panels in Fig. 5B).
Fig. 5.
GFP-hCINAP co-segregates with PSP1 but not with coilin following UV-irradiation. (A) GFP-hCINAP and PSP1 co-localization in nuclear and intranucleolar foci indicated with arrows. HeLa GFP-hCINAP fluorescence in green, PSP1 labeling in red and coilin labeling in blue. (B) Equivalent control samples. Scale bar 5 μm.
We have therefore observed that under two physiological conditions, i.e. RNA pol.II-specific transcriptional arrest and UV-induced DNA damage, hCINAP redistributes, at least partly, from its nucleoplasmic localization to different nuclear or intranucleolar compartments where, in both cases, it co-localizes with PSP1 that has relocated from disassembled Paraspeckles. It is unusual for a nucleoplasmic protein, such as hCINAP, to redistribute in this way as a previous analysis of >70 endogenous nucleoplasmic proteins has shown that most retained their original localization and were not compartmentalized upon transcriptional arrest [10].
While the differential redistribution of nucleolar components in transcriptional inhibition is now well established, our findings underline that segregation of different nuclear bodies involves a concerted process of specific redistribution of individual components into new compartments (such as the different types of perinucleolar caps) that includes the formation of new protein associations, as seen here between hCINAP and PSP1. We have not detected direct in vitro interaction between these two proteins (our unpublished observations), but transient or low affinity in vivo interaction could occur or these proteins may also associate indirectly via other partner proteins. Such interactions may take place in the nucleoplasmic fraction of these proteins in vivo, may be critical for self-assembly of functional complexes in the nucleus and may get stabilized in perinucleolar caps when transcription is inhibited.
Perinucleolar DNCs, where PSP1 and hCINAP transiently associate under conditions of RNA pol.II arrest, mostly contain proteins associated with RNA pol.II transcription [10]. Intriguingly however, both PSP1 and hCINAP also appear to have distinct topological or functional relationships with the nucleolus. PSP1 continually traffics through nucleoli, despite its distinctive steady-state enrichment within Paraspeckles, a fact that explains its original identification in the nucleolar proteome [18,20]. Additionally, Fap7, a yeast ortholog of hCINAP, has been found to be essential specifically for the cleavage of the 20S pre-rRNA from pre-40S particles and directly interacting with ribosomal protein RPS14 [21].
Acknowledgements
This work was supported by Grant YGEIA/0506/05 from the Research Promotion Foundation of Cyprus to N.S. AIL is a Wellcome Trust Principal Research Fellow.
Abbreviations
- DRB
5,6-dichloro-β-d-ribofuranosylbenzimidazole
- GFP
green fluorescent protein
- hCINAP
human coilin-interacting protein
- PSP1
Paraspeckle Protein 1
- RT-PCR
reverse transcription-polymerase chain reaction
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.febslet.2010.10.044.
References
- [1].Santama N, Ogg SC, Malekkou A, Zographos SE, Weis K, Lamond AI. Characterization of hCINAP, a novel coilin-interacting protein encoded by a transcript from the transcription factor TAFIID32 locus. J. Biol. Chem. 2005;280:36429–36441. doi: 10.1074/jbc.M501982200. [DOI] [PubMed] [Google Scholar]
- [2].Ren H, Wang L, Bennett M, Liang Y, Zheng X, Lu F, Li L, Nan J, Luo M, Eriksson S, Zhang C, Su XD. A crystal structure of human adenylate kinase 6: an adenylate kinase localised to the cell nucleus. Proc. Natl. Acad. Sci. USA. 2005;102:303–308. doi: 10.1073/pnas.0407459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zographos SE, Drakou CE, Malekkou A, Lederer CW, Hayes JM, Leonidas DD, Lamond AI, Santama N, Oikonomakos NG. hCINAP is an atypical mammalian nuclear adenylate kinase with ATPase activity: insights into its catalytic mechanism and function. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang J, Zhang F, Zheng X. Depletion of hCINAP by RNA interference causes defects in Cajal Body formation, histone transcription and cell viability. Cell. Mol. Life Sci. 2010;67:1907–1918. doi: 10.1007/s00018-010-0301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Matera GA, Shpargel KB. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Cur. Opin. Cell Biol. 2006;18:317–324. doi: 10.1016/j.ceb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- [6].Morris GE. The Cajal Body. BBA. 2008;1783:2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- [7].Pontes O, Pikaard CS. SiRNA and miRNA processing: new functions for Cajal Bodies. Cur. Opin. Genet. Dev. 2008;18:197–203. doi: 10.1016/j.gde.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Strzelecka M, Trowitzsch S, Weber G, Lührmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat. Struct. Biol. 2010;17:403–409. doi: 10.1038/nsmb.1783. [DOI] [PubMed] [Google Scholar]
- [9].Platani M, Goldberg I, Lamond AI, Swedlow JR. Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 2002;4:502–508. doi: 10.1038/ncb809. [DOI] [PubMed] [Google Scholar]
- [10].Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Almeida F, Saffrich R, Ansorge W, Carmo-Fonseca M. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol. 1998;142:899–912. doi: 10.1083/jcb.142.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to Paraspeckles in an RNA-dependent manner. Mol. Biol. Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Reynolds RC, Montgomery PO, Hughes B. Nucleolar “Caps” produced by Actinomycin D. Cancer Res. 1964;24:1269–1277. [PubMed] [Google Scholar]
- [14].Smetana K, Busch H. In: The nucleolus and nucleolar DNA in: The Cell Nucleus. Busch H, editor. Academic Press; NY: 1974. pp. 73–147. [Google Scholar]
- [15].Raska I, Ochs RL, Andrade LE, Chan EK, Burlingame R, Peebles C, Gruol D, Tan EM. Association between the nucleolus and the coiled body. J. Struct. Biol. 1990;104:120–127. doi: 10.1016/1047-8477(90)90066-l. [DOI] [PubMed] [Google Scholar]
- [16].Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis-evidence that the coiled body is a kinetic nuclear structure. J. Cell Biol. 1992;120:841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Santama N, Dotti CG, Lamond AI. Neuronal differentiation in the rat hippocampus involves a stage-specific re-organisation of subnuclear structure both in vivo and in vitro. Eur. J. Neurosci. 1996;8:892–905. doi: 10.1111/j.1460-9568.1996.tb01576.x. [DOI] [PubMed] [Google Scholar]
- [18].Fox AH, Lam YW, Leung AKL, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- [19].Cioce M, Boulon S, Matera GA, Lamond AI. UV-induced fragmentation of Cajal Bodies. J. Cell Biol. 2006;175:401–413. doi: 10.1083/jcb.200604099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Andersen JS, Lyon CE, Fox AH, Leung AKL, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- [21].Granneman S, Nandimeni MR, Baserga SJ. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol. Cell. Biol. 2005;25:10352–10364. doi: 10.1128/MCB.25.23.10352-10364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shav-Tal Y, Lee B-C, Bar-Haim S, Schori H, Zipori D. Reorganization of nuclear factors during myeloid differentiation. J. Cell. Biochem. 2001;81:379–392. doi: 10.1002/1097-4644(20010601)81:3<379::aid-jcb1052>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- [23].Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of Paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Martin C, Chen S, Maya-Mendoza A, Lovric J, Sims PFG, Jackson DA. Lamin A maintains the functional plasticity of nucleoli. J. Cell Sci. 2009;122:1551–1562. doi: 10.1242/jcs.046284. [DOI] [PMC free article] [PubMed] [Google Scholar]