Abstract
Human coilin interacting nuclear ATPase protein (hCINAP) directly interacts with coilin, a marker protein of Cajal Bodies (CBs), nuclear organelles involved in the maturation of small nuclear ribonucleoproteins UsnRNPs and snoRNPs. hCINAP has previously been designated as an adenylate kinase (AK6), but is very atypical as it exhibits unusually broad substrate specificity, structural features characteristic of ATPase/GTPase proteins (Walker motifs A and B) and also intrinsic ATPase activity. Despite its intriguing structure, unique properties and cellular localization, the enzymatic mechanism and biological function of hCINAP have remained poorly characterized. Here, we offer the first high-resolution structure of hCINAP in complex with the substrate ADP (and dADP), the structure of hCINAP with a sulfate ion bound at the AMP binding site, and the structure of the ternary complex hCINAP-Mg2+ADP-Pi. Induced fit docking calculations are used to predict the structure of the hCINAP-Mg2+ATP-AMP ternary complex. Structural analysis suggested a functional role for His79 in the Walker B motif. Kinetic analysis of mutant hCINAP-H79G indicates that His79 affects both AK and ATPase catalytic efficiency and induces homodimer formation. Finally, we show that in vivo expression of hCINAP-H79G in human cells is toxic and drastically deregulates the number and appearance of CBs in the cell nucleus. Our findings suggest that hCINAP may not simply regulate nucleotide homeostasis, but may have broader functionality, including control of CB assembly and disassembly in the nucleus of human cells.
Keywords: crystal structure, adenylate kinase 6, ATPase, coilin, Cajal bodies
INTRODUCTION
Adenylate kinase (AK; phosphotransferase; EC 2.7.4.3) is an abundant enzyme class, catalyzing the interconversion of ATP, ADP, and AMP, according to the reversible reaction: Mg2+ADP + ADP ↔ Mg2+ATP + AMP. The adenylate kinase system is able to provide ATP rapidly during intense activity and thus regarded as a reserve energy system used to regenerate ATP from ADP under energy stress conditions.
Five AK isoforms (AK1–5) with various substrate specificities and tissue distributions are known in vertebrates.1–5 These isoforms localize to the cytoplasm (AK1 and AK5) or mitochondria (AK2, AK3, and AK4). They are key enzymes in nucleotide homeostasis, maintaining the physiological nucleotide ratios in different cell compartments and cell types. Structurally, AKs are typical α/β proteins sharing a common architecture that is consistent with a β-sheet CORE domain, bearing a phosphate-binding loop (P-loop), and two flanking mobile domains called the LID and NMP-binding domains.6 hCINAP (Human Coilin Interacting Nuclear ATPase Protein), also now known as AK6 because of its AK activity,2 is a 21 kDa monomeric enzyme that has unique properties compared with the other known AKs. It exhibits unusually broad substrate specificity,2 displays intrinsic ATPase activity and has a nuclear localization.1 It also possesses a number of atypical structural features for an adenylate kinase, namely a Walker B motif and a metal-coordinating residue, highly conserved and typical among the members of the ATPase/GTPase superfamily.7–10 The enzymatic classification of hCINAP as a pure AK therefore remains open to discussion.
So far, hCINAP orthologs have been identified in yeast (Fap7), C. elegans (cAK6) and D. melanogaster (dAK6).11 In C. elegans, knockdown of the cAK6 gene results in growth suspension.12 In yeast, Fap7p13 interacts with ribosomal protein RPS14 and appears to be involved in pre-rRNA processing: a Walker B-motif mutant (D82AH84A) is unable to cleave the 20S pre-rRNA from 40S pre-rRNA particles.14 In human cells, hCINAP was shown to interact directly with the carboxy-terminal tail of coilin, a marker protein of Cajal bodies (CBs), which are nuclear organelles involved in the maturation of UsnRNPs and snoRNPs en route to their sites of function in the nucleoplasm and nucleolus.1 Furthermore, overexpression of hCINAP causes a decrease in the average number of CBs per nucleus in human cells.1 Despite these intriguing associations with RNA metabolism in the nucleus, the functional role of hCINAP is also still poorly defined.
The unique localization and structural features of hCINAP, indicating an atypical dual enzymatic activity and its undefined biological function prompted us to undertake this investigation. We present here the structural analysis of hCINAP in complex with ADP, dADP, SO42− and Mg2+ADP-PO43− and also provide a model for the hCINAP-Mg2+ATP-AMP complex based on induced fit docking calculations. These structures provide a detailed mapping of the catalytic site, identifying the residues involved in substrate recognition in conjunction with the binding of the substrates and could prove useful for additional structural studies, site-directed mutagenesis and molecular modeling/dynamics studies in order to further investigate the role of hCINAP. We also characterize the dual ATPase and AK activity of hCINAP by kinetic analysis, and with site-directed mutagenesis in residue H79 within VIVDYHG, a functional Walker B motif (hhhDXXG), and uncover the effect of this mutation on its kinetic properties. Furthermore, we demonstrate that a H79G mutation in vivo has striking effects on the formation of CBs in the nucleus of human cells.
MATERIALS AND METHODS
Cell line
HeLa cells were cultured in DMEM, containing 10% v/v fetal calf serum (Gibco/BRL), 2 mM l-glutamine and 50 U/mL of penicillin/streptomycin, and maintained at 37°C in 5% CO2.
Antibodies
The following primary antibodies were used for immunofluorescence: Mouse anti-coilin δ4 monoclonal antibody (mab) 5P10 TC at 1:50 dilution, mouse anti-PML Bodies mab P6-M3 at 1:100 (Santa Cruz Biotech.) and mouse anti-fibrillarin mab AFB01 at 1:200 (tebu-bio). Secondary antibodies (Molecular Probes) were: Alexa Fluor 555 donkey anti-mouse IgG, Alexa Fluor 568 goat anti-rabbit IgG, both at 1:1500 dilution.
Immunofluorescence
Immunofluorescence was carried as described by Santama et al.15 and labeled cells were analyzed on a Zeiss Axiovert 200M inverted fluorescence microscope, equipped with a Zeiss AxioCam MRm camera and Zeiss Axiovision 4.2 software, using a Zeiss Apochromat ×63 1.3 oil lens.
Transient transfections and statistical analysis
Exponentially growing HeLa cultures were harvested and replated onto coverslips 24 h prior to transfection. Transfections were carried out using a Ca2+/PO4−3 precipitation protocol.1 At 24 and 36 h post transfection, coverslips were retrieved for immunofluorescence and microscopic analysis. For the analysis of the CB phenotypes, random fields of cells were scored for the number of CBs per nucleus (as revealed by coilin immunostaining) from three independent experiments (n = 571 for GFP-hCINAP-H79G, n = 1439 for GFP-hCINAP-WT and n = 1072 for mock-transfected cells). Data processing was performed in Excel (Microsoft Corp.) and statistical tests undertaken in Prism (GraphPad Software, Inc.). The mean CB number (±sample standard deviation) was calculated and statistically tested by Welch’s t-test. A histogram of the number distribution was generated for each construct and the distributions compared by 2-way ANOVA. P values <0.05, 0.01 and 0.001 were assigned as significant, highly and extremely significant, respectively.
Construction of bacterial expression vectors
hCINAP cDNA was subloned as a BamHI/SalI fragment from pGEX-4T-1-hCINAP (described by Santama et al.1) into bacterial expression vector pGEX-6P-3 (Amersham Biosciences). The hCINAP-H79G mutant was generated using the QuikChange site-directed mutagenesis kit (Stratagene) and confirmed by DNA sequencing (MWG, Germany).
Bacterial expression and purification of recombinant proteins
For recombinant protein expression, competent E. coli B834(DE3)pLysS (Novagen) were transformed with pGEX-6P-3-hCINAP or pGEX-6P-3-hCINAP-H79G, cultured at 37°C until OD600 was 0.4–0.5 AU, induced with 0.5 mM isopropyl β-thiogalactopyranose (IPTG, Sigma) and grown at 18°C overnight. Cells were lysed in lysis buffer [50 mM Tris-HCl pH 8.2, 0.2M NaCl, 0.5 mM DTT, 0.5 mM PMSF, and a mixture of protease inhibitors (Roche)] and disrupted by sonication. The cell lysate was clarified (130,000g at 4°C for 30 min), the cleared supernatant was affinity purified onto a GSTrap 4B column (GE Healthcare), followed by on-column cleavage of the GST tag by injection of 3C protease, performed as described by Dian et al.16 The eluate was concentrated by ultrafiltration and further purified by gel filtration chromatography on a 150-mL Superdex 75 (GE Healthcare) column. Fractions corresponding to the WT-hCINAP monomer were pooled, concentrated to 14 mg/mL and used for crystallization experiments [Supporting Information Fig. S1(A)]. H79G was eluted as a non-separable dimer–monomer mixture [Supporting Information Fig. S1(B)]. Additionally, purified proteins were desalted by dialysis against 20 mM Tris, pH 7.5 and used for AK or ATPase assays.
AK assays
AK assays were performed on a dual-beam Cary 100 conc UV/VIS spectrophotometer. The rate of β-NADH disappearance was monitored at 340 nm by simultaneous measurement of test and reference cell absorbance. Reference samples, containing reaction mixture without hCINAP, were used to automatically subtract background absorbance, mainly attributable to the ATPase activity of pyruvate kinase and nonenzymatic ATP hydrolysis. The AK activity of hCINAP with respect to ATP was measured in the presence of 0.3 mM AMP. The final assay mixture (0.2 mL) consisted of 100 mM Tris-HCl, pH 7.5, 60 mM KCl, 0.21 mM β-NADH, 1 mM PEP, 5 mM MgCl2, 11.4 U/mL PK (Sigma), 10.6 U/mL LDH (Sigma), 20 μg hCINAP, 0.3 mM AMP and 0.01–1.0 mM ATP.
The effects of the AK-specific inhibitor, AP5A, were determined in the presence of 0.33 mM ATP, 0.3 mM AMP and 1–120 nM AP5A.
All kinetic data were analyzed with the nonlinear regression program GraFit.17
ATPase assay
ATPase activity was determined by the malachite-green assay.18 The reaction mixture (0.2 mL) contained 100 mM Tris-HCl, pH 7.5, 60 mM KCl, 5 mM MgCl2, 0.01–2 mM ATP and 20 μg wild-type or mutant enzyme. Parallel control samples, containing reaction mixture without hCINAP, were used to subtract absorbance derived mainly from nonenzymatic ATP hydrolysis. Blank samples, containing buffer with and without hCINAP, showed no absorbance difference and were used to adjust the baseline of the instrument. Reactions took place for 10 min at 30°C and were stopped by addition of the color reagent. Mixtures were allowed to stand for 10 min, and colorimetric determination of PO43− liberation was monitored at 630 nm.
Crystallization and data collection
Co-crystals of hCINAP in complex with ADP, dADP, and Mg2+ADP-PO43− (average size of 0.3–0.5 mm), were obtained at 20°C using the sitting-drop vapour diffusion technique in a buffer comprising 14 mg/mL enzyme, 0.1M HEPES pH 7.5, 1.5M Li2SO4, 0.2M NaCl, 0.5 mM DTT, 25 mM MgCl2, and 2 mM ADP or 2 mM dADP or 25 mM AP5A, respectively. Before flash freezing for data collection, crystals were transferred for 5–15 s to fresh buffer containing 25% v/v glycerol. Single crystal diffraction data were collected on the PX 10.1 beamline (SRS, Daresbury Laboratory), using a 225-mm MAR CCD detector. The crystal-to-image plate distance was 150 mm and gave a maximum resolution of 1.75 Å at the edge of the detector.
Structure determination
Integration and data reduction were performed with the programs DENZO and SCALEPACK19 or MOSFLM20 and SCALA, from CCP4 suite,21 and intensities transformed to amplitudes with TRUNCATE.22 The crystals were isomorphous with the sulfate-bound form (1RKB).2 Phases were obtained with Molrep,23 using the 1RKB structure as a molecular replacement model (Rf/σ = 11.53, Tf/σ = 35.01). Alternate cycles of manual building with the program Coot24 and refinement using the maximum likelihood target function as implemented in the program REFMAC25 improved the model phases. At this stage, water molecules were added to unidentified Fo–Fc map peaks greater than 1.0 σ by using the “water find” module of the program Coot. After an additional cycle of refinement and manual building, the ligand molecule was included in subsequent refinement cycles. ADP, dADP, Mg2+(H2O)6, PO43− and SO42− models, retrieved from the REFMAC library, were fitted into the electron density with Coot. The final model was generated by TLS refinement within REFMAC using TLS groups for the protein generated by the TLSMD web server.26 Details of data processing and refinement statistics are summarized in Table I.
Table I.
Statistics of Data Collection, Processing, and Refinement of the hCINAP Complexes
| hCINAP complex (PDB ID) | SO42− (3IIK) | ADP (3IIJ) | dADP (3IIM) | Mg2+ADP-PO43− (3IIL) |
|---|---|---|---|---|
| CCLRC PX10.1; λ (Å) | 1.04498 | 1.11665 | 0.979 | 1.04498 |
| Space group P61; Cell dimensions (Å), | 98.7, 98.7, 57.6 | 99.1, 99.1, 57.8 | 99.3, 99.3, 58.0 | 99.2, 99.2, 58.0 |
| a, b, c, α = 90°, γ = 120° | ||||
| Resolution (Å) | 34.32–1.95 | 28.3–1.76 | 25.0–2.00 | 34.52–2.00 |
| Outermost shell (Å) | 2.06–1.95 | 1.79–1.76 | 2.03–2.00 | 2.03–2.00 |
| Reflections measured | 188,312 | 152,209 | 140,350 | 144,687 |
| Unique reflections | 23,444 | 31,610 | 22,173 | 22,102 |
| Rsymma | 0.050 (0.449) | 0.044 (0.499) | 0.066 (0.470) | 0.057 (0.461) |
| Completeness % | 99.9 (100.0) | 98.1 (88.4) | 100.0 (100.0) | 99.9 (100.0) |
| σ | 25.80 (4.50) | 17.92 (3.41) | 17.80 (4.82) | 20.13 (4.25) |
| Redundancy | 8.0 (8.0) | 4.9 (4.3) | 6.3 (6.2) | 6.5 (6.5) |
| Final Rcrystb (Rfreec) % | 18.0 (20.6) | 18.1 (19.5) | 18.6 (21.6) | 17.1 (19.3) |
| No. of protein residues (residue range)d | 175 (−2 to 172) | 175 (−2 to 172) | 174 (−1 to 172) | 175 (−2 to 172) |
| No. of water molecules in final cycle | 200 | 190 | 205 | 201 |
| RMS deviation in bond lengths (Å) | 0.009 | 0.008 | 0.010 | 0.010 |
| RMS deviation in bond angles (°) | 1.1 | 1.1 | 1.2 | 1.2 |
| Average B factor (Å2) | ||||
| Protein atoms | 19.1 | 19.6 | 18.7 | 20.6 |
| Water molecules | 31.0 | 32.0 | 31.2 | 32.2 |
| Ligand molecules | 27.3 (SO4ATP site) 95.7 (SO4AMP site) |
30.2 (ADP) | 32.6 (dADP) | 33.1 (ADP) 36.6 (Mg2+) 34.1 (Pi) |
| Ramachandran (u–w) plot | Residues in most favored regions 93%, Residues in allowed regions 7% | |||
Values in parentheses are for the outermost shell.
Rsymm = ΣhΣi|I(h)–Ii(h)/ΣhΣiIi(h) where Ii(h) and I(h) are the ith and the mean measurements of the intensity of reflection h.
Rcryst = Σh|Fo–Fc|/ΣhFo, where Fo and Fc are the observed and calculated structure factors amplitudes of reflection h, respectively.
Rfree is equal to Rcryst for a randomly selected 5% subset of reflections not used in the refinement.
Residues −2 to 0 correspond to additional amino acids introduced by the cloning procedure.
PROCHECK27 was used to assess the quality of the final structure. Structures were analyzed by programs “contacts” and “angle” of the CCP4 suite, and hydrogen bonds were assigned if the distance between electronegative atoms was less than 3.3 Å and if both angles between these atoms and the preceding atoms were greater than 90°. Van der Waals (vdW) contacts were noted for non-hydrogen atoms separated by less than 4 Å.
Protein preparation for substrate docking calculations
Using the hCINAP-Mg2+ADP-PO43− X-ray complex coordinates, an initial hCINAP-Mg2+ATP-PO43− was prepared by conversion of ADP to ATP, based on the position of the extra phosphate group in the superimposed AK-AMPPNP-AMP (PDB entry 1ANK) structure.28 Wat12 and Wat8 were deleted, as the extra phosphate takes their places in coordinating to Mg2+ and forming a hydrogen bond contact with Arg109. The initial model was prepared for docking using Schrödinger’s Protein Preparation Wizard by deleting residual SO42− molecules, the Li atom, and all crystallographic water molecules beyond 5 Å of Mg2+, ATP or PO43−. Two further water molecules (Wat9 and Wat10) were also deleted to accommodate different orientations of the docked AMP molecule. Bond orders were assigned and hydrogen atoms added. Initial assignment of protonation states for basic and acidic residues and tautomeric states were based on pKa at their normal pH (7.0). Subsequent optimization of hydroxyl, His protonation states, C/N atom “flips” and side-chain O/N atom “flips” of Asn and Gln was based on optimizing hydrogen bonding patterns. His residues were assigned as neutral, either in a HIE or HID state. Any steric clashes or bad contacts were removed by a final Impref minimization using the OPLS-AA (2001) force field.29
As a validation test for our calculations, we assessed the performance of IFD starting from the hCINAP structure with a SO42− bound at the ATP-binding site (PDB ID: 3IIK) to reproduce the correct position of ADP at this site, as judged by direct comparison with the hCINAP-ADP structure (PDB: 3IIJ). For this purpose, the hCINAP-SO42− complex was prepared for docking using Schrödinger’s Protein Preparation Wizard, as above. All SO42− ions, except those bound at the ATP-binding sites, were deleted. Waters within 5 Å of the sulfate were retained in the protein preparation, but deleted in the ATP-binding site for subsequent docking to allow space for the docked ADP to bind.
Ligand preparation for docking
The AMP and ADP ligands were extracted directly from the crystal structures (PDB entries 1ANK and 3IIL, respectively), and ATP from the prepared hCINAP-Mg2+ATP-PO43− complex. Hydrogens were added, bond orders assigned and structures minimized using Macro-Model version 9.530 and the OPLS-AA (2005) force field,29,31 with water solvation effects included via the GB/SA continuum model.32
Rigid receptor docking
Initial validation of the hCINAP-Mg2+ATP-PO43− model structure and the Glide-XP (version 4.5)33 docking method34–36 was performed by re-docking of ATP to the model. Docking grids were centered on the ATP ligand and prepared with/without the PO43− ion at the AMP binding site. Glide-XP docking of AMP to the AMP binding site was performed with PO43− used as the centre for the docking grids. In each case, the grids represented the shape and properties of the binding sites with dimensions ~30 Å × 30 Å × 30 Å.
Induced fit docking
IFD calculations,37 including receptor flexibility of AMP to the AMP binding site, were performed using the Glide-XP (rigid receptor) top-ranked AMP-pose as input. Grids for the hCINAP-Mg2+ATP-AMP model had dimensions of 30 Å × 30 Å × 30 Å, centered on AMP. A maximum of 20 ligand binding poses were saved from Stage I Glide-SP docking. Stage II was a Prime induced fit, where residues within 5 Å of the AMP ligand initial poses were refined. Residues 45–54 of the NMP-binding loop were explicitly included and P-loop residues Lys16 and Thr17 explicitly excluded, due to their role in coordinating to the Mg2+. For Stage III, up to 20 structures within 30 kcal/mol of the lowest-energy structure were used for Glide-XP AMP re-docking. The resulting receptor-ligand poses were analyzed in terms of structure, ligand re-docking GlideScore (GS), and IFDScore (ligand re-docking GS + 0.05% Prime complex energy).
The docking grids for the IFD “test” calculations, that is, docking of ADP to the ATP-binding site of the hCINAP-SO42− complex, were centered on the SO42− ion. Other details of the IFD calculations are as above except that only residues within 5 Å of the initial ligand poses from Stage I were refined in Stage II; no further residues were explicitly included or excluded.
QM/MM calculations
QM/MM single point calculations using QSite 5.038 were performed on the hCINAP-Mg2+ATP-AMP model geometries from the IFD calculations to estimate changes in ESP fit atomic charges as the AMP α-phosphate approaches the ATP γ-phosphate. The QM region contained: the ATP and AMP ligands with the -CH2- groups “hydrogen-capped” in place of the riboses; any residue and water molecule directly hydrogen-bonded with the AMP and ATP α- and γ-phosphates; the hexa-coordinated Mg2+, its 3 coordinated waters and the Thr17 side chain. The B3LYP39,40 DFT method was used with the 6-31+G* basis set,41–43 whereas the OPLS-AA (2005) force field29,31 was used for the MM region.
Superimposition and structure presentation
These were performed with LSQKAB44 for hCINAP complexes and with SUPERPOSE45 for comparison between hCINAP and its homologs. Figures were prepared and rendered with PyMol.46
The coordinates of the protein complexes have been deposited to the RCSB Protein Data Bank (http://www.rcsb.org/pdb), with codes 3IIJ, 3IIK, 3IIL, and 3IIM.
RESULTS
The structures of hCINAP in complex with ADP, dADP, and SO42− : Identification of substrate binding sites
To re-examine and fully resolve the structural features of hCINAP as an atypical putative AK enzyme and probe ADP binding sites for the forward reaction (2ADP → ATP + AMP) plus the expected AMP and ATP binding sites for the bisubstrate reverse reaction (ATP + AMP → 2ADP), we provide here a series of structural analyses. We achieved the first structure of hCINAP in complex with ADP, its putative AK substrate [Figs. 1 and 2(A), Table I], at a higher resolution (1.76 Å) than the existing structure for the sulfate-bound form at 2 Å resolution (1RKB).2 Inspection of the electron density maps of the hCINAP-ADP complex shows a strong Fo–Fc accompanied by a 2Fo–Fc electron density, which can neatly accommodate one ADP molecule. The catalytic center is lined by P-loop amino acids 10–17, Arg109 from the LID domain and also His79 [Figs. 1 and 2(A)]. ADP binds in a groove located near the protein surface with the adenine ring occupying the entrance of the catalytic center and the phosphate groups orientated toward the P-Loop region. The crystal structure shows that, upon binding to hCINAP, ADP forms numerous polar and non-polar contacts with the residues of the ATP binding site (summarized in Supporting Information Table I). In agreement with the broad substrate specificity of hCINAP, the adenosine moiety of ADP participates in only three hydrogen bonds with protein residues [Fig. 2(A)].
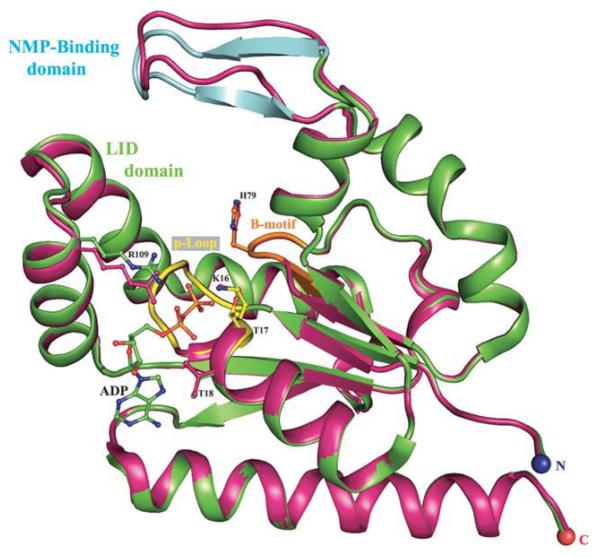
Figure 1.
Structure superimposition of hCINAP-ADP complex (green) onto the sulfate-bound form 1RKB (purple). The overall root mean square deviation (RMSD) between Cα and backbone atoms of hCINAP-ADP and 1RKB was 0.576 and 0.594 Å, respectively. The NMP binding domain (designated by cyan), which comprises amino acids 45–55 (average RMSD for Cα atoms is 2.43 Å), adopts a β strand-turn-β strand conformation. The conserved Lys16 residue adopts identical conformation in the two structures, whereas the side chain of the conserved Arg109 shifts by 1.5 Å towards the phosphate groups to stabilize the large number of negative charges. The P-loop (a.a.10–17) and the Walker B motif (a.a. 77–80) regions are indicated in yellow and orange, respectively.
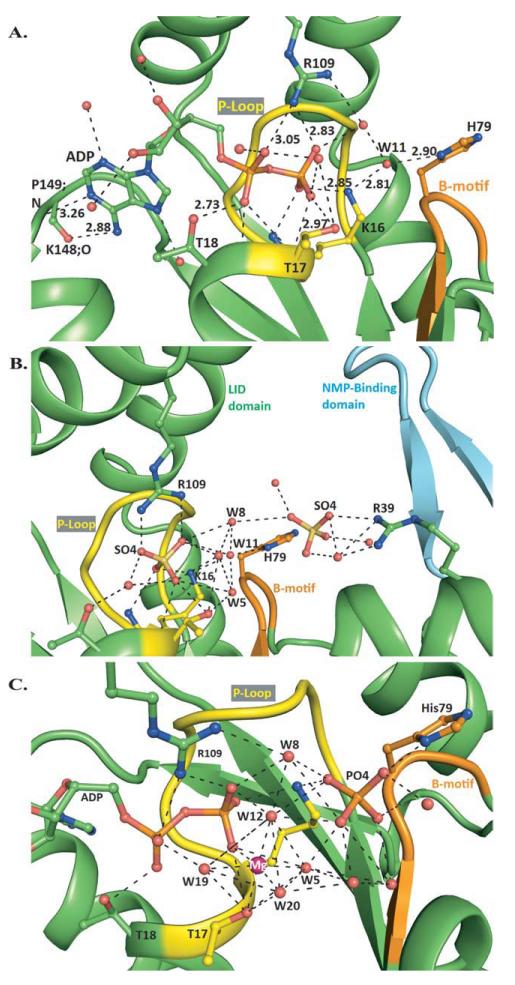
Figure 2.
Structural analysis of hCINAP complexes. A: Polar contacts of ADP in the vicinity of the Mg2+ATP binding site. Guanidine nitrogen of Arg109 from the LID domain makes polar contacts with the α- and β-phosphate oxygens, O2A and O2B. The hydroxyl group of Thr17 interacts directly with β-phosphate oxygen O3B, whereas NZ of Lys16 forms one hydrogen bond with O1B. The hydroxyl group of Thr18 interacts with O1A oxygen. The phosphate moiety makes additional polar contacts with water molecules and the main chain O/N atoms of residues Gly13, Gly15, Lys16, Thr17, and Thr18. The ribose moiety forms only two polar contacts with water molecules. The purine moiety interacts with two water molecules, while two additional polar contacts are formed between N6 and the carbonylic oxygen of Lys148 and N1 and the amide nitrogen of Pro149. B: Polar contacts of SO42− bound at the AMP binding site. The SO42− ion, bound at the AMP binding site, makes a direct polar contact with the guanidinum nitrogen NH1 and two indirect polar contacts with the NH2 via a water molecule. The imidazole ND1 of His79 is hydrogen bonded to water Wat11 (ND1-OW11; 2.82 Å), whereas nonpolar-polar contacts are formed between imidazole carbon CE1 and the sulfate oxygen O1 (CE1-O1; 3.17 Å). C: Polar contacts of Mg2+ADP and Pi, in the vicinity of the Mg2+ATP binding site. In the vicinity of the Mg2+ATP binding site, an extensive network of hydrogen bonds is created between Mg2+ADP, PO43−, P-loop residues Lys16, Thr17, Thr18, Walker B motif His79, and Arg109 from the LID-domain. The Mg2+ ion is hexa-coordinated, with the hydroxyl group of Thr17 and the Wat12 occupying two apical positions. The Wat12 and Wat8 are located in the positions that would be occupied by two of the γ-phosphate oxygens of ATP. Arg109 forms hydrogen bonds with α- and β-phosphate oxygens and Wat8 and is proposed to protect ATP from random hydrolysis. Mg2+ contributes to positive polarization of γ-phosphate and makes it more susceptible to nucleophilic attack. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
We also conducted the structural analysis of hCINAP-dADP complex at 2 Å resolution, (Table I), which showed that dADP perfectly overlaps with the ADP molecule (RMSD = 0.121 Å; data not shown), but that the N1 of adenine and the remaining sugar oxygen O3′ are not hydrogen bonded, reducing dADP interaction by four hydrogen bonds, compared to ADP (Supporting Information Table II).
In addition, in the hCINAP-ADP structure the deduced ATP binding site is bordered by a loop composed of residues 78–84. This loop links the C-terminal end of the CORE β3 strand (Asp77) with a 310 helix, and, significantly, bears a Walker B motif, 77-DYHG-80, indicating its possible role in catalysis8–10,14 (Supporting Information Fig. S2). In particular, His79 is linked with the β-phosphate in the ATP binding site by a hydrogen bond network extending from amide ND1 to Wat11, from Wat11 to P loop-Lys16 and from Lys16 back to the β-phosphate [Fig. 2(A)]. Water molecule Wat11 lies between the β-phosphate (PB) of ADP (PB-OW11; 4.5 Å) and His79 (OW11-ND1; 2.9 Å), and is hydrogen-bonded to NZ of Lys16 (2.81 Å). OW11 is inline with respect to ND1 and PB (angle ND1-OW11-PB=177.12°), suggesting that this position is of high potential for a nucleophilic attack of an oxygen on the β-phosphate of acceptor ADP during the forward AK reaction [Fig. 2(A)].
In its totality, the tertiary structure of hCINAP bound to ADP (or dADP; Fig. 1), including its ATP binding site, resembles the previously reported sulfate-bound protein structure.2 However, in the ATP site, the most important difference that we have identified is a shift by 1.5Å in the side chain of Arg109, which could be associated with a conformational change of the LID domain, possibly in order to optimize the contacts with the α- and β-phosphate groups of ADP [Figs. 1 and 2(A)]. In addition, the higher-resolution structure allowed us to build accurately the nucleoside monophosphate-binding loop (NMP-binding loop; amino acids 45–55), whose residues were interpreted as disordered in a previous structural analysis of hCINAP.2 In our structure (Fig. 1), the NMP-binding loop adopts a β strand-turn-β strand conformation and has a well-defined electron density with average B factor for all atoms 30 Å (Supporting Information Table V).
To assess whether the observed structural difference of the NMP-binding domain in this work may be due to ADP binding to the enzyme, we obtained data from the hCINAP-SO42− complex at 1.95 Å resolution, and refined to an R-factor of 0.179 (Rfree = 0.206; Table I). In our hCINAP-SO42− structure, we observed the same structural motif of the NMP-binding domain as in the ADP complex, indicating that it is independent of ligand (ADP) binding and strengthening our interpretation of the NMP-binding loop [Fig. 2(B)].
Finally, in the hCINAP-SO42− structure, in addition to a sulfate ion at the ATP-binding site, we also found a weakly bound sulfate at the AMP-binding site located between His79 and Arg39 [Fig. 2(B), Supporting Information Table III]. This position is very similar to that observed for the α-phosphate moiety of AMP in AKs from Escherichia coli (AKeco) and Sulfolobus acidocaldarius (AKas), 1ANK and 1NKS, respectively [Supporting Information Fig. S3(A)]. SO42− interacts with a conserved guanidinium group, also present in the AMP binding site of 1ANK and 1NKS (Arg36 and Arg54, respectively).47,48 These findings suggest that the location occupied by the sulfate ion (known to mimic the PO43−) represents the recognition site for the α-phosphate of AMP in hCINAP.
In conclusion, therefore, the combination of our structures allowed the structural visualization of the NMP-binding domain, the identification of the ADP binding mode, within the ATP-binding site, and suggested the location of the AMP binding site.
Mg2+ADP-PO43− binding
To further probe the AMP binding site in hCINAP and, at the same time, capture its transition intermediate, we used a known inhibitor of AKs, the bisubstrate analog bis-adenosine pentaphosphate (AP5A). We found AP5A to be a potent inhibitor of hCINAP with an IC50 of 27.03 ± 2.93 nM AP5A (Supporting Information Fig. S4).
The hCINAP structure that we obtained in the presence of AP5A [Fig. 2(C)] did not, as expected, correspond to a transition intermediate but appeared to correspond to a post-hydrolysis state. This may be explained by a technical limitation of the X-ray crystallography technique: in our experiments co-crystal formation in the presence of AP5A required 1 month, and it is therefore likely that the co-crystallized AP5A was slowly hydrolyzed enzymatically or chemically during the crystallization period. Soaking of preformed crystals with fresh AP5A solution led to crystal deterioration, indicating that the crystal complex formation was accompanied by extensive structural re-organization (as would be expected in AKs).6
The obtained structure revealed that (a) the ATP site was occupied by a Mg2+ADP complex and a phosphate group (Pi) in close contact with His79 of the Walker B motif, (b) a large shift in the backbone torsional angle of His79 occurred (ΔPhi5-67.8°), accompanied by a shift of the imidazole ring by 2.1 Å away from the ATP site in order to establish contacts with the phosphate group (Supporting Information Fig. S5), (c) the magnesium ion was hexa-coordinated with one ligand supplied by a β-phosphate oxygen and a second one supplied by the hydroxyl group of Thr17, and finally (d) ADP possessed an identical conformation to that of the ADP complex (RMSD = 0.061Å).
There is strong evidence that PO43− (as a product of hydrolysis), rather than SO42−, was bound in the presented crystal structure: (a) We could not detect a SO4−2 ion in this position or a shift in the His79 side chain in any other of the three reported structures (SO42−, ADP, dADP), but exclusively in the Mg2+ADP-PO43− structure, although the precipitant Li2SO4 was present at a concentration of 1.5 M in all four crystallization experiments. Furthermore, (b) we have carried out the co-crystallization experiments (data not shown) in the presence of Mg2+, Mg2+ADP, and Mg2+dADP, but have not obtained SO42− in this position, neither a His79 shift, nor a bound Mg2+ ion.
The PO43− at its position makes hydrogen bonds to the imidazole nitrogen of His79 (2.65 Å) and the NZ atom of Lys16 (2.83 Å). We propose that during the forward AK reaction (2ADP→ATP + AMP) the Mg2+ ion does not follow the movement of the attacking β-phosphate, but maintains its observed position and hexa-coordinated geometry, with 2 γ-phosphate oxygens of the produced ATP replacing two water molecules, most probably Wat8 and Wat12 [Fig. 2(C) and Supporting Information Table IV].
Therefore, while for technical reasons this approach did not reveal unequivocally the binding site of AMP, the post-hydrolysis structure obtained was very informative in determining the residues that provide the co-ordination for the Mg2+ ion (this is the first example of an AK with a bound Mg2+ ion) and in revealing that P loop His79 is involved in the recognition of PO43− in the structure with Mg2+ADP-PO43− present.
Prediction of the hCINAP-Mg2+ATP-AMP ternary complex by modelling
In the absence of direct crystallographic evidence for the AMP binding site (required for an AK activity) and with hCINAP already classified as an AK,2 we decided to model a hCINAP-Mg2+ATP-AMP ternary complex to predict what structural changes this would induce compared with the ADP bound complexes and to gain further insights into the nature of hCINAP if functioning as an AK.
We first validated the effectiveness of IFD for the job at hand by performing a test prediction of a nucleotide binding site, starting from a structure with initially only a small ion bound at the site. We therefore docked ADP to the ATP-binding site of the hCINAP-SO42− complex (PDB: 3IIK): ADP replaced the SO42− and the associated receptor-ligand induced-fit structural changes had to be accounted for using IFD. Results were quite satisfying. The top-3 ranked poses (poses 1–3) were tightly locked in IFD scores (−376.6 to −376.8) with all poses correctly predicting the α- and β-phosphate positions and producing close to the same receptor-ligand hydrogen bond contacts as seen in the 3IIJ crystal complex (Supporting Information Fig. S6; see a detailed account of the extensive matching of the IFD predictions with the experimental data in the figure legend). On the basis of the strong validation results outlined, we could now confidently approach the prediction of the hCINAP-Mg2+ATP-AMP complex using IFD.
Successful docking of ATP to the hCINAP-Mg2+ATP-PO43− model, prepared from the hCINAP-Mg2+ADP-PO43− X-ray complex (see Materials and Methods), required removal of the PO43− ion from the model, indicating unfavorable ATP-PO43− electrostatics with PO43− in its present position. On removal of PO43−, all ATP docking poses produced the ATP binding conformation in the model complex, with the superimposed RMSD (heavy atoms) for the top-ranked pose being 0.828 Å, compared to the starting model.
ATP binding site structural changes going from ADP to ATP are minimal with the more significant conformational changes expected in the NMP-binding domain and Walker B-motif loop in order to accommodate AMP at the AMP binding site.
Meanwhile, induced fit docking (IFD), including receptor flexibility of AMP to the AMP binding site, resulted in eight receptor-ligand binding structures, four of which made intuitive sense with the α-AMP phosphate occupying a similar position to the α-AMP phosphate in 1ANK (E. coli AK with bound AMPPNP and AMP) and 1NKS (S. acidocaldarius AK with bound ADP and AMP) structures (Table II). The hexa-coordinated Mg2+ coordination sphere is conserved for all structures. The first three structures (IFD-1–IFD-3) were also the top-ranked IFD poses. IFD-1 and IFD-2 poses with the highest IFD scores reproduce best the AMP-bound conformations in 1ANK and 1NKS, with superimposed ligand RMSDs (heavy atoms) being all less than 1 Å. They both have similar ATP-AMP inter-phosphate P–P distances (~5 Å) and form seven direct AMP-protein hydrogen bonds, which compares well with the eight hydrogen bonds formed by the homologous 1ANK and 1NKS complexes. The main structural differences between IFD-1 and IFD-2 are the length of the hydrogen bonds and the AMP α-phosphate position, which differs spatially by about 1 Å. IFD-1 (P–P = 5.12 Å), shown in Figure 3 (B), forms more favorable contacts with hCI-NAP as judged by the higher Glide Score (−10.17) compared to IFD-2 (−8.82). Compared to the hCINAP-Mg2+ADP-PO43− X-ray structure, there is a backbone shift in the NMP-binding domain (residues 45–55) with the largest shifts of 3.5–5 Å for residues 49–51. Accompanying side-chain rearrangements are mainly associated with increasing favorable contacts with AMP [Supporting Information Fig. S3(B)].
Table II.
Output Receptor-Ligand Poses (Structures) and Data from the IFD Calculcations
| Structure | P–P distance (Å)a | H-bondsb | RMSD (Å)c | Prime energy (kcal/mol) | XPG score | IFD Score |
|---|---|---|---|---|---|---|
| 1 | 5.12 | 7 | 0.880 (0.826) | −11316.3 | −10.17 | −575.98 |
| 2 | 5.05 | 7 | 0.958 (0.769) | −11319.3 | −8.82 | −574.78 |
| 3 | 4.53 | 5 | 1.429 (1.640) | −11328.3 | − 7.92 | −574.33 |
| 4 | 6.49 | 6 | 1.611 (1.367) | −11331.4 | −5.59 | −572.15 |
ATP-ADP interphosphate distances measured between P atoms.
Number of direct AMP–hCINAP hydrogen bonds.
RMSD (heavy atoms) on superimposition of AMP conformation on its conformations in the 1ANK and INKS (in parentheses) complexes.
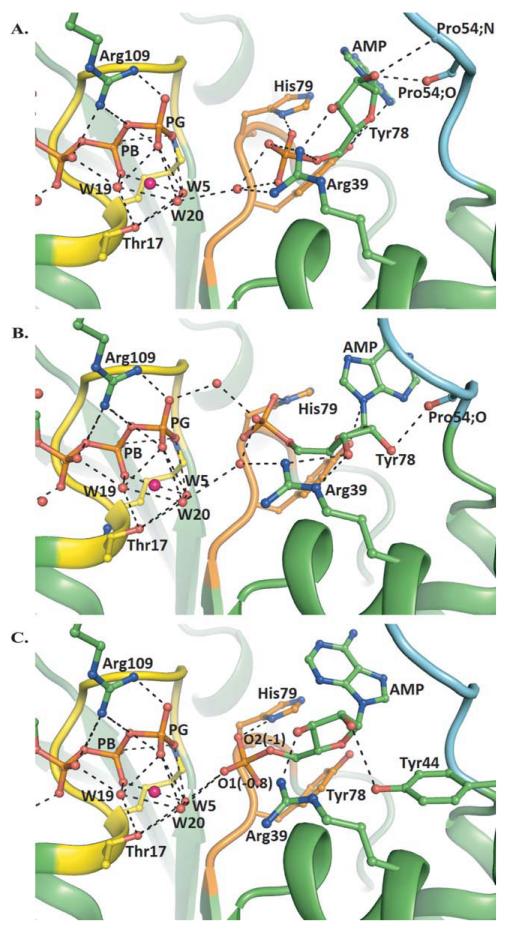
Figure 3.
Models of the hCINAP-Mg2+ATP-AMP complex from induced fit docking calculations. Predicted hCINAP-Mg2+ATP-AMP poses with different interphosphate (P–P) distances, showing the approach of a mobile AMP towards a fixed ATP. A: IFD-4, P–P distance 6.49 Å: AMP phosphate interacts with His79 side chain NH and Arg39 guanidine NH2. The Tyr78 side chain hydroxyl hydrogen-bonds with the ribose-ring oxygen. The ribose 2′- and 3′-OHs form hydrogen bonds with Arg39 guanidine NH2 (2′-OH) and backbone carbonyl and amide (3′-OH) of Pro54. B: IFD-1, P–P distance 5.12 Å: His79 forms a side-chain bond with the AMP phosphate. The Tyr78 side-chain hydroxyl hydrogen-bonds with the ribose-ring oxygen and the adeninine nitrogen N1. Water-bridging contacts of the AMP phosphate and the ATP phosphate with the Arg39 guanidine side chain (NH2) are formed. Arg39 forms a direct H-bond contact with ribose 2′-OH through side-chain guanidine NE and NH2. Ribose-3′-OH hydrogen-bonds with the backbone-carbonyl O of Pro54. C: IFD-3, P–P distance 4.53 Å: AMP phosphate now interacts with the backbone and side-chain NHs of His79, and the Mg2+-coordinated water molecules Wat5 and Wat20. Waters bridge the AMP (α-)–ATP (γ-) phosphates. Arg39 forms a direct H-bond contact with ribose 2′-hydroxyl through side chain guanidine NH2. Ribose-3′-OH hydrogen bonds with Tyr44 OH. The Tyr78 side-chain hydroxyl hydrogen-bonds with the ribose-ring oxygen. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The other two receptor-ligand poses in Table II have longer [6.49 Å; IFD-4; Fig. 3(A)] or shorter P–P distances [4.53 Å; IFD-3; Fig. 3(C)] than those of IFD-1 and IFD-2. The structures suggest a key role for water bridging in the approach of an α-AMP phosphate towards the γ-ATP phosphate, as well as some assistance from the Walker B-motif residues, Tyr78 and His79. In IFD-3, the AMP ligand is ideally placed for a phosphate transfer mechanism involving a nucleophilic attack. The case for such a pathway is accentuated by our QM/MM calculations of electrostatic potential (ESP) atomic partial charges at the different P–P distances for IFD-1, IFD-3 and IFD-4 (Supporting Information Table VI). As the AMP approaches the ATP, an AMP α-phosphate OP1 becomes hydrogen bonded with a Mg2+ coordinated water (Wat5) and this is reflected in its estimated partial charge of −0.82 for P–P distance 4.53 Å [Fig. 3(C)]. The corresponding partial charge for His79 hydrogen bonded phosphate OP2 is −0.98. Approach of a mobile AMP towards an immobile ATP is consistent with a transfer mechanism in which the AMP α-phosphate attacks the γ-phosphate of ATP, assisted by water molecules that bridge ATP γ- and AMP α-phosphates.49
Our predicted IFD model geometries and interphosphate (ATP–ADP) P–P distances are in agreement with MD predictions for the homologous E. coli AK-Mg−1-ATP-ADP complex.49 Despite this, our above successful test-validation of the IFD method and the documented successes of IFD in many other recent applications50–55 docking models obviously have to be treated with a reasonable measure of caution, like any prediction approaches. Finally, also of note in this context is a recent publication by Nucci et al.,56 which highlights the limitations of the “static” picture of water molecule positions, as given by crystal structure determination, compared with a more detailed dynamic picture, which can be produced by NMR reverse micelle technology. To address this shortcoming, retention and modeling of water molecules in our IFD calculations attempt to account for some of the dynamic water behavior associated with the approach of AMP towards ATP at different P–P distances in our proposed models.
In conclusion, therefore, the modeling overall strongly indicated that the AMP binding site would lie between the B motif and Arg39. This conclusion is further strengthened by the fact that Arg39 is a highly conserved residue in other known AKs.
An H79G mutation affects both AK and ATPase activities of hCINAP
The overall structural analysis and the proposed catalytic mechanism have highlighted the significance of certain amino acids in the P loop region (Lys16 and Thr17), the LID domain (Arg109) and Walker B motif (His79). These residues are either identical or correspond to conservative substitutions in orthologs of hCINAP across the eukaryotic phyla. Interestingly, in human adenylate kinases AK1–AK5, Lys16 and Arg109 are conserved, while Thr17 and His79, perceived as functionally critical for hCINAP, are absent. Because our structural and modelling studies indicate that H79 may be a regulatory residue that responds to substrate binding, we decided to generate an hCINAP-H79G mutant and perform kinetic analysis to evaluate its effect on catalytic activity.
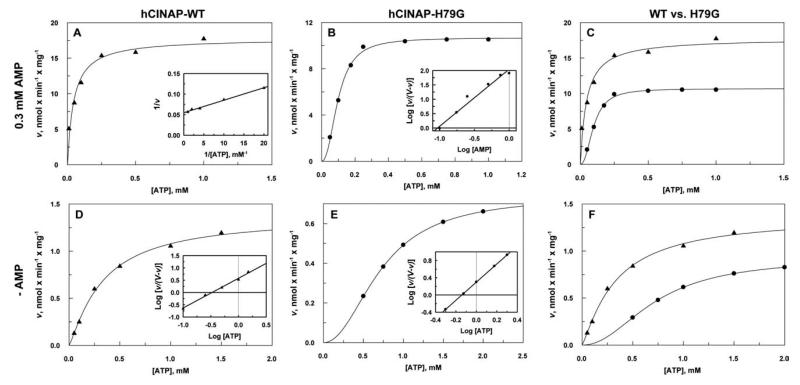
We first measured the AK activities of highly purified hCINAP and hCINAP-H79G with respect to ATP, in the presence of a fixed concentration of AMP (0.3 mM). Second, we determined ATPase hydrolysis by hCINAP-WT and hCINAP-H79G, in the absence of AMP (Table III and Fig. 4). Data from kinetic experiments are fitted using both the Hill and Michaelis–Menten equations. hCINAP-H79G data fitted the Hill equation best with nH = 1.99, whereas trying to fit data using n = 1 produced high values of standard deviations. Although the low enzyme activity made reliable measurement of rates at lower ATP concentrations (<0.01 mM) impossible, the calculated nH value is in agreement with the observed mutant-induced dimerization (Supporting Information Fig. S1), making us confident that this parameter truly reflects the effect of the mutation on enzyme characteristics.
Table III.
Summary of Kinetic Parameters
| Enzyme | Specific activity (nmol/min/mg) | kcat (s−1) | Km(μM ATP) | kcat/Km (M−1s−1) |
|---|---|---|---|---|
| Adenylate kinase avtivity (ATP + AMP↔2ADP) | ||||
| hCINAPWT | 18 ± 0.9 | 6.3 × 10−3 | 45 ± 5 | 140 |
| hCINAPH79G | 11 ± 0.16 (nH = 1.99a) | 3.7 × 10−3 | 93 ± 8a | 39 |
| ATPase activity (ATP→ADP + Pi) | ||||
| hCINAPWT | 1.37 ± 0.08 (nH = 1.21a) | 4.8 × 10−4 | 332 ± 8a | 1.45 |
| hCINAPH79G | 0.74 ± 0.04 (nH = 2.10a) | 2.5 × 10−4 | 721 ± 1a | 0.34 |
Best-fit curve obtained by fitting the data to the Hill equation. Km and nH were calculated by linear regression of the logarithmically transformed data (Hill plots).
Figure 4.
Kinetics of hCINAP-WT and hCINAP-H79G. A: hCINAP-WT adenylate kinase activity with respect to ATP at a constant concentration of AMP. Inset: Double-reciprocal plot. B: hCINAP-H79G adenylate kinase activity with respect to ATP. Inset: Hill plot. C: Comparison of the AK activities of A and B; hCINAP-WT (▲) and hCINAP-H79G (●). D: hCINAP-WT ATPase activity. Inset: Hill plot. E: hCINAP-H79G ATPase activity. Inset: Hill plot. F: Comparison of the ATPase activities of D and E; hCINAP-WT (▲) and hCINAP-H79G (●). All data are means from two experiments.
We found that in the presence of the H79 mutation (a) the AK enzymatic efficiency (kcat/Km) is reduced by 72% relative to the wild-type enzyme, and (b) the efficiency for ATP hydrolysis in the absence of AMP (ATPase activity) is reduced by 76% relative to the wild type. Furthermore, the ATPase activity of wild type hCINAP (1.45 M−1 s−1) is about 1% of its AK activity (140 M−1 s−1). Although many protein kinases exhibit ATP hydrolysis in the absence of their substrate, the high intrinsic ATPase activity of hCINAP is quite unusual for an AK. Kinetic analysis has therefore provided the evidence for a dual enzymatic activity for hCINAP that is consistent with the dual structural features of the enzyme, namely the overall folding typical of an AK and a catalytic centre characteristic for an ATPase. Additionally, for both AK and ATPase reactions the mutant enzyme displays sigmoidal kinetics (nH ~ 2), indicating that the H79G mutation causes a significant structural rearrangement that leads to protein homodimerization. In particular for the ATPase function, where a nH of 1.2 is also observed for WT-hCINAP, it is likely that the mutation mimics, in an more extreme manner, the effect of ATP binding, for example, it induces a shift from monomer to dimer.
Although the Hill model is purely mathematical and provides a convenient mean to estimate cooperativity, at the same time it makes little reference to the behavior of the enzyme.57 Therefore, a definitive mechanistic interpretation of our results is not a straightforward matter. Nevertheless, the sigmoidal saturation curve of hCINAP-WT enzyme with respect to ATP (nH = 1.2) could be explained by a sequential mechanism whereby binding of ATP to the hCINAP monomer changes its conformation, thus promoting the formation of a hybrid homodimer with only one ATP-site occupied. The interactions between the liganded and unliganded subunit can then induce such conformational changes to the unliganded partner so that the binding of a second ATP molecule can become more favorable (positive cooperativity).58 Alternatively, the cooperativity with respect to ATP could also be explained with the “mnemonical mechanism.”59 According to this mechanism, monomeric enzymes that undergo large conformational changes during their catalytic cycles can retain their active conformation (higher affinity for substrates) for a prolonged period after the catalytic reaction, owing to high energy requirements for bond reformation.
In contrast, the observed MM kinetics when hCINAPWT is tested as an AK (in the presence of saturating AMP concentration; nH = 1) suggests that the binding of AMP inhibits dimer formation or promotes a conformational change that favors ATP binding, for example, hCINAP-AMP complex binds ATP without undergoing large conformational changes.
The expression of the hCINAP-H79G mutant is toxic and causes a significant change in Cajal body organization in the nucleus
hCINAP and CB protein coilin interact directly via the latter’s 214 carboxy-terminal residues.1 Because of the effect of Walker B-motif His79 in enzymatic efficiency, affecting both AK and ATPase activity in vitro, we next examined whether expression of mutant hCINAP-H79G in vivo might have functional implications for nuclear organization, accompanied by a corresponding phenotypic effect on either coilin or CBs. GFP-tagged expression vectors of wild-type hCINAP (WT) and mutant hCINAP-H79G were constructed, tested by Western blot (data not shown) and used for transient transfections of HeLa cells.
We observed that transfection with a mutant GFP-hCINAP-H79G construct had detrimental effects both on transfection efficiency and proliferation of transfected cells: at 24 h post transfection, the proportion of hCINAP-H79G-transfected apoptotic cells (43%) was four times higher than that of wild-type GFP-hCINAP (WT) cells (11%) with extreme statistical significance, and at 36 h the proportion of apoptotic hCINAP-H79G-transfected cells was maintained very high (43% compared to 13% for WT transfected apoptotic cells) with high statistical significance (ANOVA with Tukey’s multiple comparison post-test; Supporting Information Fig. S7), indicative of mutant-specific impairment of proliferation or cell survival.
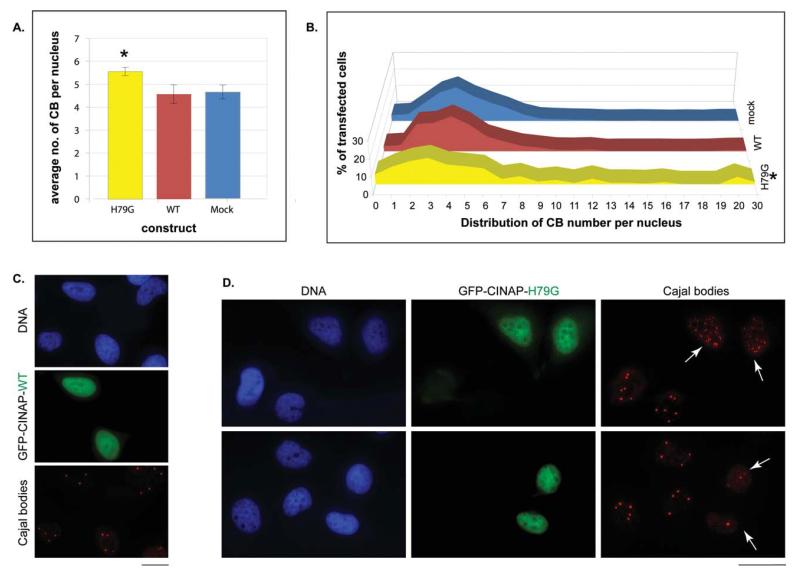
The number of CBs per nucleus, as detected with an anti-coilin antibody, was scored in cells transfected with GFP-hCINAP-WT (n = 1439), GFP-hCINAP-H79G (n = 571) and mock-transfected control cells (n = 1072). In highly-expressing GFP-hCINAP transfected cells, we observed profound changes in CB organization: the mean number of CBs per nucleus was significantly higher in cells transfected with mutant hCINAP-H79G, as compared with the mock transfected cells (Welch’s t-test, P = 0.034) [Fig. 5 (A)]. Importantly, there was a marked heterogeneity in the numbers of CBs in the nuclei of hCINAP-H79G transfected cells as evidenced by the frequency distribution displayed in Figure 5(B), so that large groups of cells either had much fewer CBs than the mean number in WT-hCINAP-expressing or mock-tranfected cells, or groups of H79G cells possessed unusually high numbers of CBs (up to 30 per nucleus) [Fig. 5(B)]. Representative examples of hCINAP-H79G-expressing cells with either a very large number of CBs [Fig. 5(D) upper panels] or a small number of CBs [Fig. 5(D) bottom panels], are shown in Figure 5. As a result of this heterogeneity, the frequency distribution of CBs in cells expressing the mutant form was significantly different overall from that of WT-hCINAP samples (two way ANOVA, P = 0.0151) and from mock-transfected samples (P = 0.0122).
Figure 5.
The overexpression of GFP-hCINAP-H79G alters Cajal Body (CB) organization (mean and distribution) in the nucleus of HeLa cells. A: Mean number of CB (±sample standard deviation) per nucleus in mock-treated cells, cells highly expressing GFP-hCINAP-WT and cells highly expressing GFP-hCINAP-H79G upon transient transfection. The mean number of CBs is significantly higher in the hCINAP-H79G mutant as compared to the mock transfected cells (Welch’s t-test, P = 0.034). B: Histogram of the distribution of Cajal Body (CB) numbers per nucleus in the same three populations of transiently transfected cells. The overall distribution of CB numbers is significantly different in hCINAP-H79G mutant samples compared to the WT control samples (P = 0.0151) and mock transfected samples (P = 0.0122, two-way ANOVA). C and D: Representative images of changed CB numbers in cells transiently transfected with GFP-hCINAP-H79G (D), as compared to cells transiently transfected with GFP-hCINAP-WT (C). Hoechst staining of nuclei (blue), GFP-fluorescence indicative of transfected cells in each case (WT or mutant H79G; green panels) and immunostaining of CBs with an anti-coilin mab (red). In the examples of mutant H79G expression given here, the CBs are either considerably increased in number in transfected cells (white arrows) as opposed to non-transfected cells in the same field (top right panel), or markedly reduced (white arrows, bottom right panel). In WT-transfected cells, the number of CBs is more homogeneous from nucleus to nucleus (C). Scale bars: 20 μm.
The same analysis was repeated examining other types of distinct subnuclear structures, namely the nucleolus, a site historically linked to CBs by Cajal himself,60 and PML Bodies (Pro Myeloid Leukemia Bodies), a nuclear domain not directly linked to CBs. No discernible phenotypic change of the numbers or overall organization of these two nuclear domains was observed in mutant vs. wild-type hCINAP-expressing cells (Supporting Information Fig. S8). This indicates that the effect of overexpression of hCINAP-H79G on CB organization (mean and distribution) is specific and not secondary to general effects of the mutant enzyme on nuclear structure.
Taken together, these data indicate that the expression of the mutant protein results in CB de-regulation (causing an elevated average number and abnormal heterogeneous distribution of CB numbers) and is toxic to the cell, resulting in enhanced apoptosis and reduced proliferation.
DISCUSSION
Although the structure of hCINAP in complex with SO42− had previously been published, the additional four structures presented in this work as well as our modeling of the hCINAP-Mg2+ATP-AMP ternary complex (a) resolve residues in the predicted AMP binding site that were previously interpreted as disordered, (b) confirm that the overall folding of hCINAP conforms to that expected for the AK family, with ATP and AMP binding sites in its catalytic centers, and (c) reveal that, in the vicinity of the ATP binding site, hCINAP possesses structural features typical of the ATPase/GTPase family rather than of AKs. These are a Walker A motif (P-loop), bearing a Mg2+-coordinating residue (Thr17), and a Walker B motif, not reported for any other known AK in any species. hCINAP is thus the first member of the adenylate kinase family (AK1–6) carrying a Walker B motif. Additionally, loop region Asp77 to Pro85, including the Walker B motif is oriented quite differently from that of the other AKs (Supporting Information Fig. S2). In all likelihood, this distinct sequence piece and its unusual orientation is important for the display of ATPase activity.
We corroborated the structural evidence by demonstrating that hCINAP has unusually high intrinsic ATPase activity, in addition to AK activity, being essentially a dual-activity AK/ATPase. The role of H79 in catalysis was found to be equally important both for the AK and ATPase reactions, as we demonstrate that the H79G mutation reduces by ~72% the AK enzymatic efficiency and by 76% the ATPase efficiency.
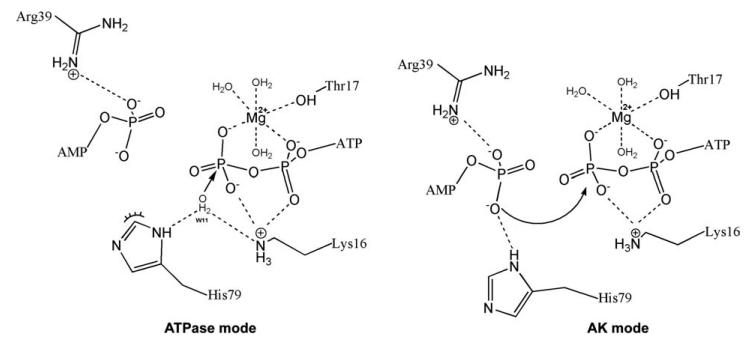
On the basis of our structural and kinetic analysis, we propose a possible mechanism that accounts for the equal participation of His79 in both ATPase and AK reactions. In our proposed model shown in Figure 6, the imidazole ring of His79 can flip between two possible orientations (rotation about a Cβ-Cγ single bond is assumed to be free61). In the first orientation (“flip” or “AK mode”), the imidazole nitrogen faces the AMP binding site and co-ordinates the α-phosphate of AMP together with Arg39 (Fig. 6 – AK mode). In the second orientation (“flop” or “ATPase mode”), the imidazole nitrogen faces the Mg+2ATP catalytic center (Fig. 6 – ATPase mode) and co-ordinates a lytic water together with Lys16 for a nucleophilic attack on the γ-phosphate of ATP, similar to the mechanism which has been proposed for myosin by Okimoto and co-workers.62
Figure 6.
A model of the role of His79 as a structural switch for the selectivity of enzymatic activity: Juxtaposition of “ATPase mode” and “AK mode.” According to the proposed ring-flip hypothesis, based on the structural analysis, the imidazole nitrogen ND1 of His79 can coordinate a lytic water molecule (together with the NZ of Lys16) during the ATPase reaction (ATP + H2O → ADP + Pi; ATPase-mode) or coordinate the α-phosphate of AMP (together with the NH1 of Arg39) during the reverse AK reaction (ATP + AMP → 2ADP; AK-mode; further details in the “Discussion” section).
Moreover, the sigmoidal kinetics of the impaired hCINAP-H79G enzyme might cause significant aberrations to the physiological adenine nucleotide ratio in the nucleus, leading to the observed phenotype in vivo.
At the same time, the relatively low in vitro catalytic activities measured, such as an AK activity of approximately 1/1000 that of AK1, bring into question whether enzymatic activity of hCINAP is its main feature in vivo. Indeed, the relatively small effect of the H79G mutation on catalytic activities, combined with its significant biological effect, raises the possibility that H79 may mediate an alternative function of hCINAP, including involvement in formation or dissolution of protein complexes and the recognition of a substrate not tested in our in vitro assays.
Intriguingly, our structural analysis beyond the catalytic center points towards possible alternative functions of hCINAP. The protein possesses an enclosed tunnel, formed by the long NMP-binding domain and the LID domain (Supporting Information Fig. S9). This tunnel, with one end exposed to the bulk solvent and the opposite end to the ATP binding site might constitute the path for the N- or C-terminus of a peptide chain. This would allow phosphorylation of a specific protein substrate (should hCINAP act as a protein kinase) or other energy-dependent processes, including the conformational correction of a misfolded target protein (should hCINAP act as a molecular chaperone). For instance, the known role of the phosphoprotein coilin in CB assembly63,64 and its interaction with hCINAP warrant further investigation of whether hCINAP may act on coilin as a kinase, a chaperone or a coordinator of its interaction in functional protein complexes.
Expression of the H79G mutant in human cells in culture was found to be toxic, reducing proliferation and increasing apoptosis compared to the wild-type enzyme (Supporting Information Fig. S7), and also exerting a profound and specific effect on CB organization in the nucleus (Fig. 5). The observed CB deregulation was detectable as a significantly altered frequency distribution compared with controls, and an abnormally wide range of between 0 and over 30 CBs/nucleus. The observed dramatic effect of the H79 mutation is in agreement with that observed for the yeast ortholog of hCINAP, FAP7.14 While the ATPase activity of FAP7 is negligible in vitro, strains harboring Fap7 K20R (Walker A motif) or D82AH84A (Walker B motif) mutations exhibit a significantly reduced growth rate compared to the wild-type strain and show that conserved amino acids predicted to be involved in nucleotide binding and NTPase activity are essential for Fap7 function in vivo. Likewise, regulation of CB assembly and disassembly during the cell cycle could be intrinsically linked either with the putative involvement of hCINAP in nucleotide homeostasis or its participation in functional protein complexes or in other phosphorylation events in the mammalian nucleus, which might affect the ATP-dependent movement65 of these highly dynamic and cell cycle-dependent structures.
In conclusion, the unique properties of hCINAP relative to other AK isoforms, including differences in structure, substrate preference, intrinsic ATPase activity and cellular localization, indicate that its function in vivo may also be exceptional among AKs. Ongoing investigation into the intriguing link between hCINAP and CB regulation might hold the key to its principal biological action.
ACKNOWLEDGMENTS
AIL is a Wellcome Trust Principal Research Fellow. Work at the Synchrotron Radiation Sources, CCLRC, Daresbury, England, MAX-lab, Lund, Sweden, and EMBL Hamburg Outstation, Germany, was supported by funding provided by the European Commission under the FP6 and FP7 Research Infrastructure Actions “Structuring the European Research Area” and “ELISA” (European Light Sources Activities).
Grant sponsor: Marie Curie European Reintegration Grant; Grant number: MERG-CT-2004-006358; Grant sponsor: Greek General Secretariat for Research and Technology; Grant number: ENTER01-EP115; Grant sponsor: EU FP7-REGPOT-2008-1 project “EUROSTRUCT”; Grant number: GA-230146; Grant sponsor: EU FP7-REGPOT-2009-1 Project “ARCADE”; Grant number: GA-245866; Grant sponsor: Research Promotion Foundation of Cyprus; Grant number: YGEIA/0506/05.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Santama N, Ogg SC, Malekkou A, Zographos SE, Weis K, Lamond AI. Characterization of hCINAP, a novel coilin-interacting protein encoded by a transcript from the transcription factor TAFIID(32) locus. J Biol Chem. 2005;280:36429–36441. doi: 10.1074/jbc.M501982200. [DOI] [PubMed] [Google Scholar]
- 2.Ren H, Wang L, Bennett M, Liang Y, Zheng X, Lu F, Li L, Nan J, Luo M, Eriksson S, Zhang C, Su XD. The crystal structure of human adenylate kinase 6: an adenylate kinase localized to the cell nucleus. Proc Natl Acad Sci USA. 2005;102:303–308. doi: 10.1073/pnas.0407459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Rompay AR, Johansson M, Karlsson A. Phosphorylation of nucleosides and nucleoside analogs by mammalian nucleoside monophosphate kinases. Pharmacol Ther. 2000;87:189–198. doi: 10.1016/s0163-7258(00)00048-6. [DOI] [PubMed] [Google Scholar]
- 4.Noma T. Dynamics of nucleotide metabolism as a supporter of life phenomena. J Med Invest. 2005;52:127–136. doi: 10.2152/jmi.52.127. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Gonzalez A, Kourembanas S, Wyatt TA, Mitsialis SA. Mutation of Murine Adenylate Kinase 7 Underlies a Primary Ciliary Dyskinesia Phenotype. Am J Respir Cell Mol Biol. 2009;40:305–313. doi: 10.1165/rcmb.2008-0102OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulz GE, Muller CW, Diederichs K. Induced-fit movements in adenylate kinases. J Mol Biol. 1990;213:627–630. doi: 10.1016/S0022-2836(05)80250-5. [DOI] [PubMed] [Google Scholar]
- 7.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989;341:209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- 9.Kjeldgaard M, Nissen P, Thirup S, Nyborg J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993;1:35–50. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- 10.Noel JP, Hamm HE, Sigler PB. The 2.2 A crystal structure of transducin-alpha complexed with GTP gamma S. Nature. 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 11.Meng G, Zhai R, Liu B, Zheng X. Identification of a novel nuclear-localized adenylate kinase from Drosophila melanogaster. Biochemistry (Mosc) 2008;73:38–43. doi: 10.1134/s0006297908010057. [DOI] [PubMed] [Google Scholar]
- 12.Zhai R, Meng G, Zhao Y, Liu B, Zhang G, Zheng X. A novel nuclear-localized protein with special adenylate kinase properties from Caenorhabditis elegans. FEBS Lett. 2006;580:3811–3817. doi: 10.1016/j.febslet.2006.05.074. [DOI] [PubMed] [Google Scholar]
- 13.Juhnke H, Charizanis C, Latifi F, Krems B, Entian KD. The essential protein fap7 is involved in the oxidative stress response of Saccharomyces cerevisiae. Mol Microbiol. 2000;35:936–948. doi: 10.1046/j.1365-2958.2000.01768.x. [DOI] [PubMed] [Google Scholar]
- 14.Granneman S, Nandineni MR, Baserga SJ. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol Cell Biol. 2005;25:10352–10364. doi: 10.1128/MCB.25.23.10352-10364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santama N, Krijnse-Locker J, Griffiths G, Noda Y, Hirokawa N, Dotti CG. KIF2beta, a new kinesin superfamily protein in non-neuronal cells, is associated with lysosomes and may be implicated in their centrifugal translocation. EMBO J. 1998;17:5855–5867. doi: 10.1093/emboj/17.20.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dian C, Eshaghi S, Urbig T, McSweeney S, Heijbel A, Salbert G, Birse D. Strategies for the purification and on-column cleavage of glutathione-S-transferase fusion target proteins. J Chromatogr B Anal Technol Biomed Life Sci. 2002;769:133–144. doi: 10.1016/s1570-0232(01)00637-7. [DOI] [PubMed] [Google Scholar]
- 17.Leatherbarrow RJ. GrafFit. Version 3.0 Erithakus Software; Staines, UK: 1992. [Google Scholar]
- 18.Geladopoulos TP, Sotiroudis TG, Evangelopoulos AE. A malachite green colorimetric assay for protein phosphatase activity. Anal Biochem. 1991;192:112–116. doi: 10.1016/0003-2697(91)90194-x. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr Part A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Jnt CCP4/ESF-EACBM Newsl Protein Crystallogr. 1992;(No.26) [Google Scholar]
- 21.COLLABORATIVE COMPUTATIONAL PROJECT N The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Part 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.French S, Wilson K. Treatment of negative intensity observations. Acta Crystallogr Sect A. 1978;34:517–525. [Google Scholar]
- 23.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr. 2006;39:109–111. [Google Scholar]
- 27.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck—a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 28.Althoff S, Zambrowicz B, Liang P, Glaser M, Phillips GN., Jr. Crystallization and preliminary X-ray analysis of Escherichia coli adenylate kinase. J Mol Biol. 1988;199:665–666. doi: 10.1016/0022-2836(88)90310-5. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen WL, Maxwell DS, TiradoRives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 30.MacroModel. version 9.5. LLC; New York, NY: 2007. Schrödinger. [Google Scholar]
- 31.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 32.Still WC, Tempczyk A, Hawley RC, Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc. 1990;112:6127–6129. [Google Scholar]
- 33.Glide. version 4.5. LLC; New York, NY: 2007. Schrödinger. [Google Scholar]
- 34.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 35.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 36.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 37.Schrödinger Suite 2007 Induced Fit Docking protocol. Glide version 4.5. LLC; New York, NY: 2007. Schrödinger. [Google Scholar]; Prime version 1.6. LLC; New York, NY: 2007. Schrödinger. [Google Scholar]
- 38.QSite. version 5.0. New York, NY: 2008. Schrödinger. [Google Scholar]
- 39.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B Condens Matter. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 40.Becke AD. Density-functional thermochemistry 3. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 41.Francl MM, Pietro WJ, Hehre WJ, Binkley JS, Gordon MS, Defrees DJ, Pople JA. Self-consistent molecular-orbital methods. A polarization-type basis set for 2nd-row elements. J Chem Phys. 1982;77:3654–3665. [Google Scholar]
- 42.Harihara PC, Pople JA. Influence of polarization functions on molecular-orbital hydrogenation energies. Theor Chim Acta. 1973;28:213–222. [Google Scholar]
- 43.Hehre WJ, Ditchfie R, Pople JA. Self-consistent molecular-orbital methods 12. Further extensions of gaussian-type basis sets for use in molecular-orbital studies of organic-molecules. J Chem Phys. 1972;56:2257. [Google Scholar]
- 44.Kabsch W. Solution for best rotation to relate 2 sets of vectors. Acta Crystallogr Sect A. 1976;32:922–923. [Google Scholar]
- 45.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMOL molecular graphics system, delano scientific. Palo Alto, CA: 2002. [Google Scholar]
- 47.Berry MB, Meador B, Bilderback T, Liang P, Glaser M, Phillips GN., Jr. The closed conformation of a highly flexible protein: the structure of E. coli adenylate kinase with bound AMP and AMPPNP. Proteins. 1994;19:183–198. doi: 10.1002/prot.340190304. [DOI] [PubMed] [Google Scholar]
- 48.Vonrhein C, Bonisch H, Schafer G, Schulz GE. The structure of a trimeric archaeal adenylate kinase. J Mol Biol. 1998;282:167–179. doi: 10.1006/jmbi.1998.2003. [DOI] [PubMed] [Google Scholar]
- 49.Krishnamurthy H, Lou H, Kimple A, Vieille C, Cukier RI. Associative mechanism for phosphoryl transfer: a molecular dynamics simulation of Escherichia coli adenylate kinase complexed with its substrates. Proteins. 2005;58:88–100. doi: 10.1002/prot.20301. [DOI] [PubMed] [Google Scholar]
- 50.Zhong H, Tran LM, Stang JL. Induced-fit docking studies of the active and inactive states of protein tyrosine kinases. J Mol Graph Model. 2009;28:336–346. doi: 10.1016/j.jmgm.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 51.Craig IR, Essex JW, Spiegel K. Ensemble docking into multiple crystallographically derived protein structures: an evaluation based on the statistical analysis of enrichments. J Chem Inf Model. 50:511–524. doi: 10.1021/ci900407c. [DOI] [PubMed] [Google Scholar]
- 52.Liao C, Park JE, Bang JK, Nicklaus MC, Lee KS. Exploring Potential Binding Modes of Small Drug-like Molecules to the Polo-Box Domain of Human Polo-like Kinase 1. ACS Med Chem Lett. 1:110–114. doi: 10.1021/ml100020e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chalmers DK, McRobb FM, Capuano B, Crosby IT, Yuriev E. Homology modeling and docking evaluation of aminergic G protein-coupled receptors. J Chem Inform Model. 2010;50:626–637. doi: 10.1021/ci900444q. [DOI] [PubMed] [Google Scholar]
- 54.Salam NK, Huang TH, Kota BP, Kim MS, Li Y, Hibbs DE. Novel PPAR-gamma agonists identified from a natural product library: a virtual screening, induced-fit docking and biological assay study. Chem Biol Drug Des. 2008;71:57–70. doi: 10.1111/j.1747-0285.2007.00606.x. [DOI] [PubMed] [Google Scholar]
- 55.Hayes JM, Skamnaki VT, Archontis G, Lamprakis C, Sarrou J, Bischler N, Skaltsounis AL, Zographos SE, Oikonomakos NG. Kinetics, in silico docking, molecular dynamics, and MM-GBSA binding studies on prototype indirubins, KT5720, and staurosporine as phosphorylase kinase ATP-binding site inhibitors: the role of water molecules examined. Proteins Struct Funct Bioinf. 2011;79:703–719. doi: 10.1002/prot.22890. [DOI] [PubMed] [Google Scholar]
- 56.Nucci NV, Pometun MS, Wand AJ. Site-resolved measurement of water-protein interactions by solution NMR. Nat Struct Mol Biol. 18:245–249. doi: 10.1038/nsmb.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol (Lond) 1910;40:iv–vii. [Google Scholar]
- 58.Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 59.Ricard J, Cornish-Bowden A. Co-operative and allosteric enzymes: 20 years on. Eur J Biochem. 1987;166:255–272. doi: 10.1111/j.1432-1033.1987.tb13510.x. [DOI] [PubMed] [Google Scholar]
- 60.Cajal SR. Un sencillo metodo de coloracion seletiva del reticulo protoplasmico y sus efectos en los diversos organos nerviosos de vertebrados y invertebrados. Trab Lab Invest Biol. 1903;2:129–221. [Google Scholar]
- 61.Ramani R, Boyd RJ. Conformational aspects of l-histidine. Can J Chem Rev Canadienne De Chimie. 1981;59:3232–3236. [Google Scholar]
- 62.Okimoto N, Yamanaka K, Ueno J, Hata M, Hoshino T, Tsuda M. Theoretical studies of the ATP hydrolysis mechanism of myosin. Biophys J. 2001;81:2786–2794. doi: 10.1016/S0006-3495(01)75921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shpargel KB, Ospina JK, Tucker KE, Hebert MD, Matera AG. Post-translational modifications of coilin control Cajal body number and composition. Mol Biol Cell. 2002;13:421a. [Google Scholar]
- 64.Shpargel KB, Ospina JK, Tucker KE, Matera AG, Hebert MD. Control of Cajal body number is mediated by the coilin C-terminus. J Cell Sci. 2003;116:303–312. doi: 10.1242/jcs.00211. [DOI] [PubMed] [Google Scholar]
- 65.Platani M, Goldberg I, Lamond AI, Swedlow JR. Cajal body dynamics and association with chromatin are ATP-dependent. Nat Cell Biol. 2002;4:502–508. doi: 10.1038/ncb809. [DOI] [PubMed] [Google Scholar]