Abstract
E7107 is a derivative of the pladienolide family of natural product spliceosome inhibitors, which targets the U2 small nuclear ribonucleoprotein (snRNP) subunit SF3b. The results of a first-in-man trial with E7107 have been reported, representing an important translational step towards the goal of modulating RNA splicing for cancer therapy.
In this issue of Clinical Cancer Research, Eskens and colleagues (1) report the results of a first-in-man clinical trial with the mRNA splicing inhibitor E7107.
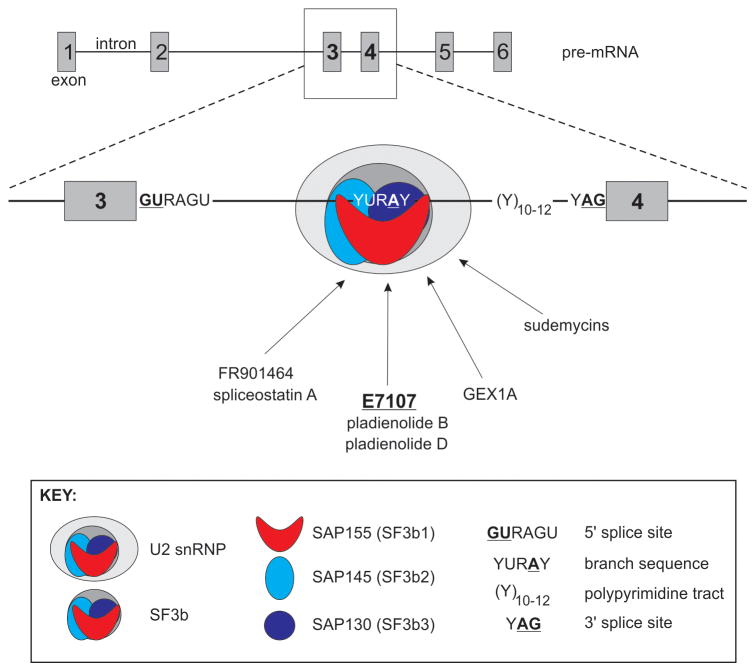
Splicing is an mRNA processing step wherein intervening introns are removed from pre-mRNAs and exons are joined. Splicing is catalyzed in the nucleus by the spliceosome, a complex consisting of the U1, U2, U4, U5, and U6 small nuclear ribonucleoproteins (snRNPs) and associated protein factors (2). Intron removal is directed by intronic motifs including the 5′ splice site, the branch sequence, the polypyrimidine tract, and the 3′ splice site, which serve as scaffolds for proper assembly of the spliceosome on newly-transcribed pre-mRNAs. The small nuclear RNA (snRNA) component of the U2 snRNP base-pairs with the branch sequence, leading to formation of the lariat structure required for intron removal. The U2 snRNP complex is composed of U2 snRNA, U2 snRNP-specific proteins, Sm proteins, and splicing factors SF3a and SF3b. SF3b is a multiprotein complex, which includes splicing associated proteins (SAP)155, SAP145, and SAP130 (also referred to as SF3b1, SF3b2, and SF3b3, respectively, Fig. 1) (3).
Fig. 1.
Mechanism-of-action schematic for E7107 and spliceosome inhibitors. E7107 inhibits splicing by blocking association of U2 snRNP with the intron branch point in pre-mRNAs. E7107 and spliceosome inhibitors bind SF3b complex core components SAP155, SA145, and SAP130.
Demarcation of intron/exon boundaries is mediated by enhancer and suppressor elements located in introns and exons. Different patterns of intron/exon demarcation can occur for a single gene, giving rise to multiple configurations of joined exons. This process, termed alternative splicing, is important for enhancing proteomic diversity. Alternative splicing can also go awry in various pathologies (4). In particular, altered splicing is a fundamental feature of cancer cells leading to activation of driver oncogenes, inactivation of tumor suppressor genes, and resistance to therapy (5, 6). Therefore, splicing inhibition or modulation could represent a promising anti-cancer strategy.
E7107 is a synthetic cycloheptylpiperazine-containing derivative of Pladienolides B and D, which are natural compounds isolated from Streptomyces platensis (7). These compounds all have anti-tumor activity in vitro and in vivo. A chemical probe approach was used to identify the cellular target of pladienolides and E7107, revealing affinity of these drugs for nuclear speckles, where splicing- and transcription-related molecules are abundant. Subsequent immunoprecipitation, crosslinking, and mass spectrometry analyses identified that pladienolides associated with a complex that contained SF3b components SAP155, SAP145, and SAP130 (7), which appears to be due to direct drug binding to SAP130 in a SAP145-dependent manner (7). Similar chemical probe approaches were used to identify SAP155, SAP145, and SAP130 as SF3b complex components targeted by the Pseudomonas anti-tumor natural product FR901464 and its synthetic derivative, spliceostatin A (8), GEX1A (herboxidiene), a natural product isolated from Streptomyces (9), and sudemycins, which are synthetic analogues based on a proposed consensus pharmacophore obtained from pladienolide B and FR901464 (Fig. 1) (10). While these findings represent major advances in target identification, definitive structural elucidation of this class of drugs in complex with SF3b remains an outstanding issue.
E7107, and perhaps other drugs in this class, appear to function mechanistically as SF3b inhibitors. For example, E7107 blocks spliceosome assembly in vitro by preventing U2 snRNP binding to the pre-mRNA branch point (11). In addition, treatment of cells with SF3b inhibitors has effects similar to knock-down of SF3b components, including accumulation of unspliced mRNAs that can be exported to the cytoplasm and translated (7, 8). One such unspliced RNA and translated protein is a variant of p27 resulting from translational read-through from exon 1 into intron 1 (8, 9)
The study by Eskens and colleagues (1) is an important advance in the field of spliceosome inhibitor development, because it represents the first human trial with this class of drugs. In this study, 40 patients were treated intravenously with E7107 for 3 consecutive weeks in a 28-day schedule with doses ranging from 0.6mg/m2–4.5mg/m2. The stated objectives of this trial were to identify dose-limiting toxicities, explore safety and tolerability, determine the pharmacokinetic profile of E7107, determine biomarkers of pharmacodynamics effect and potential efficacy, and explore antitumor activity. Pharmacokinetics of E7107 was dose-dependent. Dose limiting toxicities were seen during the first treatment cycle for 2 of the 3 patients treated at a dose of 4.5mg/m2, but 0 of the 7 patients treated at 3.0mg/m2. At 4.0 mg/m2, 1 of 6 patients required dose reduction. Based on these findings, 4.0 mg/m2 was selected as the maximum tolerable dose for further testing with 10 patients, only 1 of which required dose reduction. The most frequent side effects were nausea, vomiting, diarrhea, and fatigue. However, visual disturbance due to bilateral optic neuritis was noted as a severe toxicity in 2 patients in a companion study, leading to discontinuation of both E7107 trials. One patient in the trial reported by Eskens and colleagues (1) developed a similar optic neuritis after drug discontinuation. Although this visual disturbance was reversible, the mechanism underlying this toxicity is unknown, which represents a barrier to further clinical development. An important question is whether this may be a common on-target effect of all SF3b inhibitors, or whether this may be an effect restricted to E7107 that could be avoided with alternative SF3b inhibitors (Fig. 1).
Perhaps the most significant finding in this trial came from pharmacodynamics analysis, indicating that E7107 had in vivo effects consistent with a spliceosome inhibition mechanism of action. Properly-spliced mRNA levels of several genes that were found to be decreased by E7107 treatment in vitro (TRAPPC4, SLC25A19, and GTF2H1), were decreased in peripheral blood mononuclear cells in 38 of 40 patients. Moreover, the magnitude of this decrease was proportional to the patients’ E7107 dose. Similarly, accumulation of un-spliced pre-mRNAs for DNAJB1 and EIF4A1 were observed, although the levels of mature mRNAs for DNAJB1 and EIF4A mRNA were relatively unchanged.
Despite these pharmacodynamics results indicating on-target splicing inhibition in peripheral blood mononuclear cells, no partial or complete responses to E7107 were observed. However, a partial response was observed in one patient after drug discontinuation, and stable disease was noted for 8 patients. It is worth noting that pharmacodynamics evaluation of E7107 was restricted to blood cells. Therefore, it is not clear from this study whether E7107 was able to achieve splicing inhibition in tumor tissue. In addition, the genes selected for RT-PCR analysis in peripheral blood were selected based on large fold-changes in gene expression microarray experiments in vitro as opposed to gene selection based on known roles in cancer. It would be important to determine whether E7107 can effect a general inhibition of expression levels for important driver oncogenes or effect more specific splicing changes, such as blocking the expression of oncogenic splice variants associated with disease development, progression, and therapeutic resistance (5, 6).
This clinical study is also significant in light of recent cancer genome sequencing studies reporting recurrent mutations in proteins important for splicing regulation, including U2 snRNP core components SF3b1, SF3a1, U2AF1, and U2AF2 (12). While the mechanistic outcomes of these mutations are largely unknown, SF3b1 mutations track with poor prognosis in chronic lymphocytic leukemia (12). However, in myelodysplastic syndromes, SF3b1 mutations track with favorable prognosis (12). These dichotomous clinical findings point to possible cell-type-specific effects of mutations in spliceosome components, which may also portend organ-or cell-type specific effects of spliceosome inhibitors. This should be an important consideration for ongoing clinical development of spliceosome inhibitors, as well future trial design.
Acknowledgments
Financial Support: American Cancer Society Research Scholar Grant (RSG-12-031-01), National Institutes of Health (R01CA174777), and Department of Defense Prostate Cancer Research Program New Investigator Award (W81XWH-10-1-0353). S.M.D. is a Masonic Scholar of the Masonic Cancer Center, University of Minnesota.
Footnotes
Conflicts of Interest: none
References
- 1.Eskens FA, Ramos FJ, Burger H, O’Brien JP, Piera A, de Jonge MJ, et al. Phase I, Pharmacokinetic and Pharmacodynamic Study of the First-in-Class Spliceosome Inhibitor E7107 in Patients with Advanced Solid Tumors. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-13-0485. [DOI] [PubMed] [Google Scholar]
- 2.Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14:153–65. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 3.Golas MM, Sander B, Will CL, Luhrmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980–4. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 4.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–61. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 5.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–64. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dehm SM. mRNA splicing variants: exploiting modularity to outwit cancer therapy. Cancer Res. 2013;73:5309–14. doi: 10.1158/0008-5472.CAN-13-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nature chemical biology. 2007;3:570–5. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 8.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nature chemical biology. 2007;3:576–83. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 9.Hasegawa M, Miura T, Kuzuya K, Inoue A, Won Ki S, Horinouchi S, et al. Identification of SAP155 as the target of GEX1A (Herboxidiene), an antitumor natural product. ACS chemical biology. 2011;6:229–33. doi: 10.1021/cb100248e. [DOI] [PubMed] [Google Scholar]
- 10.Fan L, Lagisetti C, Edwards CC, Webb TR, Potter PM. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS chemical biology. 2011;6:582–9. doi: 10.1021/cb100356k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folco EG, Coil KE, Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 2011;25:440–4. doi: 10.1101/gad.2009411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cazzola M, Rossi M, Malcovati L. Biologic and clinical significance of somatic mutations of SF3B1 in myeloid and lymphoid neoplasms. Blood. 2013;121:260–9. doi: 10.1182/blood-2012-09-399725. [DOI] [PMC free article] [PubMed] [Google Scholar]