Abstract
Background
A significant proportion of children with asthma have delayed diagnosis of asthma by health care providers. Manual chart review according to established criteria is more accurate than directly using diagnosis codes, which tend to under-identify asthmatics, but chart reviews are more costly and less timely.
Objective
To evaluate the accuracy of a computational approach to asthma ascertainment, characterizing its utility and feasibility toward large-scale deployment in electronic medical records.
Methods
A natural language processing (NLP) system was developed for extracting predetermined criteria for asthma from unstructured text in electronic medical records and then inferring asthma status based on these criteria. Using manual chart reviews as a gold standard, asthma status (yes vs no) and identification date (first date of a “yes” asthma status) were determined by the NLP system.
Results
Patients were a group of children (n =112, 84% Caucasian, 49% girls) younger than 4 years (mean 2.0 years, standard deviation 1.03 years) who participated in previous studies. The NLP approach to asthma ascertainment showed sensitivity, specificity, positive predictive value, negative predictive value, and median delay in diagnosis of 84.6%, 96.5%, 88.0%, 95.4%, and 0 months, respectively; this compared favorably with diagnosis codes, at 30.8%, 93.2%, 57.1%, 82.2%, and 2.3 months, respectively.
Conclusions
Automated asthma ascertainment from electronic medical records using NLP is feasible and more accurate than traditional approaches such as diagnosis codes. Considering the difficulty of labor-intensive manual record review, NLP approaches for asthma ascertainment should be considered for improving clinical care and research, especially in large-scale efforts.
Introduction
Asthma is the most common chronic illness in childhood and is a major cause of morbidity in adults, affecting 4% to 17% of children and 7.7% of adults in the United States.1–3 Nearly 30 million Americans and 300 million people globally are estimated to be affected by asthma.4 Currently, there are no overall signs of a decreasing trend in asthma prevalence; rather, asthma continues to increase in many parts of the world.3–5 The total incremental cost of asthma to society is estimated to be $56 billion in the United States.2 In addition, compared with nonasthmatic children, asthmatics have significantly increased risks of serious or common microbial infections.6,7 Therefore, asthma, especially poorly controlled asthma, is a significant medical and economic burden to society.8
Timely identification of asthma is crucially important to mitigate this burden to society and improve the quality of life of asthmatics. Despite the availability of electronic medical records (EMRs), significant delay often occurs in diagnosing asthma, which in turn delays timely access to therapeutic interventions for asthma.9,10 For example, Molis et al reported that 179 of 276 children with asthma (65%) had a delayed diagnosis of asthma, with a median delay of 3.3 years, suggesting many asthmatic children are not diagnosed with asthma in a timely manner.11,12 This delayed diagnosis of asthma has been an impediment to asthma care and research (eg, not qualified for a priority group for H1N1 vaccination).13 In addition, the delay in asthma diagnosis might result in unnecessary treatments and evaluations such as antibiotics or chest x-ray examinations and frequent urgent care visits.9
To address these challenges, some EMR-based approaches have been used to identify asthmatics for patient care and research. For example, structured data such as diagnosis codes (eg, International Classification of Diseases, Ninth Revision [ICD-9] codesused for billing) tend to under-identify asthma cases.11,12,14 Alternatively, manual chart reviews of EMR data to ascertain asthma status are labor intensive and thus are inefficient for working with asthmatic children (eg, about 6 months for a full-time study coordinator to ascertain the asthma status for 900 children in the authors’ experience). Without the ability to promptly and accurately identify asthmatic patients, large-scale epidemiologic studies and clinical trials are limited by the expensive and lengthy cohort identification step.
To improve timely identification of asthma in children, the authors developed and validated a natural language processing (NLP) algorithm for extracting asthma-relevant information from clinical notes and other unstructured text in the EMR, thus automating the chart review process. In this exploratory study, the authors used predetermined criteria for asthma in manual medical record reviews and implemented the same criteria in the NLP algorithm for validation. The authors’ hypothesis was that NLP algorithms in the EMR would allow the identification of children with asthma in a more accurate and timely manner compared with conventional approaches such as ICD-9 codes. Decreasing the time and effort required to determine asthma status would significantly enhance the capabilities for pediatric asthma research, improve quality of asthma care through timely identification and access to effective therapy for asthma, and have a significant impact on public health.
Methods
Study Setting and Population
This study took place in Olmsted County, Minnesota, which had a 2010 census population of 124,277 (90.3% white compared with 89.4% in Minnesota and 75.1% in the United States). With the exception of a larger proportion of the working population employed in the health care industry, characteristics of Olmsted County populations were similar to those of the US white population.15–17 This allowed the authors to readily identify cases or events of interest and a community sample of study subjects representing the population of Olmsted County.
Study Design
This was a cross-sectional study and its overall design is depicted in Figure 1. Briefly, automatic NLP systems (bottom branch) were validated against comprehensive manual medical record review (top branch). As a comparison, the accuracy and timeliness of ICD-9 codes (middle branch) were also validated against medical record review. All retrieved medical documents for each subject (multiple arrows indicate multiple documents) were used to produce a subject’s asthma status as an output (single arrow indicates a single asthma status per subject).
Figure 1.
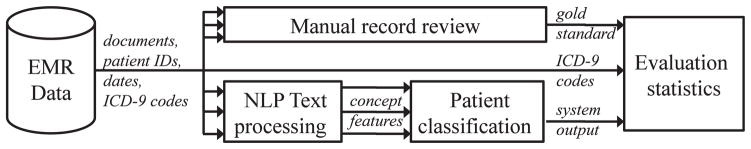
A conceptual schematic of manual (top branch) vs automatic (middle and bottom branches) asthma ascertainment. EMR, electronic medical record; ICD-9, International Classification of Diseases, Ninth Revision; NLP, natural language processing.
Study Subjects
The 112 study subjects were children younger than 4 years who were enrolled in the Mayo Clinic sick-child daycare program and their parents.18,19 The rationales for using this study cohort included (1) the availability of asthma status ascertained by predetermined criteria for asthma and ICD-9 code, (2) subjects were born after implementation of the EMR at the Mayo Clinic, and (3) a prospective follow-up including parental survey mitigating the limitations of a retrospective study. Exclusion criteria specific to this study included patients with no ICD-9 codes and patients who were recorded as being seen at hospitals other than the Mayo Clinic. This study was approved by the institutional review board for human subject research at the Mayo Clinic and Olmsted Medical Center.
Asthma Status by Medical Record Review
Given the lack of a gold standard test for ascertaining asthma status in research and clinical practice, the authors determined gold standard asthma status for subjects by comprehensive manual medical record review according to the predetermined criteria shown in Figure 2.17 These criteria infer asthma status based on a constellation of symptoms suspicious for asthma. The authors used these criteria for validating the NLP algorithm, not necessarily for promoting the criteria for research or clinical practice for the difficult problem of determining pediatric asthma status. Originally developed by Yunginger et al,17 these retrospective medical record-based asthma criteria were found to have high interobserver reliability and agreement and have been used extensively in research for asthma epidemiology.17,20–28 In the present study, probable and definite asthma types were combined because most probable asthmatics become definite over time.17,29 How to determine asthma status based on the 3 criteria displayed in Figure 2 has been previously reported.12 Although criteria 1 and 3 are concerned with individual medical events, criterion 2 (“substantial variability in symptoms from time to time …”) is concerned with the temporal relation between recurrent events. The medical record review considered the second criterion to be met when asthma symptoms or signs (ie, criteria 1 and 3) had occurred at least twice in a period of 4 weeks to 3 years.
Figure 2.
Criteria for asthma ascertainment in manual record review by a clinician. Gray highlighting indicates primary features (concepts) searched for by natural language processing. FEV1, forced expiration volume in 1 second.
In addition to asthma status, an index date of asthma was determined for all subjects. This asthma index date was defined as the earliest constellation of symptoms found in the medical record that met the predetermined criteria for asthma shown in Figure 2. How the index date is determined has been reported previously.12 It should be noted that specific criteria were used to estimate an approximate index date in cases in which the onset of asthma was described in general terms. One example is patients who were described as having asthma for “a few years.” In this study, “a few years” was consistently considered to indicate 3 years in the inferring time frame.
Asthma Status by NLP Algorithms
The authors retrieved each patient’s EMR documents that were created before the manual record review date. To automatically identify cases of asthma, the authors used a 2-step process, as indicated by the lower branch depicted in Figure 1: a text processing component (finding concepts in text that match the specified criteria) and a patient classification component (deciding the asthma status of a patient based on the available evidence). For text processing, the authors used the Mayo Clinic’s Clinical Text Analysis and Knowledge Extraction System (cTAKES) v1.3.230 and added feature extraction, relational and temporal logic, and an optional machine learning component. cTAKES analyzed text and found medical concepts according to an asthma-specific dictionary the authors provided. For example, if the NLP algorithm encountered a sentence that stated “no rales or wheezing,” it marked that the concept “wheezing” was found. It also noted that the record actually stated that the patient did not have wheezing.
Individual concepts such as “wheezing” were considered primary features for classifying patients according to their asthma status; other primary features are highlighted in Figure 2. To accurately represent the logic of the criteria, some primary features were combined into secondary features. For example, criterion 1 requires that “coughing” and “wheezing” be present. The authors required the 2 related concepts to appear in the same section of the clinical note, and this was encapsulated in a secondary feature. For criteria involving laboratory results (eg, “blood eosinophilia higher than 300/μL”), the authors looked within sentences for multiple concepts.
Patient Classification: Criteria-based Logic
With these primary and secondary features, the asthma status of each document was determined according to the criteria in Figure 2. Namely, cases of criterion 1 or 3 that repeated (recurrence of episodes of cough with wheezing) within a span of 4 weeks to 3 years (criterion 2) were considered positive asthma cases. In this way, all documents for a patient were considered jointly to determine the final asthma status of a patient. Alternatively, finding an explicit “asthma” term in a diagnosis or problem list constituted physician-diagnosed asthma, so this patient also was considered asthmatic (even without repeat symptoms).
For each patient, the asthma status and the system’s estimate of the index date (or null, if the patient was not asthmatic) were output. The latter was considered the estimated inception date.
Patient Classification: Machine Learning
In addition to a faithful implementation of the established criteria, an alternative method of obtaining asthma status from the per-document features was tested using machine learning methods, which statistically infer asthma status based on examples. The features for each document were used as a 0 or 1 indicator and were summed for each document of a patient, producing an overall frequency that each symptom was seen. The gold standard asthma status was considered the response variable, from which to learn a classifier. Note that no estimated inception date was available, because the decisions were made on the entirety of each patient’s records.
The C4.5 decision tree algorithm31 with 10-fold cross-validation in the Weka machine learning environment was used.32 The C4.5 algorithm produces an optimal decision tree given the data and asthma status per patient. The authors experimented with different parameters, including the ability to simplify (prune) the tree, because a simpler explanation might generalize better to unseen data.
Data Analysis
Automatic asthma ascertainment methods (ICD-9 codes, NLP algorithms) were analyzed against the gold standard of criteria-based medical record review. The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and F1 score (the harmonic mean of sensitivity and PPV) were calculated. In addition, the automatic methods were viewed as an alternate observer, and agreement and the Cohen unweighted κ index were calculated to determine interobserver reliability.
Further, the timeliness of automatic ascertainments was evaluated whenever an estimated inception date could be compared with an index date. For true positive cases by an automatic algorithm, the difference between the estimated inception date and the index date was measured in months. Also noted was whether the automatic algorithm identified asthma “before,” “at” the same time, or “after” the manual record review did. For false negatives, the automatic algorithm was considered to have “never” identified the correct asthma status.
Results
Study Cohort
A convenience sample of 115 children participated in previous studies on asthma,14,19 in which 84% were reported to be Caucasians, 49% were girls, the mean age was 2.0 years (standard deviation 1.03 years), and asthma was reported in 27 cases (23.5%). For this study, 3 patients were excluded because their records spanned multiple institutions. A second sample of 127 children, a subset of a previous cohort, was not fit for evaluation but served as a development set to help with the designing of the logic-based NLP algorithm.
Accuracy of Asthma Ascertainment
Sensitivity, specificity, PPV, NPV, F1 score, agreement values, and Cohen κ values are presented in Table 1 for each of the 3 approaches for retrieving cohorts: ICD-9 codes, the logic-based NLP system, and the machine learning-based NLP system.
Table 1.
Performance of various asthma ascertainment methods
| Sensitivity | Specificity | PPV | NPV | F1 score | Agreement | κ | |
|---|---|---|---|---|---|---|---|
| ICD-9 codes | 30.8% | 93.2% | 57.1% | 82.2% | 40.0% | 79.1% | 0.287 |
| NLP system (logic) | 80.8% | 95.3% | 84.0% | 94.3% | 82.4% | 92.0% | 0.772 |
| NLP system (ML) | 84.6% | 96.5% | 88.0% | 95.4% | 86.3% | 93.8% | 0.822 |
Abbreviations: ICD-9, International Classification of Diseases, Ninth Revision; ML, machine learning; NLP, natural language processing; NPV, negative predictive value; PPV, positive predictive value.
ICD-9 codes showed good specificity (93.2%) but very poor sensitivity (30.8%). Thus, many patients who were identified as asthmatics by manual record review were never coded as having asthma. The 2 NLP systems in the bottom half of Table 1 showed improvements across the board, particularly in sensitivity, PPV, F1 score, and κ values. Overall, the NLP systems captured more of the positive asthmatic cases, and did so more precisely, than ICD-9 codes.
In addition, the machine learning-based classification of patients outperformed the logic-based classification, because the harmonic mean (F1 score) increased from 82.4% to 86.3%.
Timeliness of Asthma Ascertainment
Previous work has shown that physician diagnosis is delayed or nonexistent approximately 65% of the time.12 Therefore, this study examined whether the present system’s ascertained asthma status might have similar delays. For patients with asthma (as validated by manual record review), the index dates of manually reviewed asthma cases were compared with the estimated inception dates of the automatic methods. For ICD-9 codes, the date of the earliest medical record with a code of 493 (or subcodes) was used as the estimated inception date.
Of the 112 patients in the dataset, 26 were labeled as asthmatic by manual medical record review. In comparing the timeliness of automatic identification with the gold standard, examination was restricted to these 26 cases (an asthma diagnosis cannot be delayed if the patient is not, in fact, asthmatic). In this study, the manually determined index date was considered the temporal reference point. The automatically determined estimated inception date could occur in 4 positions: after the index date (ie, a delay in automatic diagnosis), at the index date (ie, a timely automatic diagnosis), before the index date (ie, a premature automatic diagnosis), or never (ie, a missed automatic diagnosis).
Table 2 presents these qualitative categories, in which the granularity of comparisons between the estimated inception date and the index date was at 1 month. For example, “at” was defined as having an estimated inception date within 1 month of (before or after) the index date. The first row shows that most ICD-9 codes were significantly delayed, with 22 of 26 in the “after” or “never” categories (84.6%); few cases were identified in a timely fashion. Conversely, the logic-based NLP system identified only 7 of 26 asthmatics (26.9%) in these delayed-diagnosis categories. Furthermore, 14 of 26 (53.8%) were identified without delay or prematurely.
Table 2.
Timeliness of automatic asthma ascertainment methods compared with manual medical record review
| Estimated inception date | Relation to index date
|
|||
|---|---|---|---|---|
| Before | At | After | Never | |
| ICD-9 codes | 2 | 2 | 7 | 15 |
| NLP system (logic) | 5 | 14 | 3 | 4 |
Abbreviations: ICD-9, International Classification of Diseases, Ninth Revision; NLP, natural language processing.
Timeliness of diagnosis was characterized further according to the histogram displayed in Figure 3. The NLP system detected true positives in a relatively close period to the true index date (σ2 =4.97) and tended to pick out asthmatics before they were technically labeled as such (indicated by negative-valued delays, skew = −1.12). The distribution of ICD-9 code dates was more distributed (σ2 =6.98) and was more delayed on average (indicated by positive-valued delays, skew =0.29).
Figure 3.
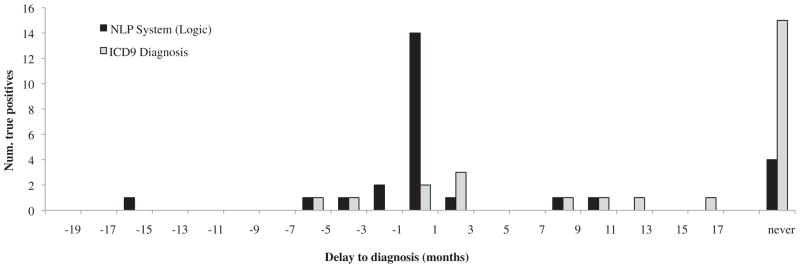
Timeliness of asthma diagnosis (histogram). The x-axis markers indicate the upper limit of a frequency bin. ICD-9, International Classification of Diseases, Ninth Revision; NLP, natural language processing.
Discussion
The results in this exploratory study show that NLP algorithms can accurately ascertain the asthma status of patients in a timely manner by using textual information in the EMR. The results show that NLP algorithms are much more accurate than commonly used ICD-9 codes for this task. The increase in sensitivity from 30.8% (ICD-9 codes) to 80.8% (NLP system with logical classification) significantly enhances data quality and, the authors suggest, is worth the additional effort.
This finding suggests that it may be misleading (although commonplace) to consider these billing codes as a stand-in for a more refined asthma ascertainment methodology. Recent work by Pacheco et al33 used ICD-9 codes alongside other structured data components, producing an automatic asthma ascertainment algorithm with excellent PPV and NPV (95% and 96%, respectively). The sensitivity of this method was not reported, which is an acceptable omission in enrolling patients for a study, but a poor fit to epidemiologic studies.
In a systematic review by Sanders and Aronsky,34 4 categories of informatics research related to asthma were identified; the present work would be categorized as an article on the “detection and diagnosis” of childhood asthma. The present study differs from and complements existing work in important ways. Previous work on detecting pediatric asthma used structured data,35 patient questionnaires,36,37 or structured data and patient questionnaires,38 but ignored NLP approaches to leveraging the information in the clinical text. Donahue et al39 performed a study of asthma identification that used structured data and clinical text and found limited uses for the clinical text. The present study is different because the clinical text was approached from a modern NLP perspective, the clinical text was not relegated to the determination of asthma severity, and sensitivity was reported. Therefore, the present study found significant use for the clinical text. Most relevant to this study is perhaps the earliest informatics study on detecting asthma40 from the Linguistic String Project. Sager et al40 applied their NLP techniques to documents and found an average sensitivity of 82.5% and a PPV of 82.1% on their test set. The present results are comparable, although the authors classified the asthma status of patients rather than individual clinical notes, and addressed the issue of time-elapsed information about chronic illness. In addition, previous techniques were not based on predetermined criteria.
It is important to note that the present results also show that the estimated inception date produced by the NLP system approximated the index date with promising accuracy compared with ICD-9 codes. This system’s lower standard deviation illustrates this; the effect would be even more pronounced if there were a means to include the undefined period of delay arising from false negatives. In addition, 14 of the positive cases (68.2% of true positives) were identified within 1 month before or after the true index date. ICD-9 codes reported only 2 positive cases (18.2% of true positives) identified in a similar span.
Several attempts have been made to quantify the delay in asthma diagnosis. Most relevant to the present study is the work by Molis et al,12 which found that physician diagnoses were delayed in 65% of their sample, with median delay of 3.3 years (or 1.4 years, excluding 57 of the 179 [32%] who were never diagnosed). In the present study, the timeliness comparison differed in that it measured the timeliness of ICD-9 codes and the NLP system, rather than physician diagnoses. In this light, ICD-9 codes had a delayed diagnosis in 22 of 26 (85%) of the sample; 15 of the 26 cases (58%) were never identified, and the median delay for remaining cases was 2.3 months. Compared with physician diagnoses, the ICD-9 codes are assigned quickly or not at all. The NLP system had a delayed diagnosis in just 7 of 26 cases (27%); the diagnoses had a median delay of 0 days regardless of whether the 4 of 26 (16%) of “never diagnosed” cases were excluded or not. Compared with physician diagnosis as discussed earlier, the NLP algorithm aligns better with the predetermined criteria for asthma and does so more promptly. This suggests that the NLP system can pick up on latent risk factors from the clinical text, accomplishing automatic and timely asthma status identification. As shown by the negative skewness, NLP algorithms may detect asthma before the index date, indicating that some early signs of asthma may trigger the identification process prematurely.
The present study findings are likely to enhance capabilities for asthma research. In addition, this NLP approach is likely to enhance quality improvement efforts in asthma, for instance, in large group practices in which it would not be feasible to conduct labor-intensive chart reviews to identify children with asthma who do not have a diagnosis of asthma. This may support quality improvement efforts in asthma care and potentially result in significant impacts on public health.
Although there is room for improvement, it is also noteworthy that the proposed methods for automatic asthma identification are accurate and timely. With relatively accurate timing information available through patient records, phenomena that involve patient care for asthma, such as the prevalence of remission and relapse, can be studied on a large scale. This would be impossible for current methods based on ICD-9 codes. Existing sophisticated, highly precise asthma ascertainment methods33 lack sensitivity and would not serve as general-purpose timing estimates.
Strengths of the present study include the use of previously reported asthma criteria for retrospective research based on medical records14 to guide the development of the NLP algorithms. Thus, the study fairly compared how the established criteria were implemented by humans vs automatic systems. In addition, this allows for future work with larger population-based cohorts to be methodologically consistent with previous findings.6,12,14,19,41 Another strength of this study is the introduction of a means for evaluating the timeliness of asthma diagnosis. Timely identification from clinical notes has not been often considered in the informatics literature but is likely to have a significant impact on research in chronic diseases. This study does have limitations that need to be considered. First, the present asthma criteria are based on medical record review, the manual record review is inherently limited by the information that is stored in the EMR, and asthma-related events reported outside a clinical visit would be missed. However, the criteria were found to have high reliability and have been used extensively in asthma epidemiologic research showing the association between asthma and known risk factors for asthma (construct validity). Second, the cohort is a modest sample of patients from a single institution. The exact algorithms and results may have limited generalizability to other institutions and EMR settings. Third, strictly speaking, comparing an NLP algorithm (designed for asthma status identification) with ICD-9 codes (designed for categorization and billing) may be incongruous; however, these are some of the best options available for research or clinical practice.
In conclusion, NLP-based algorithms have been shown to identify asthma status in a relatively timely and accurate manner and significantly outperform commonly used ICD-9 codes. With an accurate automatic method for ascertaining asthma status, larger-scale asthma epidemiologic research may be feasibly conducted. Furthermore, a promising automatic means of determining the timing of criteria-based asthma status may aid in monitoring asthma trends and aiding clinical decision support. Thus, the present results should contribute to quality improvements in asthma care by helping clinicians and health care systems identify potential asthmatics, enabling effective preventive and therapeutic interventions for asthma.
From the perspective of clinical research and care, the authors’ future work includes validating the reported association between asthma and other diseases such as microbial infections (pertussis or serious pneumococcal diseases6,7) or chronic diseases (coronary heart disease or diabetes) on a large scale. Similarly, other disease associations may be tested much more quickly on a large scale (eg, during the 2009 H1N1 novel influenza outbreak). From a medical informatics standpoint, future work includes the application of the algorithm among larger populations, the principled development of a facile user interface, extension of this work to other diseases, and an open-source distribution system.
Acknowledgments
The authors thank Drs Andrew Hashikawa and Robert Voigt and Ms Shirley Johnson for their support during the original study, Sean Murphy and Vinod Kaggal for information technologic support, and Dr Chris Derauf for review of manuscript drafts.
Funding Sources: This work was supported in part by grant ABI:0845523 from the National Science Foundation, grant 5R01LM009959 from the National Institutes of Health, grant U54 HG004028 from the National Institutes of Health Roadmap, grant 90TR000201 from SHARP 4 (principal investigator, Chute, MD, DrPH), and the Scholarly Clinician Award from Mayo Foundation (principal investigator, Young J. Juhn, MD).
Footnotes
Disclosures: Authors have nothing to disclose.
References
- 1.Eder W, Ege MJ, von Mutius E. The asthma epidemic [review] N Engl J Med. 2006;355:2226–2235. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 2.Barnett SB, Nurmagambetov TA. Costs of asthma in the United States: 2002–2007. J Allergy Clin Immunol. 2011;127:145–152. doi: 10.1016/j.jaci.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 3.Vital signs: asthma prevalence, disease characteristics, and self-management education: United States, 2001–2009. MMWR Morb Mortal Wkly Rep. 2011;60:547–552. [PubMed] [Google Scholar]
- 4.Bernsen RM, van der Wouden JC, Nagelkerke NJ, de Jongste JC. Early life circumstances and atopic disorders in childhood [review] Clin Exp Allergy. 2006;36:858–865. doi: 10.1111/j.1365-2222.2006.02518.x. [DOI] [PubMed] [Google Scholar]
- 5.Bjorksten B. Allergy priming early in life [comment] Lancet. 1999;353:167–168. doi: 10.1016/S0140-6736(05)77212-3. [DOI] [PubMed] [Google Scholar]
- 6.Juhn YJ, Kita H, Yawn BP, et al. Increased risk of serious pneumococcal disease in patients with asthma. J Allergy Clin Immunol. 2008;122:719–723. doi: 10.1016/j.jaci.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capili CR, Hettinger A, Rigelman-Hedberg N, et al. Increased risk of pertussis in patients with asthma. J Allergy Clin Immunol. 2012;129:957–963. doi: 10.1016/j.jaci.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Accordini S, Corsico AG, Braggion M, et al. The cost of persistent asthma in europe: an international population-based study in adults. Int Arch Allergy Immunol. 2013;160:93–101. doi: 10.1159/000338998. [DOI] [PubMed] [Google Scholar]
- 9.Lynch BA, Van Norman CA, Jacobson RM, Weaver AL, Juhn YJ. Impact of delay in asthma diagnosis on health care service use. Allergy Asthma Proc. 2010;31:e48–e52. doi: 10.2500/aap.2010.31.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch BA, Fenta Y, Jacobson RM, Li X, Juhn YJ. Impact of delay in asthma diagnosis on chest X-ray and antibiotic utilization by clinicians. J Asthma. 2012;49:23–28. doi: 10.3109/02770903.2011.637596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bisgaard H, Szefler S. Prevalence of asthma-like symptoms in young children. Pediatr Pulmonol. 2007;42:723–728. doi: 10.1002/ppul.20644. [DOI] [PubMed] [Google Scholar]
- 12.Molis WE, Bagniewski S, Weaver AL, Jacobson RM, Juhn YJ. Timeliness of diagnosis of asthma in children and its predictors. Allergy. 2008;63:1529–1535. doi: 10.1111/j.1398-9995.2008.01749.x. [DOI] [PubMed] [Google Scholar]
- 13.Litchfield SM. Summary recommendation for the ACIP for the use of H1N1 influenza vaccine for the 2009–2010 vaccination season. AAOHN J. 2009;57:354. doi: 10.3928/08910162-20090826-02. [DOI] [PubMed] [Google Scholar]
- 14.Juhn Y, Kung A, Voigt R, Johnson S. Characterisation of children’s asthma status by ICD-9 code and criteria-based medical record review. Prim Care Respir J. 2011;20:79–83. doi: 10.4104/pcrj.2010.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Census Bureau. 1980 and 1990 Census of Population and Housing. Washington, DC: US Census Bureau; 1983 and 1993. [Google Scholar]
- 16.Katusic SK, Colligan RC, Barbaresi WJ, Schaid DJ, Jacobsen SJ. Potential influence of migration bias in birth cohort studies. Mayo Clin Proc. 1998;73:1053–1061. doi: 10.4065/73.11.1053. [DOI] [PubMed] [Google Scholar]
- 17.Yunginger JW, Reed CE, O’Connell EJ, Melton LJ, III, O’Fallon WM, Silverstein MD. A community-based study of the epidemiology of asthma. Incidence rates, 1964–1983. Am Rev Respir Dis. 1992;146:888–894. doi: 10.1164/ajrccm/146.4.888. [DOI] [PubMed] [Google Scholar]
- 18.Voigt RG, Johnson SK, Hashikawa AH, et al. Why parents seek medical evaluations for their children with mild acute illnesses. Clin Pediatr (Phila) 2008;47:244–251. doi: 10.1177/0009922807308254. [DOI] [PubMed] [Google Scholar]
- 19.Yoo KH, Johnson SK, Voigt RG, Campeau LJ, Yawn BP, Juhn YJ. Characterization of asthma status by parent report and medical record review. J Allergy Clin Immunol. 2007;120:1468–1469. doi: 10.1016/j.jaci.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Beard CM, Yunginger JW, Reed CE, O’Connell EJ, Silverstein MD. Interobserver variability in medical record review: an epidemiological study of asthma. J Clin Epidemiol. 1992;45:1013–1020. doi: 10.1016/0895-4356(92)90117-6. [DOI] [PubMed] [Google Scholar]
- 21.Bauer BA, Reed CE, Yunginger JW, Wollan PC, Silverstein MD. Incidence and outcomes of asthma in the elderly. A population-based study in Rochester, Minnesota. Chest. 1997;111:303–310. doi: 10.1378/chest.111.2.303. [DOI] [PubMed] [Google Scholar]
- 22.Hunt LW, Jr, Silverstein MD, Reed CE, O’Connell EJ, O’Fallon WM, Yunginger JW. Accuracy of the death certificate in a population-based study of asthmatic patients. JAMA. 1993;269:1947–1952. [PubMed] [Google Scholar]
- 23.Juhn YJ, Qin R, Urm S, Katusic S, Vargas-Chanes D. The influence of neighborhood environment on the incidence of childhood asthma: a propensity score approach. J Allergy Clin Immunol. 2010;125:838–843.e2. doi: 10.1016/j.jaci.2009.12.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juhn YJ, Sauver JS, Katusic S, Vargas D, Weaver A, Yunginger J. The influence of neighborhood environment on the incidence of childhood asthma: a multilevel approach. Soc Sci Med. 2005;60:2453–2464. doi: 10.1016/j.socscimed.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 25.Juhn YJ, Weaver A, Katusic S, Yunginger J. Mode of delivery at birth and development of asthma: a population-based cohort study. J Allergy Clin Immunol. 2005;116:510–516. doi: 10.1016/j.jaci.2005.05.043. [DOI] [PubMed] [Google Scholar]
- 26.Silverstein MD, Reed CE, O’Connell EJ, Melton LJ, III, O’Fallon WM, Yunginger JW. Long-term survival of a cohort of community residents with asthma. N Engl J Med. 1994;331:1537–1541. doi: 10.1056/NEJM199412083312301. [DOI] [PubMed] [Google Scholar]
- 27.Silverstein MD, Yunginger JW, Reed CE, et al. Attained adult height after childhood asthma: effect of glucocorticoid therapy. J Allergy Clin Immunol. 1997;99:466–474. doi: 10.1016/s0091-6749(97)70072-1. [DOI] [PubMed] [Google Scholar]
- 28.Yawn BP, Yunginger JW, Wollan PC, Reed CE, Silverstein MD, Harris AG. Allergic rhinitis in Rochester, Minnesota residents with asthma: frequency and impact on health care charges. J Allergy Clin Immunol. 1999;103:54–59. doi: 10.1016/s0091-6749(99)70525-7. [DOI] [PubMed] [Google Scholar]
- 29.Juhn YJ, Kita H, Lee LA, et al. Childhood asthma and measles vaccine response. Ann Allergy Asthma Immunol. 2006;97:469–476. doi: 10.1016/S1081-1206(10)60937-4. [DOI] [PubMed] [Google Scholar]
- 30.Savova GK, Masanz JJ, Ogren PV, et al. Mayo Clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan JR. C4.5: Programs for Machine Learning. San Mateo, CA: Morgan Kaufmann; 1993. [Google Scholar]
- 32.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. SIGKDD Explor. 2009:11. [Google Scholar]
- 33.Pacheco JA, Avila PC, Thompson JA, et al. A highly specific algorithm for identifying asthma cases and controls for genome-wide association studies. AMIA Annu Symp Proc. 2009;2009:497–501. [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders DL, Aronsky D. Biomedical informatics applications for asthma care: a systematic review. J Am Med Inform Assoc. 2006;13:418–427. doi: 10.1197/jamia.M2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daley MF, Barrow J, Pearson K, et al. Identification and recall of children with chronic medical conditions for influenza vaccination. Pediatrics. 2004;113:e26–e33. doi: 10.1542/peds.113.1.e26. [DOI] [PubMed] [Google Scholar]
- 36.Kable S, Henry R, Sanson-Fisher R, Ireland M, Cockburn J. Is a computer questionnaire of childhood asthma acceptable in general practice? Fam Pract. 2006;23:88–90. doi: 10.1093/fampra/cmi079. [DOI] [PubMed] [Google Scholar]
- 37.Kable S, Henry R, Sanson-Fisher R, Ireland M, Corkrey R, Cockburn J. Childhood asthma: can computers aid detection in general practice? Br J Gen Pract. 2001;51:112–116. [PMC free article] [PubMed] [Google Scholar]
- 38.Grassi M, Villani S, Marinoni A. Classification methods for the identification of ‘case’ in epidemiological diagnosis of asthma. Eur J Epidemiol. 2001;17:19–29. doi: 10.1023/a:1010987521885. [DOI] [PubMed] [Google Scholar]
- 39.Donahue JG, Weiss ST, Goetsch MA, Livingston JM, Greineder DK, Platt R. Assessment of asthma using automated and full-text medical records. J Asthma. 1997;34:273–281. doi: 10.3109/02770909709067217. [DOI] [PubMed] [Google Scholar]
- 40.Sager N, Lyman M, Bucknall C, Nhan N, Tick LJ. Natural language processing and the representation of clinical data. J Am Med Inform Assoc. 1994;1:142–160. doi: 10.1136/jamia.1994.95236145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoo KH, Molis WE, Weaver AL, Jacobson RM, Juhn YJ. The impact of electronic medical records on timeliness of diagnosis of asthma. J Asthma. 2007;44:753–758. doi: 10.1080/02770900701645447. [DOI] [PubMed] [Google Scholar]



