Abstract
Ommatidial rotation is one of the most important events for correct patterning of the Drosophila eye. Although several signaling pathways are involved in this process, few genes have been shown to specifically affect it. One of them is nemo (nmo), which encodes a MAP-like protein kinase that regulates the rate of rotation throughout the entire process, and serves as a link between core planar cell polarity (PCP) factors and the E-cadherin–β-catenin complex. To determine more precisely the role of nmo in ommatidial rotation, live-imaging analyses in nmo mutant and wild-type early pupal eye discs were performed. We demonstrate that ommatidial rotation is not a continuous process, and that rotating and non-rotating interommatidial cells are very dynamic. Our in vivo analyses also show that nmo regulates the speed of rotation and is required in cone cells for correct ommatidial rotation, and that these cells as well as interommatidial cells are less dynamic in nmo mutants. Furthermore, microarray analyses of nmo and wild-type larval eye discs led us to identify new genes and signaling pathways related to nmo function during this process. One of them, miple, encodes the Drosophila ortholog of the midkine/pleiotrophin secreted cytokines that are involved in cell migration processes. miple is highly up-regulated in nmo mutant discs. Indeed, phenotypic analyses reveal that miple overexpression leads to ommatidial rotation defects. Genetic interaction assays suggest that miple is signaling through Ptp99A, the Drosophila ortholog of the vertebrate midkine/pleiotrophin PTPζ receptor. Accordingly, we propose that one of the roles of Nmo during ommatial rotation is to repress miple expression, which may in turn affect the dynamics in E-cadherin–β-catenin complexes.
Keywords: Nemo, Ommatidial rotation, Live-imaging, Gene expression, Miple, Drosophila eye
Introduction
The Drosophila adult eye is composed of around 800 units, or ommatidia, which are precisely oriented in mirror symmetric fashion relative to a dorsal–ventral midline, the equator. This pattern is generated during larval development in the eye imaginal disc, when ommatidial preclusters rotate 901 towards the equator adopting opposite chiral forms depending upon whether they lie dorsally or ventrally (Jenny, 2010). These patterning events closely follow a moving front of differentiation, the morphogenetic furrow (MF), which moves from posterior to anterior across the eye imaginal disc (Tomlinson and Ready, 1987). The Frizzled planar cell polarity (Fz-PCP) pathway controls the proper differentiation of R3 and R4 photoreceptors and, subsequently, the direction of ommatidial rotation (Seifert and Mlodzik, 2007). The direction of rotation depends on correct R3/R4 cell fate specification since misrotation is a common phenotype observed in loss- and gain-of-function mutants of PCP genes (Mlodzik, 1999). During this process ommatidial precursors rotate as a group, but independent of their undifferentiated, stationary neighbors, the interommatidial cells (IOCs) (Fiehler and Wolff, 2007). The exact cellular mechanisms that drive this behavior have not yet been established. In parallel to Fz-PCP signaling, which may regulate ommatidial rotation through effects on cytoskeletal elements via the Rho-Kinase Drok (Winter et al., 2001), this process is also regulated by the Epidermal growth factor receptor (Egfr) pathway (Brown and Freeman, 2003; Gaengel and Mlodzik, 2003; Strutt and Strutt, 2003). Egfr pathway members signal through both the Mitogen activated protein kinase (MAPK)/Pointed (Pnt) transcriptional cascade and Canoe (Cno) (Brown and Freeman 2003, Gaengel and Mlodzik, 2003), and also interact genetically with E-cadherin (E-cad) and N-cadherin (N-cad) during this process (Brown and Freeman, 2003; Gaengel and Mlodzik, 2003; Mirkovic and Mlodzik, 2006). Moreover, genes functionally related with cytoskeleton reorganization and cell adhesion act as downstream effectors of Egfr signaling, thus linking ommatidial rotation with cell adhesion and cytoskeleton rearrangements (Gaengel and Mlodzik, 2003; Mirkovic and Mlodzik, 2006). In addition, the cell adhesion molecules Echinoid (Ed) and Friend of Echinoid (Fred) are required at multiple steps during the ommatidial rotation process (Fetting et al., 2009), and Ed seems to be required to decrease Flamingo (one of the PCP core proteins) levels on non-rotating IOCs to permit correct rotation of ommatidial clusters (Ho et al., 2010). Other genes that have been shown to be required during ommatidial rotation are nemo (nmo), scabrous (sca) and zipper (zip) (Choi and Benzer, 1994; Chou and Chien, 2002; Escudero et al., 2007; Fiehler and Wolff, 2007, 2008; Mirkovic et al., 2011).
The Drosophila nmo gene encodes the founding member of the Nemo-like kinase (Nlk) subfamily of MAPKs (Brott et al., 1998). Nlk family members have regulatory roles in multiple developmental processes in vertebrates and invertebrates. Indeed, vertebrate NLK has been shown to participate in several signaling pathways, being activated by Transforming Growth Factor-β (TGF-β), Wnt, and IL-6 signaling (Brott et al., 1998; Ishitani et al., 1999; Kanei-Ishii et al., 2004; Kojima et al., 2005; Meneghini et al., 1999; Ohkawara et al., 2004), and to function downstream of nerve growth factor (NGF) (Ishitani et al., 2009). In addition, NLK phosphorylates and regulates the activity of several transcription factors in the nucleus such as T-cell factor (TCF)/Lymphoid enhancer factor (LEF), Signal transducer and activator of transcription 3 (STAT3), c-Myb, Smad4, the intracellular domain of Notch1 (Notch1-ICD) or Nuclear Factor-κβ (NF-κβ) through phosphorylation of its co-factor CREB binding protein (CBP) (Ishitani et al., 2010; Ishitani et al., 2003; Ishitani et al., 1999; Kanei-Ishii et al., 2004; Kojima et al., 2005; Meneghini et al., 1999; Ohkawara et al., 2004; Shi et al., 2010; Yasuda et al., 2004). In Drosophila, nmo is involved in diverse processes such as eye specification, synaptic growth, apoptosis, wing development, pair-rule patterning and circadian rhythms (Braid et al., 2010; Braid and Verheyen, 2008; Chiu et al., 2011; Merino et al., 2009; Mirkovic et al., 2002; Morillo et al., 2012; Verheyen et al., 2001; Yu et al., 2011). Moreover, it seems that nmo mediates crosstalk between multiple signaling pathways since it antagonizes Drosophila Wg signaling (Zeng and Verheyen, 2004) and attenuates BMP signaling by phosphorylating Mad during wing development (Zeng et al., 2007). Nmo was originally identified as an ommatidial rotation-specific factor (Choi and Benzer, 1994), which was subsequently shown to be essential for regulating the rate of ommatidial rotation throughout the entire process (Fiehler and Wolff, 2008; Mirkovic et al., 2011). Genetic interaction assays suggested that nmo could be functionally related to the JNK cascade during ommatidial rotation (Fiehler and Wolff, 2008; Mihaly et al., 2001). Furthermore, it has been recently demonstrated that nmo genetically interacts with several core PCP components (prickle, strabismus), members of signaling pathways (Notch, spitz, Egfr) and genes encoding cell adhesion proteins such as E-cad (shotgun) and β-catenin (armadillo) (Mirkovic et al., 2011). Indeed, it has been suggested that Nmo serves as a molecular link between core PCP factors and the E-cad–β-catenin (β-cat) complexes promoting cell motility during ommatidial rotation (Mirkovic et al., 2011).
In order to analyze more precisely the requirement of Nmo in the ommatidial rotation process, we used several strategies. In vivo analyses of wild-type and nmo mutant eye imaginal discs demonstrated that this gene regulates the speed of ommatidial rotation, as suggested from studies in fixed discs (Fiehler and Wolff, 2008). We also found that cone cell dynamics during this process is disturbed in nmo mutants and demonstrated that Nmo is required in these cells for correct ommatidial rotation. Our in vivo analyses also showed that interommatidial cells are less dynamic in nmo mutants than in wild-type discs. In addition, we performed a microarray study to identify genes that were deregulated in nmo mutant eye imaginal discs and that could be involved in ommatidial rotation. Four of the genes identified were validated and confirmed to be functionally linked to nmo by genetic interaction assays with several mutant alleles. In addition, phenotypic analyses revealed that the ommatidial rotation process is compromised when expression levels of some of those genes are modified. One of them is miple, which encodes a secreted heparin-binding protein that belongs to the midkine (MK)/pleiotrophin (PTN) family (Englund et al., 2006). In vertebrates, both MK and PTN are secreted cytokines that are implicated in many different processes, including cell migration (Muramatsu, 2010; Papadimitriou et al., 2009). Our results showed that miple overexpression causes rotation defects and that it interacts genetically with nmo and nmo-related genes, suggesting that Nmo is required to repress miple for correct ommatidial rotation.
Materials and methods
Fly stocks and genetics
Fly lines used in this study include: nmoP1 (Choi and Benzer, 1994), sev > nmo (Mirkovic et al., 2011), aosΔ7 (Freeman et al., 1992), UAS-miple (Toledano-Katchalski et al., 2007), the mthl8 allele P{Mae-UAS.6.11}mthl8F29.6 (Mukherjee et al., 2006), UAS-EgfrDN (Freeman, 1996), EgfrCO (Clifford and Schupbach, 1989). ptp99A1, shg2, arm4, cut-GAL4, iRmiple, iRLRP1, iRAlk, UAS-Dcr-2, P{EPgy2}CG32373EY21017 (named in this paper as EPCG32373) and the unc-13-4A overexpression line, EPEY04085 were obtained from the Bloomington stock center. iRmthl8, iRCG32373 and iRunc-13-4A were obtained from the Viena Drosophila RNAi Center. For UAS-mthl8 transgenic lines full length mthl8 cDNA LP02895 was subcloned into pUAST vector and flies were generated at Best-Gene Inc. (Chino Hills, USA.) by standard methods. Expression of several lines was checked by in situ hybridization with an mthl8 probe in en-GAL4/UAS-mthl8 embryos. GMR > miple, armGFP, nmoP1 and cut-GAL4, nmoP1 lines were generated by standard recombination methods. nmoDB, FRT80 (Mirkovic et al., 2011) and ey-FLP; ubiGFP, FRT80 flies were used to induce mitotic recombination for nmoDB clones analysis. armGFP was a gift of Silvia Muñoz-Descalzo (University of Cambridge, Cambridge, UK).
Live-imaging of pupal eye imaginal discs
Time-lapse imaging of pupal eye imaginal discs was performed as described (Escudero et al., 2007). Images were taken at 15 min intervals during at least 10 h in a Leica TCS SP confocal microscope. The images obtained were assembled and analyzed with ImageJ software. Measurements of IOCs apical areas were done manually with ImageJ. To quantify the number of IOCs disappearing in vivo during ommatidial rotation we followed each cell contained within the area comprised among 4 developing ommatidia from the beginning to the end of the process. IOCs that constricted their apical surface and subsequently disappeared were considered as dying cells. A total of 18 areas in armGFP control and 13 in armGFP, nmoP1 mutant discs were scored for this analysis.
Histology and immunohistochemistry
Analysis of adult retinae was performed as previously described (Tomlinson and Ready, 1987). Sections were mounted in DPX and observed through the optical microscope in dark field. At least four eyes per genotype were analyzed. For ommatidial orientation analysis, the ImageJ angle measurement tool was used. Scanning electron microscopy analysis of adult eyes was performed following the critical point dry method (Wolff, 2011) using a Philips XL-30 microscope. For immunohistochemistry, eye imaginal discs were dissected and incubated for 20 min in 4% paraformaldehyde. Pictures were taken using a Leica TCS-NT confocal laser-scanning microscope. In the case of pupal retinae, 42 h pupae were dissected and retinae were stained as previously described (Bao and Cagan, 2005). Retinae were mounted in Vectashield mounting medium (Vector) and pictures were taken in a Zeiss LSM510 microscope. The following primary antibodies were used: anti-dpERK (1:2000, Sigma, cat.# 8159) and anti-DECad (1:10, DSHB DCAD2).
Microarray analysis
Total RNA was extracted from approximately 500 eye-antenna imaginal discs of synchronized L3 larvae and purified with the mirVANA™ miRNA isolaton kit (Ambion#AM1260) following manufacturer's instructions. RNA quality analysis and quantification was performed in a Biorad Experion bioanalyzer. Three armGFP control and three armGFP, nmoP1 RNA samples were prepared and used to hybridize to Drosophila genome 2.0 Affymetrix microarrays following manufacturer's instructions (www. affymetrix.com) at the Multigenic Analysis Unit of the University of Valencia (Spain). Raw data reported in this paper have been submitted to Gene Expression Omnibus (Geo), accession GSE36127. Pre-processing of data was performed using the RMA (Robust Multi-Array) function of the affy package and differential expression analysis using the LIMMA (linear models for micro-array data) package, both from Bioconductor (www.bioconductor.com). For each gene the fold change was determined as the log2 ratio of the two compared mean intensities, so that a fold change of 2 means a 22-times increase in the expression of the corresponding gene in nmoP1 mutants. Adjustments for multiple testing were performed by using the Benjamini and Hochberg method (Benjamini, 1995). Only genes with adjusted p-values >0.05 were considered as positives.
RT-qPCR
One of the RNA samples used for the microarray analyses mentioned above and an independent one from armGFP and armGFP, nmoP1 eye imaginal discs were retrotranscribed to cDNA and used as template for RT-qPCR analyses. Total RNAs and cDNAs were also obtained from 3 independent samples of either sev-GAL4 or sev > Nmo eye imaginal discs following the same procedure. Taqman technology was used for validation of all candidate genes by RT-qPCR but nmo, for which we used the sybr-green technology. Primers and probes (Table S1) were designed from a gene region as close as possible to that corresponding to the microarray probes. Reactions using Taqman probes were performed in a LigtCycler 480 II Real-Time PCR System (Roche Applied Science) following manufacturer's instructions. α-tubulin84B was used as a reference gene for all the analyses. For nmo validation, a StepOne cycler (Applied Biosystems) was used.
Results
Live-imaging analyses of the ommatidial rotation process in wild-type and nmo mutant eye discs
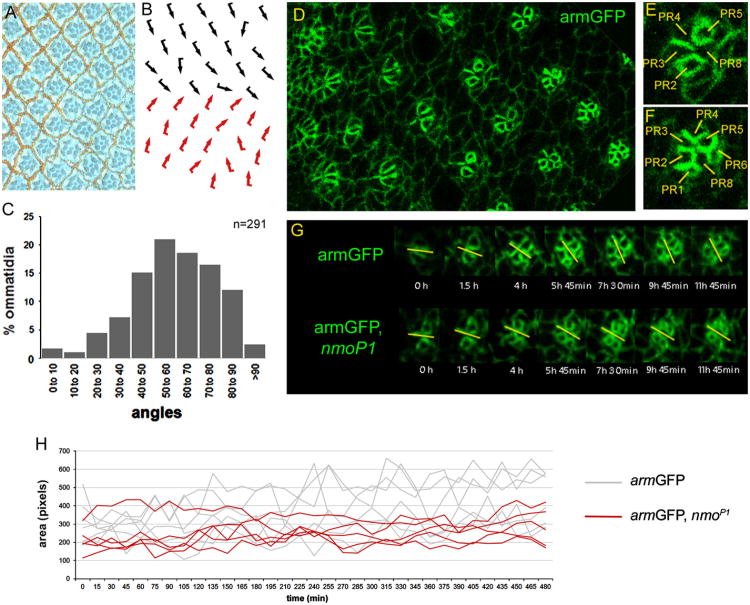
Phenotypic analyses of flies homozygous for the nmoP1 hypomorphic allele showed that all ommatidia arrested at around 45° of rotation, which led to propose that rotation might occur in two 45° steps and that nmo would be required in the second step (Choi and Benzer, 1994). However, studies performed in stained imaginal discs homozygous for the same allele suggested that the ommatidial rotation rate in nmo mutants was lower than in wild-type discs throughout the entire rotation process (Fiehler and Wolff, 2008). Supporting this hypothesis, it was shown that most ommatidia in nmoDB null mutant eyes failed to rotate at all and remained parallel to the equator (Mirkovic et al., 2011). Since Nmo overexpression caused ommatidial over-rotation, these data suggested that Nmo levels and/or activity directly correlated with the rate of rotation (Mirkovic et al., 2011). However, a deeper analysis of the cellular changes that occur during ommatidial rotation is necessary to understand how nmo is exerting its function in this process. In the last years, live-imaging techniques have been extensively used to determine how cells respond to patterning signals during development. In order to analyze in vivo potential differences in the cellular behavior during the ommatidial rotation process, we performed live-imaging analyses in wild-type and nmo mutant pupal eye imaginal discs (Escudero et al., 2007). An armGFP, nmoP1 recombinant line was generated to visualize in vivo ommatidial rotation in nmo mutants, and an armGFP line was used as a wild-type control. The experiments were perfomed with the nmoP1 hypomorphic allele since in our hands the null nmoDB allele was lethal at early pupal stages. armGFP labels apical contours of epithelial cells and allows to visualize ommatidial preclusters and to determine their orientation and developmental stage (Fig. 1D–F). First, we confirmed that the armGFP, nmoP1 line reproduced the external adult phenotypes of nmoP1 mutants both in wings and eyes (data not shown). Wings were smaller than wild-type with a rounded morphology and showed a held-out phenotype (Choi and Benzer, 1994; Verheyen et al., 2001). Eyes were externally rough and narrower than wild-type (Choi and Benzer, 1994). Tangential sections of those eyes revealed a high number of under-rotated ommatidia (Fig. 1A, B), with a mean orientation angle of 59.3° ± 3.9° with respect to the equator (Fig. 1C), thus reproducing the nmoP1 phenotype (Fiehler and Wolff, 2008).
Fig. 1.
Live-imaging analyses in eye imaginal discs reveal that nmo regulates the rate of ommatidial rotation and IOCs dynamics. (A–B) Tangential section of armGFP, nmoP1 adult eye (A) and the corresponding schematic representation with ommatidia arranged around the equator (B), with dorsal and ventral chiral forms indicated by black and red arrows, respectively. (C) Bar chart illustrating the percentage of ommatidia (y-axis) that are oriented at the angles indicated (x-axis) in armGFP, nmoP1 eyes, in which the most represented angles range from 50° to 70°. (D-F) armGFP protein localization in eye imaginal discs. A transgene with the adherens junction protein linked to GFP labels apical cell contours and outlines cell boundaries in an area of the eye imaginal disc posterior to the morphogenetic furrow (D). Magnified views of an ommatidial precluster that has initiated rotation (E), in which the five photoreceptor (PR) cells are labeled with their numbers, and an older one (F), in which almost all the PRs have been recruited. (G) Time-lapse series showing individual ommatidia during rotation after ∼ 12 h from armGFP (upper panel) and armGFP, nmoP1 (lower panel) eye imaginal discs. The yellow bars mark the orientation angle of ommatidia with respect to the equator and the time on each photogram is referred to the first image of the series. The rotation rate of armGFP, nmoP1 ommatidia is slower than that of armGFP controls. (H) Quantification of several IOCs areas (number of pixels/cell) over time in armGFP (gray lines) and armGFP, nmoP1 (red lines) eye imaginal discs. Note that fluctuations of IOCs areas are sharper in wild-type controls than in nmo mutant discs, which is consistent with the observation that apical shape changes in IOCs are reduced in such mutants.
White pupae of the corresponding genotypes were prepared and the cellular movements in eye imaginal discs were recorded (see Material and methods). Several observations could be made when analyzing the movies obtained from armGFP control discs. Our results showed for the first time in our knowledge that ommatidial rotation was not a continuous process, instead ommatidia moved forth and back until they reached their final orientation (rotation angle) (Movie 1). In addition, we demonstrated that cells in the ommatidial clusters rotate independently from the undifferentiated IOCs during this process, breaking and establishing new contacts with them (Movie 1). However, far from remaining static during the process, IOCs underwent clear shape changes independent of cell division, with continuous expansion and contraction of their apical surfaces (Fig. 1H and Movie 2). We also observed that some IOCs lying between developing ommatidial clusters, and usually not in contact with them, disappeared during the process suggesting they were suffering programmed cell death (Movie 3). We quantified the number of IOCs disappearing in an area delimitated by four developing ommatidia finding that a mean of 1.5° ± 1.2 IOCs disappeared per area (see Materials and Methods). Both apoptosis and apical cell shape changes have been demonstrated to play important roles in the dynamics of developmental processes like embryonic dorsal closure by controlling forces that drive cell movements (Blanchard et al., 2010; David et al., 2010; Solon et al., 2009; Toyama et al., 2008). Similar analyses performed in armGFP, nmoP1 discs showed that although photoreceptor recruitment in such discs occurred as in armGFP controls, ommatidia rotated at a slower pace and they stopped rotating prematurely (Fig. 1G and Movie 4). This is the first in vivo demonstration of the role of nmo in regulating the ommatidial rotation rate during the entire process, as suggested from the studies in fixed eye imaginal discs (Fiehler and Wolff, 2008). We also observed that apical shape changes in the IOCs, and in turn changes in their areas, were less evident in nmoP1 discs than in controls during the process (Fig. 1H, compare Movies 2 and 5). Besides, we did not find IOCs disappearing in nmo mutant discs when performing similar analyses to those indicated above for control discs (0 ± 0 IOCs disappearing per area, p-value < 0.0001, compare Movies 3 and 5), which is consistent with suggestions that nmo plays a role in apoptosis in the embryonic epidermis and during pupal retinae development (Mirkovic et al., 2002). Taken together, our results suggest that apical shape changes and apoptosis of IOCs, together with the remodeling of their junctions with rotating cells, could contribute to the discontinuity of the rotation process. The reduction of apical shape changes in the IOCs observed in nmoP1 mutant disc when compared to controls could be a secondary effect of the reduced ommatidial movement in such discs. Alternatively, the reduction of IOCs dynamics and death could be contributing to disturb ommatidial rotation in nmo mutants.
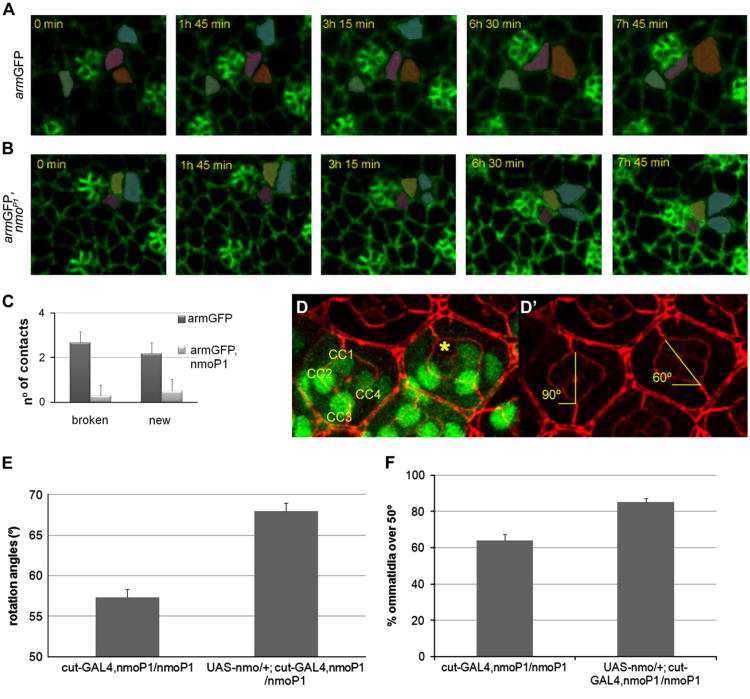
The in vivo analysis of nmo mutant discs also revealed an abnormal behavior of the cone cell precursors, which rotate together with the photoreceptor precursors and independently of their undifferentiated neighbors, the IOCs (Fiehler and Wolff, 2007). We found that in control discs the equatorial and polar cone cell precursors break and establish new contacts with neighboring IOCs until they reach their correct position in the ommatidium (Fig. 2A and Movie 2). Quantitative analyses revealed that these cells typically break 2.7 ± 1.5 contacts and establish 2.2 ± 1.6 (n = 12) new contacts in armGFP discs over a period of ∼7 h, encompassing from a stage in which R7 and anterior and posterior cone cell precursors have been recruited until equatorial and polar cone cell precursors reach their final position in the ommatidial cluster (Fig. 2C). However, these cells are significantly more static in armGFP, nmoP1 discs (Fig. 2B and Movie 5), in which they break an average of 0.3 ± 0.5 contacts and establish 0.5 ± 0.5 (n = 10) new contacts with neighboring IOCs (p-value < 0.005 in both cases) (Fig. 2C). These results suggest that the adhesive behavior of cone cell precursors is affected in nmo mutants, probably due to the reported activity of the Nmo kinase at the level of adherens junction complexes (Mirkovic et al., 2011). Next, we wondered whether this abnormal behavior of cone cells could have any consequences for the ommatidial rotation process. To address this question, we conducted a mosaic analysis by using the FRT/FLP system to generate clones of nmoDB mutant cells in pupal eye discs. We measured the degree of rotation of mosaic ommatidia with a full complement of wild-type photoreceptors but with nmo mutant cone cells. This analysis showed that while completely wild-type ommatidia rotated over 88.9° ± 2.9°, lack of nmo function in at least one cone cell disrupted the ommatidial rotation process, with ommatidia remaining at 76.7° ±6.1 ° (Fig. 2D) (22 mosaic ommatidia in a total of 25 clones analyzed, p-value<0.000001). These data indicate that nmo is also required in cone cells for correct rotation, and that the under-rotation phenotype in nmoDB eyes is in part due to its lack of function in such cells. To confirm these results, we analyzed the ommatidial rotation angles in eyes from UAS- nmo/+; cut-GAL4, nmoP1/nmoP1 flies, which expressed nmo specifically in cone cells with the cut-GAL4 driver in a nmoP1 background, and compared them to cut-GAL4, nmoP1/nmoP1 controls. The mean ommatidial rotation angle in cut-GAL4, nmoP1/nmoP1 control eyes was significantly lower than in UAS-nmo/+; cut-GAL4, nmoP1/nmoP1 eyes (57.3° ± 3.4° and 68° ± 3.2°, respectively; Fig. 2E). In addition we also observed that the percentage of ommatidia rotating over 50° was significantly higher in UAS-nmo/+; cut-GAL4, nmoP1/nmoP1 than in cut-GAL4, nmoP1/nmoP1 controls (85.1 ±2.2 % and 63.9 ± 3.6%, respectively; Fig. 2F). These results indicate that nmo expression in cone cells partially rescues the under-rotation phenotype of nmoP1 mutants. Taken together, our data demonstrate that nmo is required in cone cell precursors during ommatidial rotation.
Fig. 2.
Nmo regulates cone cell dynamics during ommatidial rotation. (A–B) Time-lapse series showing the dynamics of equatorial cone cell precursors (marked in pink) in armGFP (A) and armGFP, nmoP1 (B) eye discs over the course of ∼8 h. Time on each photogram is referred to the first image of the series. Other neighboring cells have been artificially colored to better follow the cell contacts. While in (A) the cone cell precursor breaks and establishes new contacts with neighboring cells, in (B) the cone cell precursor remains static on its initial position without breaking or forming new contacts. (C) Bar chart representing the number of cell contacts broken and established by the equatorial and polar cone cell precursors. (D-D') Confocal image of a 42 h pupal retina showing an ommatidium with wild-type cone cells (CC1 to CC4) and a mosaic ommatidium in which one of the cone cells (yellow asterisk) is mutant for nmoDB (marked by the absence of GFP staining). In both ommatidia, the whole PR complement is wild type (not shown). In (D) GFP staining (green) marks nmo+ cells, DE-Cad staining (red) shows orientation of ommatidia with respect to equator. In (D') only the DE-Cad staining is shown. The orientation angles of both ommatidia are marked in yellow. Loss of nmo function in one of the cone cells avoids complete ommatidial rotation (to 90°). (E-F) Overexpression of nmo specifically in the cone cells with the cut-GAL4 driver partially rescues the nmoP1 rotation phenotype. Quantification of orientation angles in ommatidia from UAS-nmo/+; cut-GAL4, nmoP1/nmoP1 and cut-GAL4, nmoP1/nmoP1 eyes (E) reveals a significant increase of the mean angle orientation when nmo is overexpressed in cone cells. The percentage of ommatidia with an orientation angle over 50° also significantly increases in UAS-nmo/+; cut-GAL4, nmoP1/nmoP1 eyes when compared to cut-GAL4, nmoP1/nmoP1 controls (F).
Identification of genes differentially expressed in nmo mutant eye discs
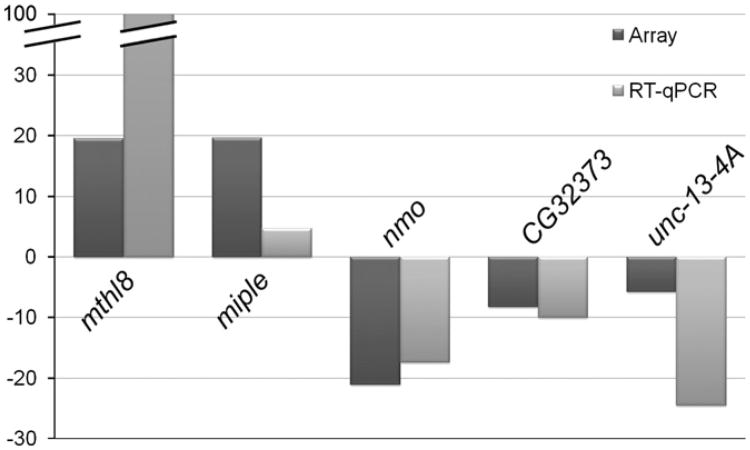
The relationship of nmo to other genes and/or pathways that could explain its exact role during ommatidial rotation is still unknown. Genetic interaction assays with bsk and TGF-β activated kinase (dTak) mutants indicated that nmo was functionally related to the JNK cascade in the eye (Fiehler and Wolff, 2008; Mihaly et al., 2001). In addition, genetic and biochemical studies have recently demonstrated a link between nmo and the Fz-PCP pathway, since Nmo physically interacts with the Stbm–Pk complex (Mirkovic et al., 2011). These experiments also showed that Nmo phosphorylates β-cat and E-cad, thus providing a potential mechanism by which Nmo could be influencing cell adhesion (Mirkovic et al., 2011). However, it has been also proposed that Nmo could regulate gene expression via its ability to phosphorylate several transcription factors and co-factors (Fiehler and Wolff, 2008). Thus, in order to identify new genes and/or pathways that could be related to nmo during ommatidial rotation we compared the expression profile of nmoP1 mutant eye imaginal discs to that of wild-type discs by using genome-wide microarray analyses. For doing so, total RNAs extracted from armGFP and armGFP, nmoP1 eye-antenna imaginal discs were used to generate cDNAs, hybridize Affymetrix Drosophila Genome 2.0 arrays (see Materials and methods) and analyze the expression profile of these genotypes. The analyses were performed with three independent RNA samples from each genotype. We thus identified 104 significantly up-regulated (50.7%) and 101 down-regulated genes (49.3%) (adjusted p-value<0.05) in nmoP1 mutants with respect to controls (see Tables S2 and S3). As expected, nmo expression was significantly reduced in nmoP1 mutant discs (it was down-regulated 21.6-fold) (Fig. 3 and Table S3). Although the function of most of the genes identified is unknown, some participate in distinct biological processes related to the ommatidial rotation process, such as cell adhesion, signaling, cytoskeleton biogenesis/organization, and carbohydrate metabolism, involved in extracellular matrix biosynthesis (Tables S2 and S3). Since nmo seems to have a role in cell adhesion during ommatidial rotation (Mirkovic et al., 2011), we chose to focus on two up-regulated and two down-regulated genes for further analyses: miple, methuselah-like 8 (mthl8), unc-13-4A and CG32373. Two main criteria were used to select these genes: (1) a high fold- change in their expression in nmoP1 mutant discs (miple and mtlh8 were up-regulated 19.6- and 19.3-fold, respectively; unc-13-4A and CG32373 were down-regulated 5.8- and 8.3-fold, respectively) (Tables S2 and S3) and (2) their possible role in cell adhesion. miple encodes the Drosophila ortholog of the vertebrate MK/PTN cytokines (Englund et al., 2006). These secreted heparin-binding proteins are implicated in several processes, including enhancement of cell growth and survival, cell migration, angiogenesis and neurite growth (Muramatsu, 2010; Papadimitriou et al., 2009). In Drosophila, miple has a role in mesoderm spreading in the embryo during gastrulation (Toledano-Katchalski et al., 2007), a process that involves collective cell migration. In addition its expression has been shown to be regulated by the Egfr ligand Spitz during eye development (Firth and Baker, 2007). mthl8 encodes a G protein-coupled receptor that has been shown to interact in a two-hybrid assay with Thrombospondin, a protein that mediates adhesion through interaction with integrins (Chanana et al., 2007). Moreover, mthl8 genetically interacts with members of the JAK/STAT pathway in the eye (Mukherjee et al., 2006). Little is known about the function of unc-13-4A and CG32373. Unc-13-4A has been shown to interact in a two-hybrid assay with Toutvelu (Stanyon et al., 2004), which participates in heparansulfate biosynthesis (Izumikawa et al., 2006; The et al., 1999) and was identified as one of the Drosophila orthologs of vertebrate proteins putatively implicated in neurotransmitter release (Lloyd et al., 2000). Finally, CG32373 encodes a protein containing an EGF-like calcium-binding conserved site and a Sushi/SCR/CCP domain, both involved in cell adhesion (de Vega et al., 2007; Nishimura et al., 2007). To confirm the microarray results for these genes, RT-qPCR analyses in nmo mutant and control eye discs were performed. In such experiments, we also analyzed nmo expression levels as a control. Our results showed that while nmo, CG32373 and unc-13-4A are significantly down-regulated in the mutants, miple and mthl8 are significantly up-regulated in the same individuals (Fig. 3), thus supporting the microarray results. The correspondence between the variations of nmo levels observed in the array and in the RT-qPCR analyses gave us a control of the reliability of the results. Therefore, we took the validated genes as candidates to be regulated by nmo during the ommatidial rotation process.
Fig. 3.
Correlation of the differential expression of candidate genes in arrays and RT-qPCR. Graphic representation of fold changes in the expression levels of nmo and four candidate genes identified in the microarray analyses of armGFP, nmoP1 eye imaginal discs. Comparisons between the fold change values obtained in RT-qPCR and microarray analyses are shown.
Candidate genes are functionally related to nmo and could have a role in the ommatidial rotation process
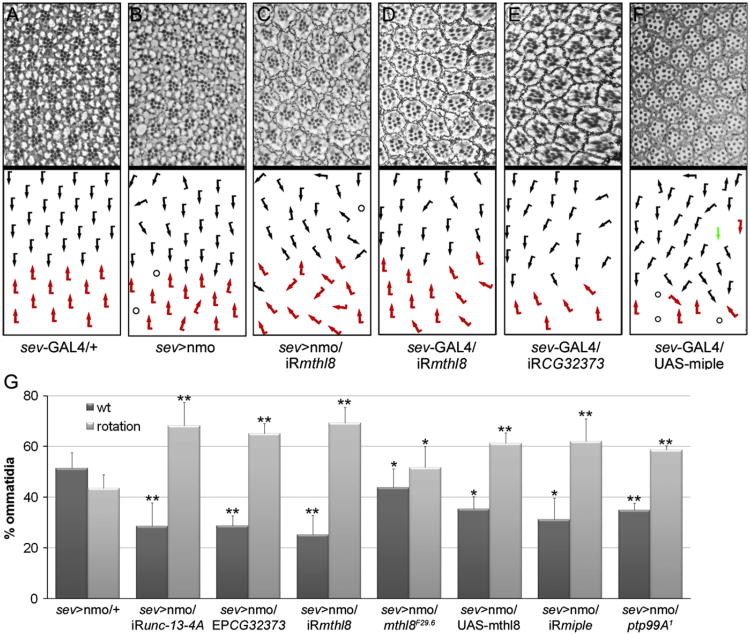
To determine the potential role of the candidate genes in ommatidial rotation or eye development we aimed to analyze the effect of their overexpression and RNAi with the sev-GAL4 and GMR-GAL4 drivers in an otherwise wild-type background. The results of these analyses are summarized in Table 1. First, we tested the unc-13-4A and CG32373 genes, both down-regulated in nmoP1 mutant discs (Fig. 3), finding that while reduction of CG32373 expression produced mild ommatidial rotation defects (Fig. 4E), as expected from the microarray results, no phenotype was observed when analyzing unc-13-4A RNAi (not shown). This result could indicate that unc-13-4A is not involved in ommatidial rotation. Alternatively, it could be that the unc-13-4A RNAi is not strong enough to reduce unc-13-4A expression to critical levels able to affect that process. According to expression results, neither unc-13-4A nor CG32373 overexpression with EP lines had any consequence for eye development (not shown). Next, we analyzed the mthl8 and miple genes, both up-regulated in nmoP1 mutant discs (Fig. 3). Consistently miple-overexpressing eyes displayed mainly ommatidial rotation defects and, less frequently, defects in photoreceptor specification (Fig. 4F). However, mthl8 overexpression (see Materials and methods) had no effect on eye development (not shown), maybe because mthl8 expression levels obtained with the transgenic lines are not high enough. When knocking-down miple and mthl8 in the eye, we found that only in the last case ommatidial rotation defects were observed (Fig. 4D). Although this result may seem contradictory for the expression results, it has been reported that both over-expression and loss of function (LOF) of genes involved in rotation, such as components of Egfr signaling, give rise to ommatidial rotation defects (Gaengel and Mlodzik, 2003).
Table 1.
Genetic interactions and phenotypic analyses of nmo targets.
| Gene | sev-GAL4 GMR-GAL4 | sev > nmo | ||
|---|---|---|---|---|
|
|
|
|||
| OE | iRNA | OE | iRNA | |
| unc-13-4 A | − | − | ∅ | e |
| CG32373 | − | + | e | ∅ |
| mthl8 | − | + | e | e |
| miple | + | - | ∅ | e |
OE, overexpression; iRNA, RNA interference; −/+, wild type/ommatidial rotation phenotype; e/∅, enhancement/no modification of the sev > nmo eye phenotype.
Fig. 4.
Candidate genes interact genetically with nmo and could have a role in ommatidial rotation. (A–F) Tangential sections of adult eyes of the indicated genotypes and the corresponding schematic representations of ommatidial orientation with respect to equator, with dorsal and ventral chiral forms indicated by black and red arrows, respectively. Circles represent ommatidia with incorrect number of PRs and the green arrow indicates a symmetric ommatidium. Experiments were performed at 29 °C in (A), (D) and (E) and at 25 °C in (B), (C) and (F). (G) Quantification of the percentage of wild type ommatidia and ommatidia with rotation defects in adult eyes of the indicated genotypes. Note that in all cases, there is a significant modification of the sev > nmo phenotype (*p-value<0.05, **p-value<0.01, Student's t-test).
Next, to confirm the functional relationship between nmo and the validated genes, we performed genetic interaction assays by examining the effect of their overexpression and RNAi knockdown on the sev > Nmo eye phenotype (Fig. 4B), which is mainly due to ommatidial rotation defects and has been shown to be dosage sensitive (Fiehler and Wolff, 2008; Mirkovic et al., 2011). The results of these genetic interactions were obtained by quantifying the percentage of ommatidia with rotation defects in the corresponding genotypes (Fig. 4G) and are indicated in Table 1. We found that the four candidate genes interact genetically with nmo. In the case of CG32373 and miple, the results of the assays were in agreement with the expression changes observed in nmoP1 mutants. The functional relationship of miple and nmo will be further confirmed and discussed below. However, the results obtained for the genetic interactions with unc-13-4A and mthl8 were somehow contradictory (Fig. 4G and Table 1). To clarify this issue, we determined the expression levels of both genes in sev > Nmo eye discs by RT-qPCR, using sev > GAL4/+ discs as controls (Fig. S1). Strikingly, we found that mthl8 is highly upregulated in sev > Nmo discs, as it happened in nmo mutants. These results indicated that mthl8 expression is dramatically affected by either reduction or increase of Nmo function, which complicates the interpretation of the genetic interaction results. Indeed, we found that both LOF and mthl8 overexpression enhanced the sev > Nmo phenotype (Fig. 4G). Since mthl8 expression is almost undetectable in wild-type eye imaginal discs by in situ hybridization (Firth and Baker, 2007; Mukherjee et al., 2006), the results obtained in the RT-qPCR analyses confirmed that mthl8 and nmo are functionally related, although the exact nature of this relation is difficult to establish. Regarding unc-13-4A, we found that the expression of this gene was not altered in nmo-overexpressing discs when compared to controls (Fig. S1), which does not give a clear explanation to the genetic interactions found between unc-13-4A and nmo.
Taken together, the results obtained in the genetic interaction assays with sev > Nmo as well as in the phenotypic analyses of flies overexpressing or with reduced expression of miple, mthl8, unc-13-4A and CG32373 suggest a role of some these genes in ommatidial rotation. However, further analyses will be required to decipher their exact role(s) and to determine their functional relationship to nmo in this context.
Miple is functionally linked to ptp99A and with members of the E-cad–β-cat complex
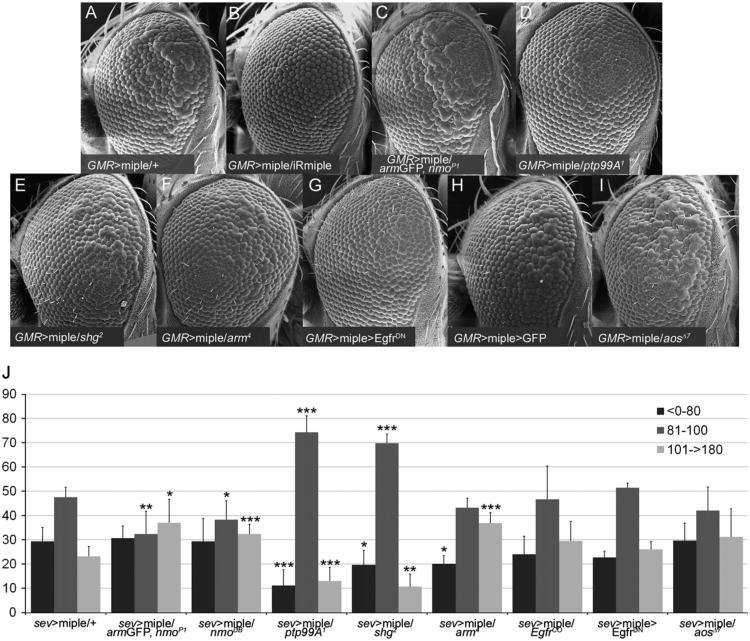
As mentioned above, Miple is the Drosophila ortholog of the vertebrate MK/PTN cytokines, which participate in several processes involving cell migration (Muramatsu, 2010; Papadimitriou et al., 2009). These cytokines use different receptors to exert their function including PTPζ, Alk, LRP1, N-Syndecan and integrins ανβ3 (Muramatsu, 2010; Papadimitriou et al., 2009). One of them, PTPζ, is a chondroitin sulfate proteoglycan that binds to both MK and PTN with high affinity and is recognized through the heparin-binding sites of these proteins. Its intracellular domain exhibits protein tyrosine (Tyr) phosphatase activity and has been shown to interact with β-cat in vertebrates to promote dephosphorylation of its Tyr residues (Meng et al., 2000). Indeed, it seems that PTN binding to this receptor inhibits the phosphatase activity of PTPζ, eventually inducing Tyr phosphorylation of β-cat and causing a disruption of the E-cad–β-cat complex (CCC) stability and cell adhesion (Meng et al., 2000; Perez-Pinera et al., 2006). Interestingly, Nmo can phosphorylate both Arm, the Drosophila ortholog of β-cat, and E-cad (Mirkovic et al., 2011). β-cat phosphorylation by Nmo occurs preferentially in three Ser/Thr residues in the C-terminal region of the protein. Although this phosphorylation did not affect CCC formation in vitro, it was shown to be biologically relevant for ommatidial rotation (Mirkovic et al., 2011). Considering this, and to determine whether the molecular mechanisms underlying Miple function during ommatidial rotation in Drosophila could be similar to those in vertebrates, we performed genetic interaction assays to identify candidate genes that could be acting downstream of miple. Although the sev-GAL4/UAS-miple flies displayed a clear rotation phenotype, it was externally too weak to be clearly modified. Therefore, we generated a GMR > miple recombinant line that showed a clear rough eye phenotype, which in tangential sections displayed rotation defects but mainly defects in photoreceptor recruitment (Fig. 5 A and not shown). The GMR > miple phenotype was dosage sensitive, as it was markedly suppressed by reducing miple expression with a UAS-IRmiple line (Fig. 5A,B). Our results showed that dosage reduction of nmo with the nmoP1 and nmoDB alleles was able to enhance the eye roughness of GMR > miple flies (Fig. 5C and data not shown), thus supporting the functional relationship between both genes. Next, mutant alleles of several candidate genes, including those encoding putative Miple receptors and downstream effectors as well as genes functionally related to nmo in the ommatidial rotation process were tested for interactions with GMR > miple (Fig. 5). Ptp99A was described as the putative Drosophila ortholog of the PTPζ receptor. Ptp99A is involved in motor axon guidance in the Drosophila embryo (Desai et al., 1996), although no phenotypic analyses have been performed in mutant larvae. Therefore, we wondered whether ptp99A could be functionally linked to miple during eye development. Our results showed that reduction of ptp99A dosage (with the ptp99A1 allele) markedly suppressed the eye roughness of GMR > miple flies (Fig. 5D). Similar analyses with mutant alleles for genes encoding other putative Miple receptors such as Alk (Bazigou et al., 2007) and CG33087, which encodes the Drosophila ortholog of LRP1, yielded negative results (data not shown). To get further insight into the potential downstream effectors of miple function, we tested whether the GMR > miple eye phenotype was sensitive to endogenous levels of arm and shotgun (shg). Our results showed that the arm4 and shg2 null alleles were dominant suppressors of that phenotype (Fig. 5E,F), suggesting that Miple function is linked to components of the CCC. Signaling pathways downstream of vertebrate MK/PKN activation include MAPKs as important components (reviewed in Kadomatsu and Muramatsu, 2004). Consistent with this, miple overexpression activates MAPK during Drosophila embryonic mesoderm development (Toledano-Katchalski et al., 2007). Activation of Egfr signaling also leads to MAPK activation and this pathway is involved in ommatidial rotation (Brown and Freeman, 2003; Gaengel and Mlodzik, 2003; Strutt and Strutt, 2003). Indeed, it has been suggested that nmo could be regulating the rate of rotation through the Egfr pathway (Brown and Freeman, 2003; Choi and Benzer, 1994; Gaengel and Mlodzik, 2003; Mirkovic et al., 2011). To check for a potential relationship between miple and the Egfr pathway during rotation, we first analyzed MAPK activation by dp-ERK staining in GMR > miple eye imaginal discs but these experiments did not provide consistent results due to high signal variability both in discs overexpressing Miple and in controls (data not shown). We therefore tested for genetic interactions between GMR > miple and components of the Egfr pathway. We found that the GMR > miple eye phenotype was suppressed by down-regulation of Egfr signaling, both by expressing a dominant negative form of the receptor (EgfrDN) (Fig. 5G) or with the EgfrCO mutant allele (data not shown), and enhanced when up-regulating Egfr signaling by dosage reduction of the aos gene (using the aosΔ7 allele, Fig. 5I).
Fig. 5.
miple interacts genetically with nmo, ptp99A and members of the CCC in the OR context. (A–I) Scanning electron microscope images of female adult eyes showing the external phenotype of miple overexpression with the GMR-GAL4 driver (GMR > miple). This phenotype (A) is dominantly suppressed by an UAS-IR miple line (B), the ptp99A1 (D), shg2 (E) and arm4 (F), and by down-regulation of Egfr signaling with a UAS-EgfrDN transgene (G). In contrast, the GMR > miple phenotype is enhanced by nmoP1 (C) and aosΔ7 (I). Note that this phenotype is not modified by GFP overexpression (H). All experiments were performed at 25 °C. (J) Graphic representation of genetic interactions with the sev > miple ommatidial rotation phenotype. A quantification of the ommatidial orientation angles for each genotype is represented. The different angles have been grouped in three categories: > 0°–80° (under-rotated ommatidia), 81°–100° (wild type ommatidia) and 101°- > 180° (over-rotated ommatidia). Asterisks indicate statistically significant modification of the sev > miple phenotype for a given ommatidial orientation category (*p-value<0.05, **p-value<0.01, ***p-value<0.005, Student's t-test).
To determine whether all the observed genetic interactions were relevant for the ommatidial rotation process, we subsequently repeated the experiments with the sev > miple line but only testing the interacting alleles. In this case, we analyzed tangential sections of eyes with the corresponding genotypes by measuring ommatidial rotation angles (Fig. 5J). These analyses confirmed the genetic interactions found between miple and nmo, ptp99A and the CCC components, since dosage reduction of these genes was able to modify the ommatidial rotation phenotype of sev > miple eyes (Fig. 5J). However, we found that components of the Egfr pathway did not significantly modify the ommatidial rotation phenotype of sev > miple eyes (Fig. 5 J), thus suggesting that the interactions observed when using the GMR > miple line could be affecting photoreceptor recruitment, a process that has also been shown to be regulated by Egfr and is altered in the GMR > miple line. It would be interesting in the future to investigate this possibility.
Taken together, the results obtained in the genetic interaction assays confirmed the functional relationship between miple and nmo. Moreover, the interactions found between miple and arm and shg are in agreement with the previous results in which null alleles of both genes were strong enhancers of the sev > Nmo phenotype (Mirkovic et al., 2011). As expected from the micro-array results the two genes interact genetically with nmo and miple in opposite directions, thus confirming that Nmo is required to inhibit miple expression. Our data also suggest that Ptp99A could be acting as a Miple receptor during eye development and points to a conservation of the MK/PTN signaling mechanisms between Drosophila and vertebrates. Interestingly, we also observed that the ptp99A1 allele significantly enhanced the ommatidial rotation phenotype of sev > Nmo (Fig. 4G), confirming that these genes are functionally linked during the process.
Discussion
Multicellular movements are essential in multiple morphogenetic processes. Among them, ommatidial rotation (OR) in the Drosophila eye is an example of a highly coordinated cell motility process, which is necessary to achieve the regular arrangement of retinal cells. The Nmo kinase is an important player during the entire rotation process, probably regulating the activity of the E-cad–β-cat complex as well as integrating signals from several pathways such as Fz-PCP, N and Egfr (Mirkovic et al., 2011). Here we demonstrate that Nmo is required in and regulates cone cell dynamics during OR, and that it could be also modulating IOCs death during the process. In addition, we have identified new OR genes whose expression is dependent on nmo activity, thus discovering new molecular mechanisms and regulatory pathways operating downstream of nmo during the process.
Live imaging reveals cone cell requirements of nmo
Our live-imaging analyses of pupal eye imaginal discs demonstrate that almost the complete OR process can be tracked, thus making it possible to analyze the behavior of individual cell types involved in OR. These analyses revealed for the first time that OR is not a continuous process. It was previously reported that ommatidial clusters, which contain photoreceptor and cone cell precursors, move independently of the undifferentiated stationary IOCs (Fiehler and Wolff, 2008). Thus intercellular contacts between both subsets of cells likely need to be constantly remodeled to enable OR without disrupting the integrity of the epithelium, as suggested (Fiehler and Wolff, 2008). Our results indicate that ommatidial clusters move forth and back during the process, probably as a consequence of the constant remodeling of cell contacts between the preclusters and the stationary IOCs. Although it could be a secondary effect of the ommatidial clusters rotation, one possibility could be that the contractile movements of the IOCs might generate forces able to pull and push the rotating clusters. This is an interesting hypothesis that would be worth to check in the future. In addition, programmed cell death might provide part of the forces affecting OR, similar to what happens during embryonic dorsal closure (Toyama et al., 2008). Our in vivo analyses of pupal eye discs homozygous for the nmoP1 allele confirm that nmo is required throughout the entire OR process (Mirkovic et al., 2011). Consistent with this, we found that the rate of OR in nmoP1 mutants is lower than in controls at any point of the process. Our results suggest that nmo could be affecting OR through regulation of several distinct cellular aspects. First, we find that nmo regulates cone cell dynamics, which are very static when nmo function is reduced. In particular, the number of contacts they establish/break with surrounding cells in nmo mutant discs is significantly lower than in controls. Our mosaic analyses in pupal discs confirm that nmo is required in cone cells for correct OR. Moreover, the rotation defects in nmo mutants are partially rescued by expressing Nmo specifically in cone cells. Although a recent study already demonstrated a role of the cone cells in ommatidial rotation (Fetting et al., 2009), this is the first evidence of Nmo requirement in these cells during the process. Second, the absence or reduction of IOCs programmed cell death during OR in nmoP1 mutants suggests that Nmo is required to eliminate surplus cells and supports a dynamic role for apoptosis during this process. Finally, we also find a reduction of IOCs apical shape changes in nmo mutants with respect to controls. One possibility could be that this is a secondary effect of the reduced rate of OR in the mutants. However, an alternative hypothesis could be that nmo might regulate the contractility of these cells, and in turn the forces they are contributing during retinal development. Supporting this hypothesis, it has been reported that nmo regulates the activity of the CCC by directly phosphorylating β-cat (Mirkovic et al., 2011), and that zip1 suppresses the OR phenotype of sev > Nmo eyes (Fiehler and Wolff, 2008). These results suggest that Nmo could be acting upstream of the actin–myosin contractility by modulating polarized remodeling of adherens junctions (Mirkovic et al., 2011), and could support a role of Nmo in regulating the adhesive properties of CCs, as suggested by its involvement in the dynamics of these cells.
A differential expression screen for Nmo targets
Our differential expression analyses in eye imaginal discs revealed that Nmo regulates the expression of several genes that encode cell adhesion and signaling molecules, among others. Preliminary data of four candidate genes (miple, mthl8, unc13-4-A and CG32373) indicate that some of them interact genetically with nmo and that their deregulation causes OR defects, supporting the validity of the microarray results. Interestingly, one gene identified in these analyses, four wheel drive (fwd), which was upregulated in nmoP1 mutant discs (see Table S2), has been recently isolated as a dominant modifier of a gain-of-function eye phenotype of the Fz-PCP core components Diego (Dgo) and Prickle (Pk) (Weber et al., 2012), further supporting a functional relationship between nmo and the Fz-PCP pathway (Mirkovic et al., 2011). However, in this study we have mainly focused on the analysis of miple function during OR. We demonstrate that miple overexpression leads to OR defects, consistent with the finding that it is highly up-regulated in nmoP1 mutant eye discs. Interestingly, one of the vertebrate orthologs of miple, PTN, is involved in the modification of cell adhesiveness (Perez-Pinera et al., 2006). Both PTN and MK, the second vertebrate ortholog of miple, contain a thrombospondin type I repeat homologous domain, and belong to the thrombospondin superfamily of adhesion molecules (Kilpelainen et al., 2000). In addition, the Mthl8 receptor has been shown to interact with Thrombospondin in a two hybrid assay (Giot et al., 2003), and two additional members of this family, m-spondin (mspo) and fat-spondin, were also identified in the microarray analyses as being significantly up- and down-regulated, respectively, in nmoP1 mutants (see Tables S2 and S3). Taken together, these results suggest that members of the thrombospondin superfamily could be important during OR and support the role of Nmo in regulating cell adhesion.
A possible role of miple in CCC regulation
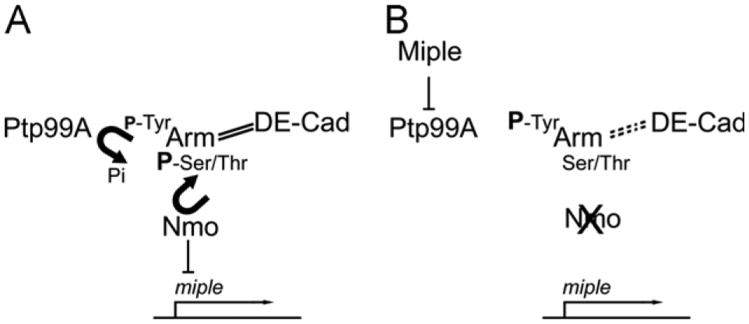
To get further insight into the potential role of miple during OR, we tested whether its function was sensitive to endogenous levels of several candidate genes. We found that miple interacts genetically with ptp99A, which encodes the Drosophila ortholog of the PTPζ receptor, thus suggesting that miple signaling in Drosophila could be similar molecular mechanisms as its vertebrate counterparts. The genetic interaction between miple and the CCC members arm and shg also indicates that it could be participating in the remodeling of adherens junctions in the eye, as has been suggested in vertebrates (Perez-Pinera et al., 2006). The mechanism by which miple could affect OR remains however unclear. As described above, Nmo phosphorylates β-cat in Ser/Thr residues upon binding to PCP core components (Mirkovic et al., 2011), a process that could hinder phosphorylation of Tyr residues thus stabilizing the CCC (Fig. 6A). We propose that in absence of nmo function these Tyr residues would be exposed to phosphorylation leading to CCC destabilization. Interestingly, it has been shown that the PTPζ receptor in vertebrates is able to promote dephosphorylation of β-cat Tyr residues, and this activity is inhibited after PTN binding (Meng et al., 2000; Perez-Pinera et al., 2006). A similar situation could exist in the Drosophila eye. Here, in wild-type, where miple expression is repressed, β-cat would be phosphorylated by Nmo on Ser/Thr residues and this, in cooperation with Ptp99A activity, would lead to low levels of Arm phosphorylation in Tyr residues and to CCC stabilization (Fig. 6A). Upon miple up-regulation in nmo mutants, the phosphatase activity of Ptp99A could be inhibited, thus contributing to an increase of Tyr phosphorylation of Arm, to CCC destabilization (Fig. 6B) and in turn to rotation defects. Since it is unclear which Nmo expressing cells are also targets of Miple function and considering that ommatidial clusters rotate independently from the surrounding IOCs, this model could be probably applied to the interface between rotating and non-rotating cells. Alternatively, it has been proposed that Nmo could regulate the rate of rotation independently of the PCP complexes through Egfr and/or N signaling (Mirkovic et al., 2011). Our results show however that miple does not interact with components of the Egfr pathway at the OR level thus discarding this pathway as a link between miple and nmo during this process.
Fig. 6.
Model for the effect of loss of nmo function on CCC destabilization mediated by Miple during ommatidial rotation. (A) In a wild-type situation, Nmo inhibits miple expression and phosphorylates Arm in Ser/Thr residues thus stabilizing the CCC and hindering phosphorylation of Tyr residues by other kinases. The levels of Tyr phosphorylation could be also lowered by the phosphatase activity of Ptp99A. (B) In nmo mutants, miple expression is activated and the Miple protein binds to the Ptp99A receptor, inhibiting its phosphatase activity against the Tyr residues of Arm and leading to CCC destabilization. Solid and dashed lines between Arm and DE-cad represent stabilization and destabilization of the complex, respectively. P represents phosphorylation of the corresponding amino acid, and its size correlates with phosphorylation levels.
Finally, an interesting question is how miple expression could be regulated by nmo in the Drosophila eye. Regarding this, it has been shown that MK expression in vertebrates is regulated by NF-κβ (You et al., 2008), a transcription factor whose activity is in turn negatively regulated by NLK, the vertebrate ortholog of Nmo, through phosphorylation of its co-factor CREB binding protein (CBP). Our preliminary results demonstrate that dosage reduction of nejire (nej), which encodes the Drosophila ortholog of CBP, dominantly modifies the eye phenotypes produced by overexpression of either nmo or miple (VM-S and NP, unpublished results). These data suggest a potential mechanism by which nmo could be regulating miple expression and would explain miple up-regulation in nmoP1 mutant eye discs.
Supplementary Material
Movie S1. Live-imaging of the ommatidial rotation process in armGFP pupal eye imaginal discs. 15 min time-lapse confocal images of an armGFP pupal eye imaginal disc starting at the beginning of rotation (upper ommatidium) until rotation is almost complete. Anterior is left and posterior is right. The different developmental stages of the ommatidium as well as cell divisions suffered by IOCs can be observed. A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.02.006
Movie S2. Dynamics of cone cell precursors in armGFP pupal eye imaginal discs. 15 min time-lapse confocal images of an armGFP pupal imaginal disc. Arrow heads point to cell contacts that are being broken or established between cone cell precursors and IOCs while the cone cells are adopting their final position in the developing ommatidium. Apical shape changes (expansions and contractions) suffered by the IOCs during the ommatidial rotation can be also observed. A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.02.006.
Movie S3. Programmed cell death of IOCs during ommatidial rotation in armGFP pupal eye imaginal discs. 15 min time-lapse confocal images of an armGFP pupal imaginal disc. Note that several IOCs (marked with arrow heads) suffer a constriction of their apical surface until they disappear. As a result of this, contacts between neighboring cells are restructured. A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.02.006.
Movie S4. Live-imaging of the ommatidial rotation process in armGFP, nmoP1 pupal eye imaginal discs. 15 min time-lapse confocal images of an armGFP, nmoP1 ommatidium starting at the beginning of rotation and during a period similar to Movie 1. Note that ommatidial rotation occurs at slower pace than in armGFP discs and that a premature stop of rotation is observed although the ommatidium develops correctly. A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.02.006.
Movie S5. Dynamics of cone cell precursors in armGFP, nmoP1 pupal eye imaginal discs. 15 min time-lapse confocal images of an armGFP, nmoP1 pupal imaginal disc. Arrow heads point to cell contacts that are being broken or established between cone cell precursors and IOCs while the cone cells are adopting their final position in the developing ommatidium. Note that cone cell precursors in nmo mutant discs are more static than in armGFP controls (compare to Movie 2), since less contacts are being broken/established. A reduction of apical shape changes in IOCs can also be observed in nmo mutant discs compared to controls. A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.02.006.
Acknowledgments
We are grateful to L M. Escudero and R Jonhson for their precious advices on experimental procedures, to M. I. Galindo, E. Verheyen, S. Muñoz-Descalzo, T. Volk, M. Zeidler, to the Bloomington Stock Center and the Vienna Drosophila RNAi Center for kindly providing fly stocks, and to the Drosophila Genomics Resource Center for providing the mthl8 cDNA clone. Confocal and scanning electron microscopy was performed at the SCSIE (Universitat de València) and the Microscopy Shared Resource Facility of the MSSM. This work was supported by a short-term EMBO fellowship and a Journal of Cell Science travel fellowship to VM.-S. and by Grants from Ministerio de Educación y Ciencia (BFU2007-63213) and Conselleria d'Educació, Formació i Ocupació (PROMETEO/2010/081) to N.P.
Footnotes
Appendix A. Supporting information: Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2013.02.006.
References
- Bao S, Cagan R. Preferential adhesion mediated by Hibris and Roughest regulates morphogenesis and patterning in the Drosophila eye. Dev Cell. 2005;8:925–935. doi: 10.1016/j.devcel.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Bazigou E, Apitz H, Johansson J, Loren CE, Hirst EM, Chen PL, Palmer RH, Salecker I. Anterograde Jelly belly and Alk receptor tyrosine kinase signaling mediates retinal axon targeting in Drosophila. Cell. 2007;128:961–975. doi: 10.1016/j.cell.2007.02.024. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Blanchard GB, Murugesu S, Adams RJ, Martinez-Arias A, Gorfinkiel N. Cytoskeletal dynamics and supracellular organisation of cell shape fluctuations during dorsal closure. Development. 2010;137:2743–2752. doi: 10.1242/dev.045872. [DOI] [PubMed] [Google Scholar]
- Braid LR, Lee W, Uetrecht AC, Swarup S, Papaianni G, Heiler A, Verheyen EM. Nemo phosphorylates even-skipped and promotes Eve-mediated repression of odd-skipped in even parasegments during Drosophila embryogenesis. Dev Biol. 2010;343:178–189. doi: 10.1016/j.ydbio.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Braid LR, Verheyen EM. Drosophila nemo promotes eye specification directed by the retinal determination gene network. Genetics. 2008;180:283–299. doi: 10.1534/genetics.108.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brott BK, Pinsky BA, Erikson RL. Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc Natl Acad Sci USA. 1998;95:963–968. doi: 10.1073/pnas.95.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Freeman M. Egfr signalling defines a protective function for ommatidial orientation in the Drosophila eye. Development. 2003;130:5401–5412. doi: 10.1242/dev.00773. [DOI] [PubMed] [Google Scholar]
- Clifford RJ, Schupbach T. Coordinately and differentially mutable activities of torpedo, the Drosophila melanogaster homolog of the vertebrate EGF receptor gene. Genetics. 1989;123:771–787. doi: 10.1093/genetics/123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanana B, Graf R, Koledachkina T, Pflanz R, Vorbruggen G. AlphaPS2 integrin-mediated muscle attachment in Drosophila requires the ECM protein Thrombospondin. Mech Dev. 2007;124:463–475. doi: 10.1016/j.mod.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Chiu JC, Ko HW, Edery I. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell. 2011;145:357–370. doi: 10.1016/j.cell.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KW, Benzer S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell. 1994;78:125–136. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- Chou YH, Chien CT. Scabrous controls ommatidial rotation in the Drosophila compound eye. Dev Cell. 2002;3:839–850. doi: 10.1016/s1534-5807(02)00362-3. [DOI] [PubMed] [Google Scholar]
- David DJ, Tishkina A, Harris TJ. The PAR complex regulates pulsed actomyosin contractions during amnioserosa apical constriction in Drosophila. Development. 2010;137:1645–1655. doi: 10.1242/dev.044107. [DOI] [PubMed] [Google Scholar]
- Desai CJ, Gindhart JG, Jr, Goldstein LS, Zinn K. Receptor tyrosine phosphatases are required for motor axon guidance in the Drosophila embryo. Cell. 1996;84:599–609. doi: 10.1016/s0092-8674(00)81035-1. [DOI] [PubMed] [Google Scholar]
- de Vega S, Iwamoto T, Nakamura T, Hozumi K, McKnight DA, Fisher LW, et al. TM14 is a new member of the fibulin family (fibulin-7) that interacts with extracellular matrix molecules and is active for cell binding. J Biol Chem. 2007;282:30878–30888. doi: 10.1074/jbc.M705847200. [DOI] [PubMed] [Google Scholar]
- Englund C, Birve A, Falileeva L, Grabbe C, Palmer RH. Miple1 and miple2 encode a family of MK/PTN homologues in Drosophila melanogaster. Dev Genes Evol. 2006;216:10–18. doi: 10.1007/s00427-005-0025-8. [DOI] [PubMed] [Google Scholar]
- Escudero LM, Bischoff M, Freeman M. Myosin II regulates complex cellular arrangement and epithelial architecture in Drosophila. Dev Cell. 2007;13:717–729. doi: 10.1016/j.devcel.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Fetting JL, Spencer SA, Wolff T. The cell adhesion molecules Echinoid and Friend of Echinoid coordinate cell adhesion and cell signaling to regulate the fidelity of ommatidial rotation in the Drosophila eye. Development. 2009;136:3323–3333. doi: 10.1242/dev.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehler RW, Wolff T. Drosophila Myosin II, Zipper, is essential for ommatidial rotation. Dev Biol. 2007;310:348–362. doi: 10.1016/j.ydbio.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehler RW, Wolff T. Nemo is required in a subset of photoreceptors to regulate the speed of ommatidial rotation. Dev Biol. 2008;313:533–544. doi: 10.1016/j.ydbio.2007.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth LC, Baker NE. Spitz from the retina regulates genes transcribed in the second mitotic wave, peripodial epithelium, glia and plasmatocytes of the Drosophila eye imaginal disc. Dev Biol. 2007;307:521–538. doi: 10.1016/j.ydbio.2007.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Freeman M, Klambt C, Goodman CS, Rubin GM. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell. 1992;69:963–975. doi: 10.1016/0092-8674(92)90615-j. [DOI] [PubMed] [Google Scholar]
- Gaengel K, Mlodzik M. Egfr signaling regulates ommatidial rotation and cell motility in the Drosophila eye via MAPK/Pnt signaling and the Ras effector Canoe/AF6. Development. 2003;130:5413–5423. doi: 10.1242/dev.00759. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Ho YH, Lien MT, Lin CM, Wei SY, Chang LH, Hsu JC. Echinoid regulates Flamingo endocytosis to control ommatidial rotation in the Drosophila eye. Development. 2010;137:745–754. doi: 10.1242/dev.040238. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Hirao T, Suzuki M, Isoda M, Ishitani S, Harigaya K, Kitagawa M, Matsumoto K, Itoh M. Nemo-like kinase suppresses Notch signaling by interfering with formation of the Notch active transcriptional complex. Nat Cell Biol. 2010;12:278–285. doi: 10.1038/ncb2028. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Ishitani S, Matsumoto K, Itoh M. Nemo-like kinase is involved in NGF-induced neurite outgrowth via phosphorylating MAP1B and paxillin. J Neurochem. 2009;111:1104–1118. doi: 10.1111/j.1471-4159.2009.06400.x. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Ninomiya-Tsuji J, Matsumoto K. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:1379–1389. doi: 10.1128/MCB.23.4.1379-1389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T, Ninomiya-Tsuji J, Nagai S, Nishita M, Meneghini M, Barker N, et al. The TAK1–NLK–MAPK-related pathway antagonizes signaling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- Izumikawa T, Egusa N, Taniguchi F, Sugahara K, Kitagawa H. Heparan sulfate polymerization in Drosophila. J Biol Chem. 2006;281:1929–1934. doi: 10.1074/jbc.M509138200. [DOI] [PubMed] [Google Scholar]
- Jenny A. Planar cell polarity signaling in the Drosophila eye. Curr Top Dev Biol. 2010;93:189–227. doi: 10.1016/B978-0-12-385044-7.00007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanei-Ishii C, Ninomiya-Tsuji J, Tanikawa J, Nomura T, Ishitani T, Kishida S, et al. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 2004;18:816–829. doi: 10.1101/gad.1170604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpelainen I, Kaksonen M, Kinnunen T, Avikainen H, Fath M, Linhardt RJ, et al. Heparin-binding growth-associated molecule contains two heparin-binding beta-sheet domains that are homologous to the thrombospondin type I repeat. J Biol Chem. 2000;275:13564–13570. doi: 10.1074/jbc.275.18.13564. [DOI] [PubMed] [Google Scholar]
- Kojima H, Sasaki T, Ishitani T, Iemura S, Zhao H, Kaneko S, et al. STAT3 regulates Nemo-like kinase by mediating its interaction with IL-6-stimulated TGFbeta-activated kinase 1 for STAT3 Ser-727 phosphorylation. Proc Natl Acad Sci USA. 2005;102:4524–4529. doi: 10.1073/pnas.0500679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd TE, Verstreken P, Ostrin EJ, Phillippi A, Lichtarge O, Bellen HJ. A genome-wide search for synaptic vesicle cycle proteins in Drosophila. Neuron. 2000;26:45–50. doi: 10.1016/s0896-6273(00)81136-8. [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Ishitani T, Carter JC, Hisamoto N, Ninomiya-Tsuji J, Thorpe CJ, et al. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399:793–797. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- Meng K, Rodriguez-Pena A, Dimitrov T, Chen W, Yamin M, Noda M, et al. Pleiotrophin signals increased tyrosine phosphorylation of beta beta-catenin through inactivation of the intrinsic catalytic activity of the receptor-type protein tyrosine phosphatase beta/zeta. Proc Natl Acad Sci USA. 2000;97:2603–2608. doi: 10.1073/pnas.020487997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino C, Penney J, Gonzalez M, Tsurudome K, Moujahidine M, O'Connor MB, et al. Nemo kinase interacts with Mad to coordinate synaptic growth at the Drosophila neuromuscular junction. J Cell Biol. 2009;185:713–725. doi: 10.1083/jcb.200809127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaly J, Kockel L, Gaengel K, Weber U, Bohmann D, Mlodzik M. The role of the Drosophila TAK homologue dTAK during development. Mech Dev. 2001;102:67–79. doi: 10.1016/s0925-4773(01)00285-4. [DOI] [PubMed] [Google Scholar]
- Mirkovic I, Charish K, Gorski SM, McKnight K, Verheyen EM. Drosophila nemo is an essential gene involved in the regulation of programmed cell death. Mech Dev. 2002;119:9–20. doi: 10.1016/s0925-4773(02)00289-7. [DOI] [PubMed] [Google Scholar]
- Mirkovic I, Gault WJ, Rahnama M, Jenny A, Gaengel K, Bessette D, et al. Nemo kinase phosphorylates beta-catenin to promote ommatidial rotation and connects core PCP factors to E-cadherin-beta-catenin. Nat Struct Mol Biol. 2011;18:665–672. doi: 10.1038/nsmb.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovic I, Mlodzik M. Cooperative activities of drosophila DE-cadherin and DN-cadherin regulate the cell motility process of ommatidial rotation. Development. 2006;133:3283–3293. doi: 10.1242/dev.02468. [DOI] [PubMed] [Google Scholar]
- Mlodzik M. Planar polarity in the Drosophila eye: a multifaceted view of signaling specificity and cross-talk. EMBO J. 1999;18:6873–6879. doi: 10.1093/emboj/18.24.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo SA, Braid LR, Verheyen EM, Rebay I. Nemo phosphorylates Eyes absent and enhances output from the Eyasine oculis transcriptional complex during Drosophila retinal determination. Dev Biol. 2012 doi: 10.1016/j.ydbio.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee T, Schafer U, Zeidler MP. Identification of Drosophila genes modulating Janus kinase/signal transducer and activator of transcription signal transduction. Genetics. 2006;172:1683–1697. doi: 10.1534/genetics.105.046904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T. Midkine, a heparin-binding cytokine with multiple roles in development, repair and diseases. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:410–425. doi: 10.2183/pjab.86.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Shimojima M, Tohya Y, Miyazawa T. Molecular cloning of a cDNA encoding the feline CD62L. J Vet Med Sci. 2007;69:81–84. doi: 10.1292/jvms.69.81. [DOI] [PubMed] [Google Scholar]
- Ohkawara B, Shirakabe K, Hyodo-Miura J, Matsuo R, Ueno N, Matsumoto K, et al. Role of the TAK1–NLK–STAT3 pathway in TGF-beta-mediated mesoderm induction. Genes Dev. 2004;18:381–386. doi: 10.1101/gad.1166904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitriou E, Mikelis C, Lampropoulou E, Koutsioumpa M, Theochari K, Tsirmoula S, et al. Roles of pleiotrophin in tumor growth and angiogenesis. Eur Cytokine Networks. 2009;20:180–190. doi: 10.1684/ecn.2009.0172. [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P, Alcantara S, Dimitrov T, Vega JA, Deuel TF. Pleiotrophin disrupts calcium-dependent homophilic cell–cell adhesion and initiates an epithelial–mesenchymal transition. Proc Natl Acad Sci USA. 2006;103:17795–17800. doi: 10.1073/pnas.0607299103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert JR, Mlodzik M. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet. 2007;8:126–138. doi: 10.1038/nrg2042. [DOI] [PubMed] [Google Scholar]
- Shi Y, Ye K, Wu H, Sun Y, Shi H, Huo K. Human SMAD4 is phosphorylated at Thr9 and Ser138 by interacting with NLK. Mol Cell Biochem. 2010;333:293–298. doi: 10.1007/s11010-009-0230-2. [DOI] [PubMed] [Google Scholar]
- Solon J, Kaya-Copur A, Colombelli J, Brunner D. Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell. 2009;137:1331–1342. doi: 10.1016/j.cell.2009.03.050. [DOI] [PubMed] [Google Scholar]
- Stanyon CA, Liu G, Mangiola BA, Patel N, Giot L, Kuang B, et al. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H, Strutt D. EGF signaling and ommatidial rotation in the Drosophila eye. Curr Biol. 2003;13:1451–1457. doi: 10.1016/s0960-9822(03)00545-1. [DOI] [PubMed] [Google Scholar]
- The I, Bellaiche Y, Perrimon N. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol Cell. 1999;4:633–639. doi: 10.1016/s1097-2765(00)80214-2. [DOI] [PubMed] [Google Scholar]
- Toledano-Katchalski H, Nir R, Volohonsky G, Volk T. Post-transcriptional repression of the Drosophila midkine and pleiotrophin homolog miple by HOW is essential for correct mesoderm spreading. Development. 2007;134:3473–3481. doi: 10.1242/dev.006080. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Neuronal differentiation in Drosophila ommatidium. Dev Biol. 1987;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- Toyama Y, Peralta XG, Wells AR, Kiehart DP, Edwards GS. Apoptotic force and tissue dynamics during Drosophila embryogenesis. Science. 2008;321:1683–1686. doi: 10.1126/science.1157052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen EM, Mirkovic I, MacLean SJ, Langmann C, Andrews BC, MacKinnon C. The tissue polarity gene nemo carries out multiple roles in patterning during Drosophila development. Mech Dev. 2001;101:119–132. doi: 10.1016/s0925-4773(00)00574-8. [DOI] [PubMed] [Google Scholar]
- Weber U, Gault WJ, Olguin P, Serysheva E, Mlodzik M. Novel regulators of planar cell polarity: a genetic analysis in Drosophila. Genetics. 2012;191:155–162. doi: 10.1534/genetics.111.137190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter CG, Wang B, Ballew A, Royou A, Karess R, Axelrod JD, et al. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell. 2001;105:81–91. doi: 10.1016/s0092-8674(01)00298-7. [DOI] [PubMed] [Google Scholar]
- Wolff T. Preparation of Drosophila eye specimens for scanning electron microscopy. Cold Spring Harb Protoc. 2011;2011:1383–1385. doi: 10.1101/pdb.prot066506. [DOI] [PubMed] [Google Scholar]
- Yasuda J, Yokoo H, Yamada T, Kitabayashi I, Sekiya T, Ichikawa H. Nemo-like kinase suppresses a wide range of transcription factors, including nuclear factor-kappaB. Cancer Sci. 2004;95:52–57. doi: 10.1111/j.1349-7006.2004.tb03170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Dong Y, Kong X, Beckett LA, Gandour-Edwards R, Melamed J. Midkine is a NF-kappaB-inducible gene that supports prostate cancer cell survival. BMC Med Genomics. 2008;1:6. doi: 10.1186/1755-8794-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Houl JH, Hardin PE. NEMO kinase contributes to core period determination by slowing the pace of the Drosophila circadian oscillator. Curr Biol. 2011;21:756–761. doi: 10.1016/j.cub.2011.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YA, Rahnama M, Wang S, Sosu-Sedzorme W, Verheyen EM. Drosophila Nemo antagonizes BMP signaling by phosphorylation of Mad and inhibition of its nuclear accumulation. Development. 2007;134:2061–2071. doi: 10.1242/dev.02853. [DOI] [PubMed] [Google Scholar]
- Zeng YA, Verheyen EM. Nemo is an inducible antagonist of Wingless signaling during Drosophila wing development. Development. 2004;131:2911–2920. doi: 10.1242/dev.01177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Live-imaging of the ommatidial rotation process in armGFP pupal eye imaginal discs. 15 min time-lapse confocal images of an armGFP pupal eye imaginal disc starting at the beginning of rotation (upper ommatidium) until rotation is almost complete. Anterior is left and posterior is right. The different developmental stages of the ommatidium as well as cell divisions suffered by IOCs can be observed. A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.02.006
Movie S2. Dynamics of cone cell precursors in armGFP pupal eye imaginal discs. 15 min time-lapse confocal images of an armGFP pupal imaginal disc. Arrow heads point to cell contacts that are being broken or established between cone cell precursors and IOCs while the cone cells are adopting their final position in the developing ommatidium. Apical shape changes (expansions and contractions) suffered by the IOCs during the ommatidial rotation can be also observed. A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.02.006.
Movie S3. Programmed cell death of IOCs during ommatidial rotation in armGFP pupal eye imaginal discs. 15 min time-lapse confocal images of an armGFP pupal imaginal disc. Note that several IOCs (marked with arrow heads) suffer a constriction of their apical surface until they disappear. As a result of this, contacts between neighboring cells are restructured. A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.02.006.
Movie S4. Live-imaging of the ommatidial rotation process in armGFP, nmoP1 pupal eye imaginal discs. 15 min time-lapse confocal images of an armGFP, nmoP1 ommatidium starting at the beginning of rotation and during a period similar to Movie 1. Note that ommatidial rotation occurs at slower pace than in armGFP discs and that a premature stop of rotation is observed although the ommatidium develops correctly. A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.02.006.
Movie S5. Dynamics of cone cell precursors in armGFP, nmoP1 pupal eye imaginal discs. 15 min time-lapse confocal images of an armGFP, nmoP1 pupal imaginal disc. Arrow heads point to cell contacts that are being broken or established between cone cell precursors and IOCs while the cone cells are adopting their final position in the developing ommatidium. Note that cone cell precursors in nmo mutant discs are more static than in armGFP controls (compare to Movie 2), since less contacts are being broken/established. A reduction of apical shape changes in IOCs can also be observed in nmo mutant discs compared to controls. A video clip is available online. Supplementary material related to this article can be found online at http://dx.doi.org/10.1016/j.ydbio.2013.02.006.