Abstract
Amyotrophic lateral sclerosis (ALS) is the third most common adult-onset neurodegenerative disease. Individuals with ALS rapidly progress to paralysis and die from respiratory failure within 3 to 5 years after symptom onset. Epidemiological factors explain only a modest amount of the risk for ALS. However, there is growing evidence of a strong genetic component to both familial and sporadic ALS risk. The International Consortium on Amyotrophic Lateral Sclerosis Genetics was established to bring together existing genome-wide association cohorts and identify sporadic ALS susceptibility and age at symptom onset loci. Here, we report the results of a meta-analysis of the International Consortium on Amyotrophic Lateral Sclerosis Genetics genome-wide association samples, consisting of 4243 ALS cases and 5112 controls from 13 European ancestry cohorts from across the United States and Europe. Eight genomic regions provided evidence of association with ALS, including 9p21.2 (rs3849942, odds ratio [R] = 1.21; p = 4.41 × 10−7), 17p11.2 (rs7477, OR = 1.30; p = 2.89 × 10−7), and 19p13 (rs12608932, OR = 1.37, p = 1.29 × 10−7). Six genomic regions were associated with age at onset of ALS. The strongest evidence for an age of onset locus was observed at 1p34.1, with comparable evidence at rs3011225 (R2partial = 0.0061; p = 6.59 × 10−8) and rs803675 (R2partial = 0.0060; p = 6.96 × 10−8). These associations were consistent across all 13 cohorts. For rs3011225, individuals with at least 1 copy of the minor allele had an earlier average age of onset of over 2 years. Identifying the underlying pathways influencing susceptibility to and age at onset of ALS may provide insight into the pathogenic mechanisms and motivate new pharmacologic targets for this fatal neurodegenerative disease.
Keywords: Amyotrophic lateral sclerosis, Genome-wide association study, Age at onset
1. Introduction
Amyotrophic lateral sclerosis (ALS) is the third most common adult-onset neurodegenerative disease, affecting approximately 10,000 Americans annually (Hirtz et al., 2007). Disease progression is rapid and results in progressive paralysis and death from respiratory failure, usually within 3 to 5 years. The current estimated incidence is between 2.2 and 2.8 per 100,000 (Logroscino et al., 2008; Traynor et al., 1999; Worms, 2001; Zoccolella et al., 2006) with a lifetime prevalence of about 1 in 300 (Johnston et al., 2006). Risk factors for ALS include male gender, increasing age, positive family history of ALS, and smoking (Armon, 2009; Siddique et al., 1998). Males tend to have a slightly earlier age of onset than females (approximately 65 and approximately 67 years of age, respectively) and a male-to-female ratio of approximately 1.6 that varies with age (Manjaly et al., 2010). Approximately 5%–10% of individuals diagnosed with ALS have a family history (i.e., familial ALS), while the remaining 90%–95% do not report a family history and are classified as sporadic ALS (Al-Chalabi and Lewis, 2011; Chiò et al., 2008; Fallis and Hardiman, 2009; Fang et al., 2009; Hanby et al., 2011; Logroscino et al., 2005; Traynor et al., 1999). Of the other posited risk factors, smoking provides the strongest epidemiological factor (Weisskopf et al., 2010); other factors such as exercise, heavy metal and occupational exposures, remain controversial. Taken together, these epidemiologic risk factors do not explain much of the variation in the risk for ALS, age of onset of motor neuron dysfunction, or survival time once diagnosed with ALS.
The increased familial risk and the identification of genetic variation that increases risk of ALS are consistent with ALS being a genetic trait. Analyses of multiplex families have identified mutations in several genes that are causative for ALS, including but not limited to SOD1 (Rosen et al., 1993, 1994), VAPB (Nishimura et al., 2004), TARDBP (Sreedharan et al., 2008), FUS (Kwiatkowski et al., 2009; Vance et al., 2009), OPTN (Maruyama et al., 2010), VCP (Johnson et al., 2010), and UBQLN2 (Deng et al., 2011). Genome-wide association studies (GWAS) that have been completed for sporadic ALS have identified loci of interest such as DPP6 (Cronin et al., 2008; van Es et al., 2008), ITPR2 (van Es et al., 2007), SUNC1 (Chiò et al., 2009), UNC13a (van Es et al., 2009), and 9p21.2 (Laaksovirta et al., 2010; Shatunov et al., 2010; van Es et al., 2009). Among these, only the association signal on chromosome 9p21 has been consistently replicated across studies. Building upon these GWAS results, it was recently reported that a large hexanucleotide repeat expansion of the C9orf72 gene underlies this locus in a portion of both sporadic and familial ALS (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Thus, the C9orf72 result underscores the importance of well-powered GWAS in ALS.
Efforts to find genetic factors that influence ALS phenotypes are also under way, but have yet to yield unambiguous results. Variants within KIFAP3 have been implicated in modifying patient survival time from diagnosis of ALS (Landers et al., 2009), but these variants were not replicated in an independent cohort (Traynor et al., 2010). Similarly CHGB, TXNRD1, APOE, and VEGF variants and a 50-base pair promoter deletion of SOD1, have been associated with modifying age of ALS onset (Gros-Louis et al., 2009 [CHGB], Mitchell et al., 2009 [TXNRD1], Li et al., 2004 [APOE], Oosthuyse et al., 2001 [VEGF]). Age of ALS onset shows evidence for heritability in studies of multiple families with SOD1 mutations from a single founder (Fogh et al., 2007), but none of these loci are considered established. The amyotrophic lateral sclerosis online database (ALSOD; alsod.iop.kcl.ac.uk/) and the ALSGene database (www.alsgene.org/) provide an up-to-date summary of ALS genetic risk factors for familial and sporadic ALS (Lill et al., 2011; Wroe et al., 2008).
There are multiple plausible reasons for the modest number of established sporadic ALS, age of onset, and susceptibility loci. These reasons parallel the relatively modest number of weak epidemiologic risk factors that are considered established. One might hypothesize that the genetic influences on sporadic ALS are weak. However, the lack of strong epidemiologic factors, the existence of known genetic influences on familial ALS, twin studies (Al-Chalabi et al., 2010), and the existence of sporadic ALS predisposing variants suggest that genetic factors exist. Alternatively, one might hypothesize that the lack of established associations may be the result of modest statistical power, given that ALS is a relatively rare disease and most loci likely have modest effects.
The International Consortium on Amyotrophic Lateral Sclerosis Genetics (ALSGEN; https://alsgen.phs.wfubmc.edu/public/index.cfm) was established to combine resources to maximize the power to identify ALS susceptibility loci and genetic variation that influences age at symptom onset. Here, we report the results of a joint and meta-analysis of the ALSGEN GWAS samples focused on predisposition to ALS and ALS age of onset.
2. Methods
2.1. Subjects
Samples were received from a total of 13 sources spanning the United States and Europe, including Belgium, France, the Netherlands, Ireland, Italy, Sweden, and the United Kingdom, with the Netherlands and Ireland each contributing 2 separate cohorts (Supplementary Table 1). All cases met the El Escorial criteria for probable or definite ALS. Controls were received from all sources except the French cohort. With the exception of samples from Northwestern University, all samples have been described in previous publications (Chiò et al., 2009; Cronin et al., 2008; Landers et al., 2009; Schymick et al., 2007; Traynor et al., 2010; Valdmanis et al., 2007; van Es et al., 2007, van Es et al., 2009). The Northwestern sporadic ALS are age and ethnicity matched cases are from the Neurologic Diseases Registry, with spousal or community controls. All samples were of self-reported Caucasian ethnicity and of European or European-American ancestry.
2.2. Genotyping
All of the contributed samples were genotyped on 1 of the Illumina (San Diego, CA, USA) single nucleotide polymorphism (SNP) chips (see Supplementary Table 1); 254,145 SNPs common across the various SNP genotyping arrays and that passed quality control were analyzed in these cohorts.
2.3. Statistical analysis
A principal-component analysis of all 254,145 autosomal SNPs that pass the highest standards of quality control (i.e., low missingness, no differential missingness between cases and controls, no departure from Hardy–Weinberg Equilibrium expectations, relatedness, gender ambiguities, heterozygosity outliers, samples with low call rates (< 95%) and samples contributing investigators excluded in their previously published analyses) was computed. Two principal components were identified that reduced the overall inflation factor and were included as covariates in all subsequent modeling. Additional principal components did not further reduce inflation factor nor substantively change any inferences reported. The principal-component analysis identified 20 individuals (9 cases, 11 controls) that were modest genetic outliers. Analyses were completed with and without these outliers and the results were comparable. Results reported here exclude these 20 individuals.
To test for an association between an individual SNP and ALS, both a joint analysis of the entire ALSGEN cohort and a meta-analysis of the individual cohorts were computed. The joint analysis provides greater statistical power for SNPs with less frequent alleles, particularly for additive and recessive genetic models. In contrast, the meta-analysis is asymptotically as powerful as the joint analysis for more common alleles but may be additionally more robust to intercohort heterogeneity. In the meta-analysis, tests for additive or recessive genetic models may not be computed for low frequency SNPs (see Joint Analysis section 2.4.).
2.4. Joint analysis
To test for an association between an individual SNP and ALS, a logistic regression model was computed as implemented in the software SNPLash (https://www.phs.wfubmc.edu/public/bios/gene/downloads.cfm). Specifically, the logistic regression model contained the first 2 principal components to adjust for potential population structure, cohort, and the individual SNP parameterized under 1 of the 3 a priori genetic models (i.e., dominant, additive, recessive). The cohort adjustment was parameterized as a series of 12 indicator variables, 1 less than the total number of cohorts. The additive genetic model is the primary inference unless there is significant evidence of departure from an additive genetic model (p < 0.05). To increase the robustness of the tests of association, the additive and recessive genetic models required at least 10 and 30 individuals homozygous for the minor allele, respectively. The inflation factor was computed as the mean of the χ2 statistic for the additive genetic model and was compared with its theoretic mean and variance of 1 and 2, respectively. All analyses were repeated stratifying by gender and testing for the gender-by-SNP interaction.
2.5. Meta-analysis
The meta-analysis was computed using the same logistic regression model as the joint analysis except that it was computed within each cohort and did not include the cohort indicator variables. Because the sample size for each individual cohort is significantly less than that of the entire ALSGEN cohort, more SNPs will fail to meet the additive and recessive model count criteria for the number of homozygotes of the minor allele.
The evidence of association from each cohort was combined using the weighted inverse normal method. Here let ni and Zi denote the sample size and standard normal random variant from the Wald test for cohort i. Then,
has a standard normal distribution under the null hypothesis of no association. A test for heterogeneity of association across the ALSGEN cohorts was also computed. As in the joint analysis, all meta-analyses were repeated stratifying by gender and testing for the gender-by-SNP interaction. Results of the joint and meta-analyses were comparable except when models (e.g. additive and recessive models) could not be computed due to low counts.
2.6. Multilocus modeling
To investigate the joint effects of these loci on ALS case/control status and ALS age of onset, 2 stepwise regression models were computed. Specifically for ALS case/control status, stepwise modeling (forward selection with backward elimination) was computed under a logistic regression model using the 200 most statistically significant SNPs for ALS case/control status and an entry and exit criteria of p < 0.00001. From the final model, the area under the receiver operator characteristic curve (i.e., C-statistic) was computed. To reduce the bias of the C-statistic due to the computations using the discovery sample, a 10-fold cross-validation estimate of the C-statistic was computed and the mean and 95% confidence interval based on these 10 samples was computed. The stepwise modeling for age of ALS onset was done similarly except the analysis was completed under a linear regression model and the partial R2 for each SNP was computed. The partial R2 estimates the proportion of the variation in age of ALS onset; the individual SNP explains adjusting for the principal components, center, and the other SNPs in the model.
The C9orf72 region is known to harbor risk variants for ALS. Thus, 3 additional analyses were completed to test whether any locus across the genome might have a differential effect depending on the presence or absence of the C9orf72 risk variants (i.e., statistical interaction). The C9orf72 risk variant was modeled using the surrogate SNP rs2453556 identified in the above analyses. The first interaction analysis tested each SNP in the GWAS for evidence of a statistical interaction with C9orf72 by testing for significant heterogeneity of the odds ratio between individuals with and without the risk allele at rs2453556; the approach is implemented in the program Metal (www.sph.umich.edu/csg/abecasis/metal). The second analysis simply stratified the data by the presence or absence of the risk allele at rs2454556 and recomputed the GWAS. Finally, the third analysis computed a case-only test of an interaction. Here, a GWAS was computed using a logistic regression model with the principal components as covariates and the binary outcome defined as the presence versus absence of the risk allele at rs2454556. This case-only analysis is statistically more powerful than the test for heterogeneity of the odds ratio under the strong assumption that the loci are otherwise uncorrelated. By Mendel’s Laws, loci on different chromosomes are inherited independently and therefore the assumptions of the analysis are appropriate.
3. Results
3.1. Sample and preliminary analysis
The ALSGEN sample reported here consisted of 4243 cases and 5112 controls from 13 European ancestry cohorts from across the United States and Europe (Supplementary Table 1). Overall, females were less prevalent in both cases (41%) and controls (42%). The mean age of onset was 57.5 ± 12.9 years of age. Median survival time once diagnosed with ALS was 3.2 years. The site of symptom onset was relatively similar across cohorts, with 67% being limb, 28% being bulbar, and 5% unknown or unreported. The total number of autosomal SNPs that passed quality control inspection in all cohorts was 254,145. Ignoring the correlation among the SNPs due to linkage disequilibrium, a Bonferroni correction factor for 254,145 SNPs is 2.0 × 10−7.
The first 2 principal components captured sufficient genetic variation to reduce the inflation factor in the joint case/control analysis to 1.04 and the joint age-of-onset analysis to 1.01. These inflation factors, the lack of polymorphisms with a known European gradient in allele frequency among the top statistical associations, and the P–P plot (Supplementary Figs. 1 and 2) suggest that potential biases due to admixture are unlikely to have been a significant problem in our analysis.
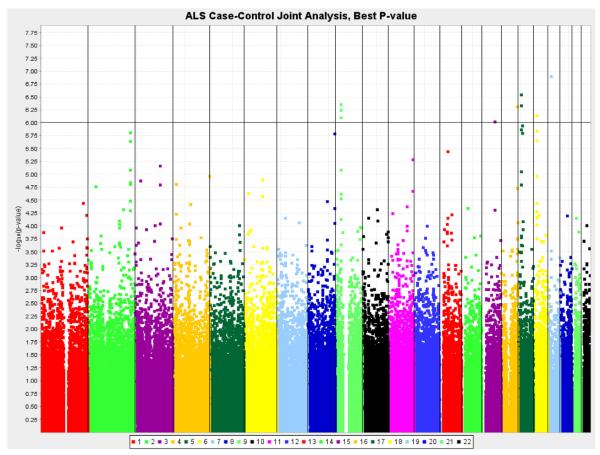
Eight genomic regions provided evidence of association with ALS (p < 1.0 × 10−6; Table 1, Fig. 1), though none of these achieved significance after correction for multiple testing. The joint analysis and the meta-analyses are strongly correlated and provide comparable evidence of association for these 8 regions. There is no evidence of heterogeneity of the odds ratio across the ALSGEN sample for these 8 regions, and no suggestive evidence of heterogeneity across the entire genome (p < 1.0 × 10−5).
Table 1.
Case-control outcome adjusted for admixture (first 2 principal components) and center
| SNP | Chr | Position (Mb) |
Gene | Minor allele |
MAF (case) |
MAF (control) |
Joint p value | Meta p value | OR | 95% CI |
|---|---|---|---|---|---|---|---|---|---|---|
| rs2303565 | 2q35 | 219.254 | STK36 | C | 0.47 | 0.43 | 1.56E-06 (a) | 6.18E-07 (a) | 1.17 | (1.10–1.24) |
| rs1344642 | 2q35 | 219.264 | STK36 | A | 0.47 | 0.43 | 2.30E-06 (a) | 9.92E-07 (a) | 1.17 | (1.10–1.25) |
| rs2814707 | 9p21.2 | 27.526 | C9orf72a, MOBKL2Ba | A | 0.26 | 0.23 | 5.70E-07 (d) | 8.39E-07 (d) | 1.21 | (1.11–1.33) |
| rs3849942 | 9p21.2 | 27.533 | C9orf72a, MOBKL2Ba | A | 0.26 | 0.23 | 4.41E-07 (d) | 6.47E-07 (d) | 1.21 | (1.11–1.33) |
| rs2453556 | 9p21.2 | 27.576 | C9orf72a | G | 0.44 | 0.40 | 8.12E-07 (a) | 6.95E-07 (a) | 1.16 | (1.09–1.24) |
| rs1971791 | 15q23 | 68.363 | None within 150 kb | G | 0.49 | 0.47 | 9.70E-07 (r) | 2.11E-06 (r) | 1.26 | (1.14–1.40) |
| rs8056742 | 16q24.1 | 83.650 | KIAA0513 | C | 0.13 | 0.11 | 4.91E-07 (a) | 2.94E-06 (d) | 1.27 | (1.14–1.41) |
| rs2109262 | 17p11.2 | 16.125 | PIGL | C | 0.47 | 0.51 | 4.67E-07 (a) | 4.12E-07 (d) | 0.78 | (0.71–0.87) |
| rs7477 | 17p11.2 | 16.187 | CENPV | T | 0.52 | 0.48 | 2.89E-07 (r) | 3.05E-07 (r) | 1.30 | (1.16–1.44) |
| rs2006933 | 17q11.2 | 23.625 | FLJ40504a | G | 0.23 | 0.25 | 1.15E-06 (d) | 8.60E-07 (d) | 0.79 | (0.73–0.86) |
| rs11082762 | 18q11.2 | 19.620 | LAMA3 | A | 0.40 | 0.36 | 7.26E-07 (a) | 1.78E-06 (a) | 1.16 | (1.09–1.24) |
| rs12608932 | 19p13.11 | 17.614 | UNC13A | C | 0.38 | 0.34 | 1.29E-07 (a) | 4.96E-08 (r) | 1.37 | (1.21–1.56) |
SNPs are listed that have p < 1 × 10−6 in either the joint analysis or meta-analysis. For joint analysis, the additive model is presented unless the test for lack-of-fit to an additive model was significant (p < 0.05); (a) additive, (d) dominant, (r) recessive. For meta-analysis, the minimum p value of the 3 genetic models is used, noting that quality control standards were applied to ensure only appropriate samples sizes were included in the meta-analysis for each genetic model. There was no evidence for heterogeneity of the odds ratio across centers (p = 0.08, 0.09, and, > 0.20 for rs3849942, rs2814707, and remaining SNPs in the table, respectively). OR shown is from meta-analysis with 95% CI boundaries. Positions are from build 36.3.
Key: Chr, chromosome; CI, confidence interval; kb, kilobases; MAF, minor allele frequency; Mb, megabase; OR, odds ratio; SNP, single nucleotide polymorphism.
Less than 20 kb away.
Fig. 1.
Results of the genome-wide association analysis contrasting amyotrophic lateral sclerosis (ALS) cases versus controls (joint analysis).
Multiple SNPs in the chromosomal region 9p21.2 showed evidence of association with ALS (Table 1). The SNPs rs3849942 (odds ratio [OR] = 1.21; p = 4.41 × 10−7), rs2814707 (OR = 1.21; p = 5.70 × 10−7), and rs2453556 (OR = 1.16, p = 8.12 × 10−7) span nearly 50 kilobases, are relatively common and provide comparable evidence of association. The linkage disequilibrium is strong between rs3849942 and rs2814707 (r2 = 0.98) but is very modest between rs2814707 and rs2453556 (r2 = 0.20) and rs3849942 and rs2453556 (r2 = 0.20). In these SNPs the minor allele was associated with an increased risk of ALS. During the course of this study, the genetic lesion underlying this locus was cloned (DeJesus-Hernandez et al., 2011; Renton et al., 2011), and it was not feasible to screen the samples included in the current study for the pathogenic hexanucleotide repeat expansion.
Two regions on chromosome 17 exhibited evidence of association with ALS (Table 2). In the gene-rich 17p11.2 region, rs2109262 (OR = 0.78, p = 4.67 × 10−7) near PIGL and rs7477 (OR = 1.30; p = 2.89 × 10−7) near CENPV showed increased risk for the minor and major alleles, respectively. On the other arm of chromosome 17, the 17q11.1 region (rs2006933, OR = 0.79, p = 1.15 × 10−6) was also associated with ALS. In addition, chromosomal regions 15q23 (rs1971791, OR = 1.26, p = 9.70 × 10−7), 16q24.1 (rs8056742, OR = 1.27, p = 4.91 × 10−7), and 18q11.2 (rs11082762, OR = 1.16, p = 7.26 × 10−7) all showed increased risk with the minor allele.
Table 2.
Stepwise logistic regression starting with top 200 SNPs from joint case-control analysis, adjusted for admixture (first 2 principal components) and center
| SNP | Chr | Position (Mb) |
Minor allele |
MAF (cases) |
MAF (controls) |
Single locus p valuea |
Model | Stepwise p value |
OR | 95% CI |
|---|---|---|---|---|---|---|---|---|---|---|
| rs4331558 | 2p21 | 41.935 | C | 0.16 | 0.18 | 1.75E-05 | Dom | 6.01E-06 | 0.81 | (0.73–0.88) |
| rs2303565 | 2q35 | 219.254 | C | 0.47 | 0.43 | 1.56E-06 | Add | 2.91E-07 | 1.17 | (1.10–1.25) |
| rs1320900 | 3q22.1 | 133.196 | A | 0.31 | 0.34 | 6.92E-06 | Dom | 1.39E-06 | 0.81 | (0.74–0.88 |
| rs4917300 | 8q24.3 | 143.12 | T | 0.49 | 0.46 | 1.65E-06 | Rec | 1.16E-05 | 1.26 | (1.14–1.40) |
| rs2453556 | 9p21.2 | 27.576 | G | 0.44 | 0.40 | 8.12E-07 | Add | 2.93E-07 | 1.18 | (1.11–1.25) |
| rs1971791 | 15q23 | 68.363 | G | 0.49 | 0.47 | 9.70E-07 | Rec | 2.00E-06 | 1.28 | (1.16–1.42) |
| rs8056742 | 16q24.1 | 83.650 | C | 0.13 | 0.11 | 4.91E-07 | Add | 8.68E-07 | 1.26 | (1.15–1.39) |
| rs7477 | 17p11.2 | 16.187 | T | 0.52 | 0.48 | 2.89E-07 | Rec | 7.27E-07 | 1.29 | (1.16–1.42) |
| rs9909055 | 17q11.2 | 23.620 | T | 0.22 | 0.25 | 1.60E-06 | Dom | 1.36E-06 | 0.81 | (0.74–0.88) |
| rs2337186 | 18q11.2 | 19.566 | G | 0.37 | 0.33 | 2.24E-06 | Add | 1.16E-06 | 1.17 | (1.10–1.25) |
| rs12608932 | 19p13.11 | 17.614 | C | 0.38 | 0.34 | 1.29E-07 | Add | 8.08E-08 | 1.19 | (1.12–1.27) |
Area under receiver operating characteristic curve for model without SNPs, C-statistic = 0.591. Difference in C-statistics for model adjusting for center and admixture versus model adjusting for center, admixture, and SNPs in table = 0.051. Model entry and exit criteria: 1 × 10−5. Area under receiver operating characteristic curve, C-statistic = 0.642, 10-fold cross-validation C-statistic = 0.635.
Key: Add, additive; Chr, chromosome; CI, confidence interval; Dom, dominant; kb, kilobases; MAF, minor allele frequency; Mb, megabase; OR, odds ratio; Rec, recessive; SNP, single nucleotide polymorphism.
Single locus p value reflects statistical significance of the SNP in the joint analysis.
The strongest evidence of association in the entire ALSGEN cohort was for rs12608932 (19p13, OR = 1.37, p = 1.29 × 10−7). This region contains the UNC13a, an association previously reported in a subset of the ALSGEN samples (van Es et al., 2009). After removing these samples, the estimated odds ratio is in the same direction as the original association (Supplementary Table 2), but the remaining cases (715) and controls (717) only had 0.17 power to detect the effect previously reported and did not provide statistically significant evidence of association with ALS (OR = 1.08, p = 0.29).
Previous studies have suggested a number of candidate loci for association with ALS. Given the overlap in samples between the previous reports and the ALSGEN consortium data, analyses were computed on the independent set of samples (i.e., the samples used in the initial report were excluded from our analysis, see Supplementary Table 2). Three SNPs do provide suggestive evidence of association: rs6700125 on chromosome 1 near the FGGY gene, rs1541160 also on chromosome 1 near the KIFAP3 gene, and rs10260404 on chromosome 7 near the DPP6 gene. However, none of these associations remain significant after adjusting for multiple comparisons. Thus, these data do not provide strong corroborating evidence of association at these loci. Supplementary Table 3 summarizes the evidence of association in all of the ALSGEN samples (i.e., including the originally published discovery cohorts for that SNP). Three loci had very low statistical power (power < 0.20) to detect an OR = 1.1, while 4 were well powered power to detect an association (> 0.80).
Adjusting for the evidence of association at strongly associated loci can improve the statistical power to detect additional loci, while removing the redundancy among SNPs in linkage disequilibrium. The result of the exploratory stepwise logistic regression modeling continued to provide evidence for 5 of the 8 regions in Table 1 (Table 2). Most notable among the loci that showed increased evidence of association with ALS, when adjusting for the other loci, was 2q35 (rs2303565, OR = 1.17, p = 2.91 × 10−7). Considering these 13 regions jointly under a logistic regression model yielded a modest increase in the area under the receiver operator characteristic curve of ΔC = 0.051. Although significant and important, the value of ΔC = 0.051 indicates these 13 loci do not explain a large proportion of ALS susceptibility. Two regions, 15q23 and 17q11.1, with evidence of association in the single locus models no longer provide strong evidence of association in the stepwise analysis (p > 1 × 10−5). Supplementary Fig. 3 plots the evidence of association with ALS in the top statistically associated regions and Supplementary Table 4 reports the top 2000 statistical associations for ALS from the joint and meta-analyses.
3.2. Genome-wide tests for interactions with C9orf72 region
To explore whether there might be any evidence for an interaction between the C9orf72 region and the rest of the genome, 3 analyses were computed: (1) a genome-wide scan for heterogeneity of the odds ratio that compared individuals with versus without the risk allele for C9orf72 at surrogate SNP rs2453556, (2) a stratified analysis stratified on the presence or absence of the risk allele at rs2453556, and (3) a case-only genome-wide scan that contrasted individuals with versus without the risk allele at rs2453556. No individual SNP exhibited genome-wide evidence of a differential effect of the locus depending on the presence or absence of the rs2453556 risk allele (Supplementary Table 5, Supplementary Fig. 4). Interestingly, there was suggestive evidence of a differential effect on chromosome 21q22 at rs2838568 (p = 3.1 × 10−6) that was modestly supported in the stratified and case-only analyses. In the stratified analysis, no SNP in either strata (presence of risk allele or absence of risk allele) exhibited genome-wide significance for an association with ALS (Supplementary Tables 6 and 7, Supplementary Figs. 5 and 6). In the subset of cases and controls with no copies of the risk allele at the rs2453556 locus, the same chromosome 21q22 locus, rs2838568, that provided suggestive evidence of heterogeneity of the odds ratio exhibited suggestive evidence of association with ALS (p = 2.0 × 10−7, OR = 0.76). In this subset of individuals, no other loci provided this level of suggestive evidence of an interaction (Supplementary Table 5) with suggestive evidence of association with ALS (Supplementary Table 6, Supplementary Fig. 5). In the subset of cases and controls that have at least 1 copy of the risk allele for rs2453556, the observed suggestive associations on chromosome 3 and 19 (Supplementary Table 7) are not supported by evidence of an interaction (Supplementary Table 5). Finally, in the case-only genome-wide association analysis to test for any potential interaction between SNPs across the genome and c9orf72, no loci provided evidence of a significant interaction with c9orf72 as measured by rs2453556 (Supplementary Table 8, Supplementary Fig. 7). The chromosome 21q22 locus exhibited modest evidence of an interaction (rs2838568, p = 1.5 × 10−5). Thus, no loci provides genome-wide evidence of an interaction with C9orf72 but the potential interaction between the c9orf72 and chromosome 21q22 loci merits further evaluation in independent cohorts.
3.3. Age of ALS onset analysis
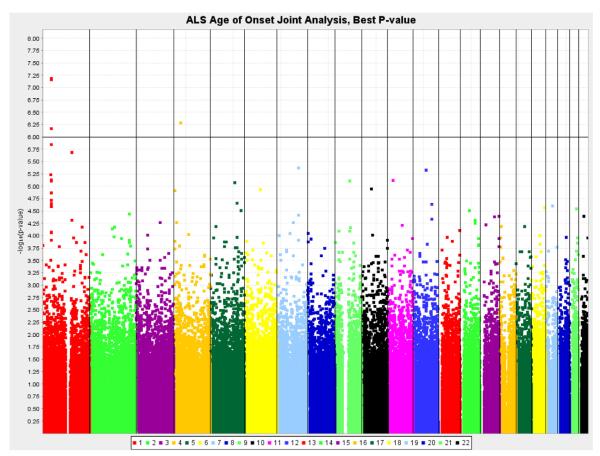
Six genomic regions were associated with age at onset of ALS with ps below 5 × 10−6 (Table 3, Fig. 2). The strongest evidence was observed at 1p34.1, with comparable evidence at rs3011225 (R2partial = 0.0061; p = 6.59 × 10−8) and rs803675 (R2partial = 0.0060; p = 6.96 × 10−8). Fig. 3 shows the consistency of the association across all cohorts under a dominant genetic model. For rs3011225, individuals with at least 1 copy of the minor allele had an earlier average age of onset of over 2 years. This difference is nearly equivalent to the median survival time once diagnosed with ALS. This pattern of association and genetic model was evident at multiple 1p34 loci due to linkage disequilibrium (Fig. 4, Supplementary Fig. 8). In addition to 1p34 region, chromosomal regions 1q22 (rs2364403, p = 2.06 × 10−6) and 12q15 (rs2904524, p = 4.65 × 10−6) show evidence of association. The strength of the association at 4p14 and 8p21 differed between the joint and the meta-analysis, because the recessive genetic model was not computed in some cohorts with a small number of individuals homozygous for the minor allele.
Table 3.
Age of onset outcome adjusted for admixture (first 2 principal components) and center
| SNP | Chr | Position (Mb) |
Gene | Minor allele |
MAF | Joint p value | Meta p value | Genotypic mean ± SE |
||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | Aa | aa | ||||||||
| rs2819332 | 1p34.2 | 43.778 | PTPRF | A | 0.23 | 5.87E-06 (d) | 4.91E-06 (d) | 58.22 ± 0.25 | 56.31 ± 0.33 | 57.21 ± 0.80 |
| rs304303 | 1p34.1 | 43.951 | ST3GAL3 | A | 0.27 | 1.41E-06 (a) | 2.90E-06 (a) | 58.30 ± 0.26 | 56.78 ± 0.31 | 55.37 ± 0.71 |
| rs3011225 | 1p34.1 | 44.092 | ST3GAL3 | G | 0.22 | 6.59E-08 (d) | 1.65E-07 (d) | 58.33 ± 0.25 | 56.16 ± 0.33 | 56.33 ± 0.85 |
| rs2906457 | 1p34.1 | 44.111 | ST3GAL3 | T | 0.27 | 6.86E-07 (a) | 1.08E-06 (a) | 58.36 ± 0.26 | 56.57 ± 0.31 | 55.76 ± 0.70 |
| rs803675 | 1p34.1 | 44.126 | ST3GAL3 | C | 0.21 | 6.96E-08 (d) | 1.46E-07 (d) | 58.29 ± 0.24 | 56.08 ± 0.34 | 56.49 ± 0.89 |
| rs2364403 | 1q22 | 154.194 | ARHGEF2 | A | 0.20 | 2.06E-06 (d) | 1.28E-06 (d) | 58.19 ± 0.24 | 56.16 ± 0.34 | 57.18 ± 0.99 |
| rs12651329 | 4p14 | 35.756 | ARAP2 | C | 0.39 | 5.17E-07 (r) | 1.66E-05 (r) | 57.77 ± 0.32 | 58.02 ± 0.28 | 55.21 ± 0.49 |
| rs6956741 | 7q31.1 | 111.882 | IFRD1 | G | 0.06 | 4.27E-06 (a) | 2.43E-06 (d) | 57.18 ± 0.21 | 59.66 ± 0.58 | 64.31 ± 2.88 |
| rs10503672 | 8p21.3 | 20.032 | SLC18A1a | T | 0.26 | 9.92E-04 (r) | 2.75E-06 (r) | 57.68 ± 0.26 | 57.64 ± 0.31 | 55.11 ± 0.75 |
| rs2904524 | 12q15 | 68.976 | CNOT2 | T | 0.13 | 4.65E-06 (a) | 1.20E-05 (d) | 58.01 ± 0.22 | 55.89 ± 0.40 | 55.83 ± 1.43 |
SNPs are listed with p < 5 × 10−6 in either the joint analysis or meta-analysis. For joint analysis, the additive model is presented unless the test for lack of fit to an additive model was significant (p < 0.05); (a) additive, (d) dominant, (r) recessive. For meta-analysis, the minimum p value of the 3 genetic models is used, noting that quality control standards were applied to ensure only appropriate samples sizes were included in the meta-analysis for each genetic model. Genotypic means and standard errors (SE) are based on the residuals from a linear regression of age of onset on the first 2 principal components and center. Positions are from build 36.3.
Key: Chr, chromosome; kb, kilobases; MAF, minor allele frequency; Mb, megabase; SNP, single nucleotide polymorphism.
Less than 20 kb away.
Fig. 2.
Results of the genome-wide association analysis for age of amyotrophic lateral sclerosis (ALS) onset (joint analysis).
Fig. 3.
Chromosome 1 top hit from age of onset, joint analysis.
Fig. 4.
Age of onset chromosome 1p34.1 region (dominant model p values).
The 10 candidate loci previously reported for association with ALS were examined for association with age of onset (Supplementary Table 9). Only rs12608932 in UNC13A (19p13, R2partial = 0.002, p = 0.004) remained significant after adjusting for multiple comparisons. Three other SNPs provided suggestive evidence of association (p = 0.05). In this sample of 4243 ALS cases, the power to detect 0.1%, 0.2%, and 0.3% of the variation in age of onset is 0.54, 0.83, and 0.95, respectively. Thus, this sample has sufficient power to detect even small variation in age of onset.
The stepwise modeling provided evidence of association with age at onset at 8 loci across the genome (p < 1.0 × 10−5; Table 4). All of the SNPs identified in the multilocus modeling were among the top 200 SNPs in the age of ALS onset single SNP association analysis. Four of the 8 loci showed stronger evidence of association after adjusting for the other loci (rs2364403, rs4147719, rs251018, and rs2904524); 3 loci had comparable evidence of association (rs3011225, rs6956741, and rs7047865) and 1 had reduced evidence (rs12651329). After adjusting for the principal components and center, these 8 loci explain 5% of the variation in age of ALS onset (partial R2 = 0.05). However, no 1 locus dominates the other as after adjusting for principal components, center, and the other loci; each locus individually explains approximately 0.5% of the variation (partial R2 range, 0.0042–0.0073).
Table 4.
Stepwise linear regression starting with top 200 SNPs from joint age of onset analysis, adjusted for admixture (first 2 principal components) and center
| SNP | Chr | Position (Mb) | Minor allele | MAF (cases) | Single locus p valuea |
Stepwise p value |
Genotypic mean ± SD |
Partial R2 |
||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | Aa | aa | ||||||||
| rs3011225 | 1p34.1 | 44.092 | G | 0.22 | 6.59E-08 (d) | 8.39E-08 | 58.33 ± 0.2 | 56.16 ± 0.3 | 56.33 ± 0.9 | 0.0073 |
| rs2364403 | 1q22 | 154.194 | A | 0.20 | 2.06E-06 (d) | 2.83E-07 | 58.19 ± 0.2 | 56.16 ± 0.3 | 57.18 ± 1.0 | 0.0056 |
| rs4147719 | 2q33.3 | 206.715 | G | 0.31 | 3.68E-05 (a) | 5.74E-06 | 56.66 ± 0.3 | 58.09 ± 0.3 | 58.85 ± 0.6 | 0.0042 |
| rs12651329 | 4p14 | 35.756 | C | 0.39 | 5.17E-07 (r) | 4.36E-06 | 57.77 ± 0.3 | 58.02 ± 0.3 | 55.21 ± 0.5 | 0.0063 |
| rs251018 | 5q31.3 | 140.882 | C | 0.47 | 2.18E-05 (a) | 2.88E-06 | 56.27 ± 0.4 | 57.74 ± 0.3 | 58.54 ± 0.4 | 0.0045 |
| rs6956741 | 7q31.1 | 111.882 | G | 0.06 | 4.27E-06 (a) | 6.24E-06 | 57.18 ± 0.2 | 59.66 ± 0.6 | 64.31 ± 2.9 | 0.0053 |
| rs7047865 | 9q21.13 | 77.856 | G | 0.41 | 7.78E-06 (a) | 3.26E-06 | 58.42 ± 0.3 | 57.39 ± 0.3 | 55.87 ± 0.5 | 0.0050 |
| rs2904524 | 12q15 | 68.976 | T | 0.13 | 4.65E-06 (a) | 3.43E-07 | 58.01 ± 0.2 | 55.89 ± 0.4 | 55.83 ± 1.4 | 0.0052 |
Partial R2 for all SNPs in table = 0.0450; partial R2 is adjusted for admixture (first 2 principal components) and center. Model entry and exit criteria: 1 × 10−5.
Key: Chr, chromosome; MAF, minor allele frequency; Mb, ; SNP, single nucleotide polymorphism.
Single locus p-value reflects statistical significance of the SNP in the joint analysis.
Supplementary Table 10 reports the top 2000 statistical associations for age of ALS onset from the joint and meta-analyses.
4. Discussion
In this study, the ALSGEN Consortium performed a large meta-analysis and joint-analysis of 4252 ALS cases and 5123 controls in an attempt to find genetic variants altering susceptibility to disease and age of ALS onset. The cases and controls were drawn from 7 populations of European descent that were genotyped as part of 7 different GWAS of ALS (Chiò et al., 2009; Cronin et al., 2008; Landers et al., 2009; Schymick et al., 2007; Traynor et al., 2010; van Es et al., 2007, 2008, 2009).
The most novel finding in this study is the identification of SNPs on chromosome 1p34.1 that significantly influence age of symptom onset among patients. This finding is important for a number of reasons. First, although it appears that the clinical syndrome of ALS is inherently heterogeneous, it remains possible that these diverse initiating events converge on a single or small number of final common pathways that result in motor neuron degeneration. The finding of a locus that influences age at symptom onset may represent a step toward identification of these final common pathways. Identifying such a final common pathway would be particularly important for therapeutic development as ameliorating therapeutic interventions aimed at this pathway would potentially benefit all patients, and not just the smaller subset that carry that mutations or risk variants in a particular gene.
Second, although a delay of symptom onset of 2.5 years is not large in absolute terms, it is highly significant in the context of ALS where it is comparable to median survival time once diagnosed with ALS (Chiò et al., 2009). By comparison, a chemotherapeutic agent that had an effect comparable to median survival time for a terminal cancer patient would be considered a major therapeutic advance. The specific functional variants or even the most plausible gene causing the earlier age of onset have not been explicitly identified in this analysis. Further, the biological pathways these genes influence have not been identified. Thus, it is possible that pharmacological manipulation of the biological pathways ultimately identified from these discoveries may translate into an even greater delay in the onset of ALS. Despite this potential, much remains unknown about this chromosome 1p locus. Although the pattern of association is highly consistent in magnitude in each of the 13 cohorts, additional work is needed to localize the effect because the region spans nearly 750 kb and contains multiple genes of unclear functional significance to age of ALS onset. It is interesting to note that none of these SNPs appear to alter expression of neighboring genes based on a previously published mapping of expression quantitative trait loci in 4 regions of the brain (Gibbs et al., 2010). In addition, it is possible that these particular variants exert their effect uniquely within the spinal cord, rather than in the brain.
The 2 most statistically significant associations for developing ALS identified in this study were on 9p21 and 19p13.3. Both of these loci had been previously identified in a subset of the samples reported here. The analyses of samples not part of those previous publications (715 cases and 717 controls) were underpowered (power approximately 0.20) and were not able to replicate the associations. However, the genetic lesion underlying the chromosome 9p21 locus has recently been identified (DeJesus-Hernandez et al., 2011; Renton et al., 2011).
In contrast to chromosome 9p21 and 19p13.3, other previously described risk variants such as DPP6 (Cronin et al., 2008; van Es et al., 2009), ITPR2 (van Es et al., 2007, 2008), FGGY (Dunckley et al., 2007), and SUNC1 (Chiò et al., 2008) were not significantly associated with risk of ALS in this meta-analysis of sporadic ALS GWASs. Their lack of association highlights the importance of well-powered, independent replication studies to establish an association with disease.
Taken together, the results of our meta-analysis and previously published GWAS point toward considerable genetic heterogeneity existing within the ALS clinical phenotype (Chiò et al., 2009). ALS is increasingly recognized not as a single disease entity, but rather as a group of diseases unified by their primary effect on motor neurons. The existence of such heterogeneity is supported by the observation of multiple distinct genetic causes of familial ALS. The effect of this clinical and genetic heterogeneity is to significantly diminish the power of association studies and suggests that a combination of complementary approaches (e.g., biological pathways, expression quantitative trait locus [eQTL] analysis) and increased sample sizes are required to identify causative loci.
Our study has several limitations. First and foremost, though the cohort size consisted of several thousand cases and controls, experience from other neurodegenerative diseases shows that even larger sample sizes are required to reliably detect loci (Seshadri et al., 2010). Fortunately the falling cost coupled with increased coverage of the genome makes such large-scale projects both feasible and likely more productive. Second, there is considerable overlap between the samples reported in our current study and previous reports. The analyses only of samples that are independent of the previous reports are underpowered for the ALS susceptibility effect sizes. Third, our approach is an agnostic scan of modest coverage. Such an approach can be complimented by more informed methods with targeted content, such as disease-related chip platforms developed for immune and metabolic disorders or by exome sequencing. The current GWAS technology is primarily powered for common variants and should be complemented by emerging exome or whole genome sequencing technologies. Fourth, the current study only considered SNP variation and not other sources of structural variation or epigenetic variation that may be critically important for ALS.
The results reported here are based on the subset of SNPs that passed stringent quality control in all cohorts. Interestingly, association analyses based on imputation of the genotype data to HapMap3 SNPs using the program IMPUTE did not provide any novel associations (data not shown).
In summary, this meta-analysis has identified a novel locus on chromosome 1p that may influence age at symptom onset. Although much work remains to unravel the mechanism or pathway involved in the chromosome 1 locus, this may represent the first identification of a component of the final common pathway of motor neuron degeneration.
4.1. Electronic resources
Package: SNPLash; Authors: Richard T. Guy, Matt L. Stiegert, Joshua D. Grab, and Carl D. Langefeld. URL: www.phs.wfubmc.edu/public/bios/gene/downloads.cfm. Table 4.
Supplementary Material
Acknowledgements
The ALSGEN consortium and related work was supported by the ALS Association. Analysis and computing resources were provided by the Wake Forest School of Medicine Center for Public Health Genomics. Additional funding included: the Packard Center for ALS Research atHopkins, and Microsoft Research, the Intramural Research Program of the NIA (Z01-AG000949-02) and NINDS; the European Community’s Health Seven Framework Programme (FP7/2007-2013) under grant agreement number 259867 (EUROMOTOR); National Institute for Health Research (NIHR) Dementia Biomedical Research Unit at South London; Maudsley NHS Foundation Trust; King’s College, London; Medical Research Council; Motor Neuron Disease Association (UK); American ALS Association; the Heaton-Ellis Trust; the Psychiatry Research Trust; the Muscular Dystrophy Association (United States); The Irish Motor Neurone Disease Research Foundation; the Health Research Board of Ireland; the Irish Institute of Clinical Neuroscience Travel Award, the Les Turner ALS Foundation; National Institute of Neurological Disorders and Stroke (Grants NS050641, ES014469); ALS Therapy Alliance (CVS pharmacies); the Vena E. Schaff ALS Research Fund; the Harold Post Research Professorship; the Herbert and Florence C. Wenske Foundation; the David C. Asselin, M.D. Memorial Fund; the Help America Foundation; Ride for Life; Federazione Italiana Giuoco Calcio, Fondazione Vialli e Mauro per la Sclerosi Laterale Amiotrofica onlus, Ministero della Salute (Ricerca Sanitaria Finalizzata, 2007), Regione Piemonte (Progetti Finalizzati 2003 and 2004); the Myasthenia Gravis Foundation, FIGC, and AriSLA; the Rudolf Magnus Institute Young Talent Fellowship; the Swedish Brain Power; the Bertil Hållsten Brain Research Foundation; and the Swedish Science Council.
All protocols and data were approved by their respective institutional review boards or study equivalent. Additional details are provided in the Supplemental Table on authors.
Footnotes
Disclosure statement There were no actual or potential conflicts declared for the following authors (see funding sources in the acknowledgements section): Kreshnik B. Ahmeti, Peter M. Andersen, Jennifer Armstrong, Anne Birve, Hylke M. Blauw, Robert H. Brown, Lucie Bruijn, Wenjie Chen, Shawn E. Christopher, Mary C. Comeau, Simon Cronin, Frank P. Diekstra, Athina Soraya Gkazi, Jonathan D. Glass, Josh D. Grab, Ewout J. Groen, Jonathan L. Haines, Orla Hardiman, Scott Heller, Jie Huang, Wu-Yen Hung, James M. Jaworski, Ashley Jones, Humanira Khan, John E. Landers, Carl D. Langefeld, P. Nigel Leigh, Miranda C. Marion, Judith Melki, Russell L. McLaughlin, Vincent Meininger, Jack W. Miller, Gabriele Mora, Margaret A. Pericak-Vance, Evadnie Rampersaud, Wim Robberecht, Laurie P. Russell, Francios Salachas, Christiaan G. Saris, Aleksey Shatunov, Christopher E. Shaw, Nailah Siddique, Teepu Siddique, Bradley N. Smith, Simon Topp, Caroline Vance, Philip van Damme, Leonard H. van den Berg, Michael A. van Es, Paul W. van Vught, Jan H. Veldink, Yi Yang, and JG Zheng.
In addition the authors make the following disclosures. Lucie Bruijn is Chief Scientist and Vice President of Research for ALS Association (www.alsa.org), which provided partial funding for this work. Ammar Al-Chalabi is a consultant for Cytokinetics Inc and Biogen Idec. He receives royalties for the books Genetics of Complex Human Diseases and The Brain, and has a visiting position at Cold Spring Harbor Laboratory where he receives an honorarium for running a course on genetics of complex human diseases. Senda Ajroud-Driss received compensation from Pfizer for being on their advisory board. Robert Sufit has no conflicts of interest regarding this manuscript. He is on a DSMB for Pfizer for cancer pain treated with tanuzemab and on a speakers bureau for Avanir for Pseudobulbar Affect treated with Nuedexta ™. Adraino Chio is a consultant for Cytokinetics Inc and Biogen Idec. Bryan J. Traynor has a patent pending on the discovery of the hexanucleotide repeat expansion of the C9orf72 gene.
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.neurobiolaging.2012.07.017.
References
- Al-Chalabi A, Fang F, Hanby MF, Leigh PN, Shaw CE, Ye W, Rijsdijk F. An estimate of amyotrophic lateral sclerosis heritability using twin data. J. Neurol. Neurosurg. Psychiatry. 2010;81:1324–1326. doi: 10.1136/jnnp.2010.207464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A, Lewis CM. Modelling the effects of penetrance and family size on rates of sporadic and familial disease. Hum. Hered. 2011;71:281–288. doi: 10.1159/000330167. [DOI] [PubMed] [Google Scholar]
- Armon C. Smoking may be considered an established risk factor for sporadic ALS. Neurology. 2009;73:1693–1698. doi: 10.1212/WNL.0b013e3181c1df48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Schymick JC, Restagno G, Scholz SW, Lombardo F, Lai SL, Mora G, Fung HC, Britton A, Arepalli S, Gibbs JR, Nalls M, Berger S, Kwee LC, Oddone EZ, Ding J, Crews C, Rafferty I, Washecka N, Hernandez D, Ferrucci L, Bandinelli S, Guralnik J, Macciardi F, Torri F, Lupoli S, Chanock SJ, Thomas G, Hunter DJ, Gieger C, Wichmann HE, Calvo A, Mutani R, Battistini S, Giannini F, Caponnetto C, Mancardi GL, La Bella V, Valentino F, Monsurrò MR, Tedeschi G, Marinou K, Sabatelli M, Conte A, Mandrioli J, Sola P, Salvi F, Bartolomei I, Siciliano G, Carlesi C, Orrell RW, Talbot K, Simmons Z, Connor J, Pioro EP, Dunkley T, Stephan DA, Kasperaviciute D, Fisher EM, Jabonka S, Sendtner M, Beck M, Bruijn L, Rothstein J, Schmidt S, Singleton A, Hardy J, Traynor BJ. A two-stage genome-wide association study of sporadic amyotrophic lateral sclerosis. Hum. Mol. Genet. 2009;18:1524–1532. doi: 10.1093/hmg/ddp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Traynor BJ, Lombardo F, Fimognari M, Calvo A, Ghiglione P, Mutani R, Restagno G. Prevalence of SOD1 mutations in the Italian ALS population. Neurology. 2008;70:533–537. doi: 10.1212/01.wnl.0000299187.90432.3f. [DOI] [PubMed] [Google Scholar]
- Cronin S, Berger S, Ding J, Schymick JC, Washecka N, Hernandez DG, Greenway MJ, Bradley DG, Traynor BJ, Hardiman O. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum. Mol. Genet. 2008;1:768–774. doi: 10.1093/hmg/ddm361. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Huentelman MJ, Craig DW, Pearson JV, Szelinger S, Joshipura K, Halperin RF, Stamper C, Jensen KR, Letizia D, Hesterlee SE, Pestronk A, Levine T, Bertorini T, Graves MC, Mozaffar T, Jackson CE, Bosch P, McVey A, Dick A, Barohn R, Lomen-Hoerth C, Rosenfeld J, O’Connor DT, Zhang K, Crook R, Ryberg H, Hutton M, Katz J, Simpson EP, Mitsumoto H, Bowser R, Miller RG, Appel SH, Stephan DA. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N Engl J. Med. 2007;23:822–823. doi: 10.1056/NEJMoa070174. [DOI] [PubMed] [Google Scholar]
- Fallis BA, Hardiman O. Aggregation of neurodegenerative disease in ALS kindreds. Amyotroph. Lateral Scler. 2009;10:95–98. doi: 10.1080/17482960802209664. [DOI] [PubMed] [Google Scholar]
- Fang F, Kamel F, Lichtenstein P, Bellocco R, Sparén P, Sandler DP, Ye W. Familial aggregation of amyotrophic lateral sclerosis. Ann. Neurol. 2009;66:94–99. doi: 10.1002/ana.21580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogh I, Rijsdijk F, Andersen PM, Sham PC, Knight J, Neale B, McKenna-Yasek D, Silani V, Brown RH, Jr., Powell JF, Al-Chalabi A. Age at onset in sod1-mediated amyotrophic lateral sclerosis shows familiality. Neurogenetics. 2007;8:235–236. doi: 10.1007/s10048-007-0092-2. [DOI] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, Arepalli S, Dillman A, Rafferty IP, Troncoso J, Johnson R, Zielke HR, Ferrucci L, Longo DL, Cookson MR, Singleton AB. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros-Louis F, Andersen PM, Dupre N, Urushitani M, Dion P, Souchon F, D’Amour M, Camu W, Meininger V, Bouchard JP, Rouleau GA, Julien JP. Chromogranin B P413L variant as risk factor and modifier of disease onset for amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U. S. A. 2009;106:21777–21782. doi: 10.1073/pnas.0902174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanby MF, Scott KM, Scotton W, Wijesekera L, Mole T, Ellis CE, Leigh PN, Shaw CE, Al-Chalabi A. The risk to relatives of patients with sporadic amyotrophic lateral sclerosis. Brain. 2011;134:3454–3457. doi: 10.1093/brain/awr248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirtz D, Thurman DJ, Gwinn-Hardy K, Mohamed M, Chaudhuri AR, Zalutsky R. How common are the “common” neurologic disorders? Neurology. 2007;68:326–337. doi: 10.1212/01.wnl.0000252807.38124.a3. [DOI] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurrò MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G, ITALSGEN Consortium. Galassi G, Scholz SW, Taylor JP, Restagno G, Chiò A, Traynor BJ. Exome sequencing reveals VCP mutations as a cause of familial ALS [erratum in: 2011;69:397.] Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Stanton BR, Turner MR, Gray R, Blunt AH, Butt D, Ampong MA, Shaw CE, Leigh PN, Al-Chalabi A. Amyotrophic lateral sclerosis in an urban setting: a population based study of inner city London. J. Neurol. 2006;253:1642–1643. doi: 10.1007/s00415-006-0195-y. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr., Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Laaksovirta H, Peuralinna T, Schymick JC, Scholz SW, Lai SL, Myllykangas L, Sulkava R, Jansson L, Hernandez DG, Gibbs JR, Nalls MA, Heckerman D, Tienari PJ, Traynor BJ. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol. 2010;9:978–985. doi: 10.1016/S1474-4422(10)70184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers JE, Melki J, Meininger V, Glass JD, van den Berg LH, van Es MA, Sapp PC, van Vught PW, McKenna-Yasek DM, Blauw HM, Cho TJ, Polak M, Shi L, Wills AM, Broom WJ, Ticozzi N, Silani V, Ozoguz A, Rodriguez-Leyva I, Veldink JH, Ivinson AJ, Saris CG, Hosler BA, Barnes-Nessa A, Couture N, Wokke JH, Kwiatkowski TJ, Jr., Ophoff RA, Cronin S, Hardiman O, Diekstra FP, Leigh PN, Shaw CE, Simpson CL, Hansen VK, Powell JF, Corcia P, Salachas F, Heath S, Galan P, Georges F, Horvitz HR, Lathrop M, Purcell S, Al-Chalabi A, Brown RH., Jr. Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9004–9009. doi: 10.1073/pnas.0812937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YJ, Pericak-Vance MA, Haines JL, Siddique N, McKenna-Yasek D, Hung WY, Sapp P, Allen CI, Chen W, Hosler B, Saunders AM, Dellefave LM, Brown RH, Siddique T. Apolipoprotein E is associated with age at onset of amyotrophic lateral sclerosis. Neurogenetics. 2004;5:209–213. doi: 10.1007/s10048-004-0193-0. [DOI] [PubMed] [Google Scholar]
- Lill CM, Abel O, Bertram L, Al-Chalabi A. Keeping up with genetic discoveries in amyotrophic lateral sclerosis: the ALSoD and ALSGene databases. Amyotroph. Lateral Scler. 2011;12:238–249. doi: 10.3109/17482968.2011.584629. [DOI] [PubMed] [Google Scholar]
- Logroscino G, Beghi E, Zoccolella S, Palagano R, Fraddosio A, Simone IL, Lamberti P, Lepore V, Serlenga L, SLAP Registry Incidence of amyotrophic lateral sclerosis in southern Italy: a population based study. J. Neurol. Neurosurg. Psychiatry. 2005;76:1094–1098. doi: 10.1136/jnnp.2004.039180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logroscino G, Traynor BJ, Hardiman O, Chio A, Couratier P, Mitchell JD, Swingler RJ, Beghi E, EURALS Descriptive epidemiology of amyotrophic lateral sclerosis: new evidence and unsolved issues. J. Neurol. Neurosurg. Psychiatry. 2008;79:6–11. doi: 10.1136/jnnp.2006.104828. [DOI] [PubMed] [Google Scholar]
- Manjaly ZR, Scott KM, Abhinav K, Wijesekera L, Ganesalingam J, Goldstein LH, Janssen A, Dougherty A, Willey E, Stanton BR, Turner MR, Ampong MA, Sakel M, Orrell RW, Howard R, Shaw CE, Leigh PN, Al-Chalabi A. The sex ratio in amyotrophic lateral sclerosis: A population based study. Amyotroph. Lateral Scler. 2010;11:439–442. doi: 10.3109/17482961003610853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, Suzuki N, Aoki M, Kawata A, Hirai T, Kato T, Ogasawara K, Hirano A, Takumi T, Kusaka H, Hagiwara K, Kaji R, Kawakami H. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- Mitchell J, Morris A, de Belleroche J. Thioredoxin reductase 1 haplotypes modify familial amyotrophic lateral sclerosis onset. Free Radic. Biol. Med. 2009;46:202–211. doi: 10.1016/j.freeradbiomed.2008.09.041. [DOI] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, Skehel P, Zatz M. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosthuyse B, Moons L, Storkebaum E, Beck H, Nuyens D, Brusselmans K, Van Dorpe J, Hellings P, Gorselink M, Heymans S, Theilmeier G, Dewerchin M, Laudenbach V, Vermylen P, Raat H, Acker T, Vleminckx V, Van Den Bosch L, Cashman N, Fujisawa H, Drost MR, Sciot R, Bruyninckx F, Hicklin DJ, Ince C, Gressens P, Lupu F, Plate KH, Robberecht W, Herbert JM, Collen D, Carmeliet P. Deletion of the hypoxia-response element in the vascular endothelial growth factor promoter causes motor neuron degeneration. Nat. Genet. 2001;28:131–138. doi: 10.1038/88842. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M, ITALSGEN Consortium. Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Bowling AC, Patterson D, Usdin TB, Sapp P, Mezey E, McKenna-Yasek D, O’Regan J, Rahmani Z, Ferrante RJ, et al. A frequent ala 4 to val superoxide dismutase-1 mutation is associated with a rapidly progressive familial amyotrophic lateral sclerosis. Hum. Mol. Genet. 1994;3:981–987. doi: 10.1093/hmg/3.6.981. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van den Bergh R, Hung W, Bird T, Deng G, Mulder DW, Smyth C, Laing NG, Soriano E, Pericak-Vance MA, Haines J, Rouleau GA, Gusella JS, Horvitz HR, Brown RH., Jr. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;4:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Schymick JC, Scholz SW, Fung HC, Britton A, Arepalli S, Gibbs JR, Lombardo F, Matarin M, Kasperaviciute D, Hernandez DG, Crews C, Bruijn L, Rothstein J, Mora G, Restagno G, Chiò A, Singleton A, Hardy J, Traynor BJ. Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2007;6:322–328. doi: 10.1016/S1474-4422(07)70037-6. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EM, Ramirez-Lorca R, Debette S, Longstreth WT, Jr., Janssens AC, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MM, CHARGE Consortium. GERAD1 Consortium. EADI1 Consortium Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatunov A, Mok K, Newhouse S, Weale ME, Smith B, Vance C, Johnson L, Veldink JH, van Es MA, van den Berg LH, Robberecht W, Van Damme P, Hardiman O, Farmer AE, Lewis CM, Butler AW, Abel O, Andersen PM, Fogh I, Silani V, Chiò A, Traynor BJ, Melki J, Meininger V, Landers JE, McGuffin P, Glass JD, Pall H, Leigh PN, Hardy J, Brown RH, Jr., Powell JF, Orrell RW, Morrison KE, Shaw PJ, Shaw CE, Al-Chalabi A. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol. 2010;9:986–994. doi: 10.1016/S1474-4422(10)70197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique T, Pericak-Vance MA, Caliendo J, Hong ST, Hung WY, Kaplan J, McKenna-Yasek D, Rimmler JB, Sapp P, Saunders AM, Scott WK, Siddique N, Haines JL, Brown RH. Lack of association between apolipoprotein E genotype and sporadic amyotrophic lateral sclerosis. Neurogenetics. 1998;1:213–216. doi: 10.1007/s100480050031. [DOI] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynor BJ, Codd MB, Corr B, Forde C, Frost E, Hardiman O. Incidence and prevalence of ALS in Ireland, 1995–1997: a population-based study. Neurology. 1999;52:504–509. doi: 10.1212/wnl.52.3.504. [DOI] [PubMed] [Google Scholar]
- Traynor BJ, Nalls M, Lai SL, Gibbs RJ, Schymick JC, Arepalli S, Hernandez D, van der Brug MP, Johnson JO, Dillman A, Cookson M, Moglia C, Calvo A, Restagno G, Mora G, Chiò A. Kinesin-associated protein 3 (KIFAP3) has no effect on survival in a population-based cohort of ALS patients. Proc. Natl. Acad. Sci. U. S. A. 2010;6:12335–12338. doi: 10.1073/pnas.0914079107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdmanis PN, Dupre N, Bouchard JP, Camu W, Salachas F, Meininger V, Strong M, Rouleau GA. Three families with amyotrophic lateral sclerosis and frontotemporal dementia with evidence of linkage to chromosome 9p. Arch. Neurol. 2007;64:240–245. doi: 10.1001/archneur.64.2.240. [DOI] [PubMed] [Google Scholar]
- van Es MA, Van Vught PW, Blauw HM, Franke L, Saris CG, Andersen PM, Van Den Bosch L, de Jong SW, van’t Slot R, Birve A, Lemmens R, de Jong V, Baas F, Schelhaas HJ, Sleegers K, Van Broeckhoven C, Wokke JH, Wijmenga C, Robberecht W, Veldink JH, Ophoff RA, van den Berg LH. ITPR2 as a susceptibility gene in sporadic amyotrophic lateral sclerosis: a genome-wide association study. Lancet Neurol. 2007;6:869–877. doi: 10.1016/S1474-4422(07)70222-3. [DOI] [PubMed] [Google Scholar]
- van Es MA, van Vught PW, Blauw HM, Franke L, Saris CG, Van den Bosch L, de Jong SW, de Jong V, Baas F, van’t Slot R, Lemmens R, Schelhaas HJ, Birve A, Sleegers K, Van Broeckhoven C, Schymick JC, Traynor BJ, Wokke JH, Wijmenga C, Robberecht W, Andersen PM, Veldink JH, Ophoff RA, van den Berg LH. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat. Genet. 2008;40:29–31. doi: 10.1038/ng.2007.52. [DOI] [PubMed] [Google Scholar]
- van Es MA, Veldink JH, Saris CG, Blauw HM, van Vught PW, Birve A, Lemmens R, Schelhaas HJ, Groen EJ, Huisman MH, van der Kooi AJ, de Visser M, Dahlberg C, Estrada K, Rivadeneira F, Hofman A, Zwarts MJ, van Doormaal PT, Rujescu D, Strengman E, Giegling I, Muglia P, Tomik B, Slowik A, Uitter-linden AG, Hendrich C, Waibel S, Meyer T, Ludolph AC, Glass JD, Purcell S, Cichon S, Nöthen MM, Wichmann HE, Schreiber S, Vermeulen SH, Kiemeney LA, Wokke JH, Cronin S, McLaughlin RL, Hardiman O, Fumoto K, Pasterkamp RJ, Meininger V, Melki J, Leigh PN, Shaw CE, Landers JE, Al-Chalabi A, Brown RH, Jr., Robberecht W, Andersen PM, Ophoff RA, van den Berg LH. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat. Genet. 2009;41:1083–1087. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisskopf MG, Gallo V, O’Reilly EJ, Vineis P, Ascherio A. Smoking may be considered an established risk factor for sporadic ALS. Neurology. 2010;74:1927–1928. [PubMed] [Google Scholar]
- Worms PM. The epidemiology of motor neuron diseases: a review of recent studies. J. Neurol. Sci. 2001;191:3–9. doi: 10.1016/s0022-510x(01)00630-x. [DOI] [PubMed] [Google Scholar]
- Wroe R, Wai-Ling Butler A, Andersen PM, Powell JF, Al-Chalabi A. ALSOD: the Amyotrophic Lateral Sclerosis Online Database. Amyotroph. Lateral Scler. 2008;9:249–250. doi: 10.1080/17482960802146106. [DOI] [PubMed] [Google Scholar]
- Zoccolella S, Beghi E, Palagano G, Fraddosio A, Samarelli V, Lamberti P, Lepore V, Serlenga L, Logroscino G, SLAP Registry Signs and symptoms at diagnosis of amyotrophic lateral sclerosis: a population-based study in southern Italy. Eur. J. Neurol. 2006;13:789–792. doi: 10.1111/j.1468-1331.2006.01384.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.