Abstract
The CAPRI and CASP prediction experiments have demonstrated the power of community wide tests of methodology in assessing the current state of the art and spurring progress in the very challenging areas of protein docking and structure prediction. We sought to bring the power of community wide experiments to bear on a very challenging protein design problem that provides a complementary but equally fundamental test of current understanding of protein-binding thermodynamics. We have generated a number of designed protein-protein interfaces with very favorable computed binding energies but which do not appear to be formed in experiments, suggesting there may be important physical chemistry missing in the energy calculations. 28 research groups took up the challenge of determining what is missing: we provided structures of 87 designed complexes and 120 naturally occurring complexes and asked participants to identify energetic contributions and/or structural features that distinguish between the two sets. The community found that electrostatics and solvation terms partially distinguish the designs from the natural complexes, largely due to the non-polar character of the designed interactions. Beyond this polarity difference, the community found that the designed binding surfaces were on average structurally less embedded in the designed monomers, suggesting that backbone conformational rigidity at the designed surface is important for realization of the designed function. These results can be used to improve computational design strategies, but there is still much to be learned; for example, one designed complex, which does form in experiments, was classified by all metrics as a non-binder.
Introduction
Protein-protein interactions underlie all biological processes. Despite the availability of many co-crystal structures of complexes, there is still not a complete understanding of the energetics of protein association, and this limits our ability to consistently predict the structures of complexes from monomers, predict the energetic effects of mutations at protein interfaces, and engineer high-affinity and –specificity interactions. An improved understanding of binding energetics therefore holds the key to resolving some of the most important problems in protein biophysics and molecular biology.
A recently developed method for de novo binder design produced two proteins that interacted with a sterically hindered surface on Spanish influenza hemagglutinin (SC1918/H1 HA; hereafter referred to as HA)1. Following in vitro evolution 2-4 mutations in the periphery of each of these interfaces improved binding to low nanomolar dissociation constants and one of the proteins inhibited HA function. However, 71 other designed proteins predicted to bind did not experimentally interact with HA. The Baker group has had similar low success rates with other de novo interface design problems (to be published), highlighting limitations in the understanding of protein-binding energetics and their repercussions for the ability to design novel protein functions. More sensitive experimental detection methods could identify additional binders in this set (the current method requires dissociation constants better than 10μM and binding off-rates less than 10 s-1); but the ability to computationally generate high-affinity interactions is vital for engineering new protein functions.
We asked the protein-docking community to help identify what was missing in our protein-modeling calculations. This paper describes the benchmark tests we established and summarizes the insights from the many interface-modeling experts who took up the challenge.
Results
A protein-interface design benchmark
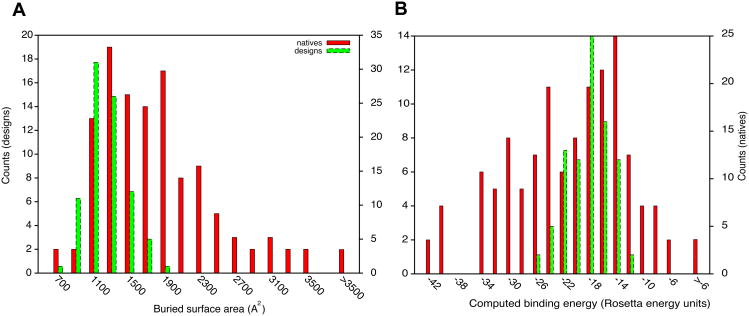
The computational interface design protocol consists of (i) pre-computing a set of high-affinity amino acid residue interactions with the target surface; (ii) redesigning natural protein scaffolds to incorporate a number of these amino acids; and (iii) designing the remainder of the interface to enhance binding affinity1. This protocol can produce protein complexes with computed binding characteristics that rival natural complexes. For instance, the distributions of interface buried-surface areas and computed binding energies of designed and naturally occurring protein complexes overlap (Fig. 1; Table S1). In many cases, designed protein complexes show more favorable values than do natural complexes. This is despite the fact that the vast majority of the designed complexes do not experimentally bind. The discrepancy between prediction and experiment is the focus of this study: our goal is to identify the missing components in binding-energy calculations to improve both our ability to design high affinity interfaces and, more generally, our understanding of protein-association thermodynamics.
Figure 1.
Natural and designed complexes have similar overall properties. (A) buried surface area at the interface; (B) computed binding energy. Computed binding energies are reasonably correlated with experimentally determined dissociation constants (Pearson correlation r=0.53 ref. 15). All plots were produced using gnuplot 4.4 and enhanced with Adobe Illustrator. In all figures, native refers to natural complexes in the docking benchmark8.
We set out to identify thermodynamic components of binding that are poorly modeled and could be the underlying cause of the low success rate of de novo binder design. In a preliminary experiment, a set of 20 designed binders of several targets that did not show detectable binding to their targets were provided to participants in the community-wide experiment on the Critical Assessment of Predicted Interactions (CAPRI)3, alongside one experimentally determined but, at that time, unpublished co-crystal structure of two proteins that bound with a low-nanomolar dissociation constant4. The participants were asked to rank the 21 complexes according to their propensity to bind in the modeled or experimentally determined binding mode. In this preliminary experiment, only two of 28 participating groups (Groups 1 and 6) clearly identified the co-crystal structure as the true binder – performance that is not significantly different from chance at 5% confidence (to be discussed in the next Special Issue on CAPRI). These results suggested that the task of identifying complexes that are likely to bind is non-trivial, and that a larger scale community wide investigation could provide considerable insight into this problem.
To set up a benchmark for a more comprehensive community wide investigation into the elements that are missing in our evaluation of binding thermodynamics, we prepared a set of 87 designed proteins targeting three different proteins of interest (models available as Supplemental Data and plasmids encoding genes for expressing the designs using yeast cell-surface display are available through http://www.addgene.com). The three target proteins were Spanish influenza HA (62% of the designed complexes; chains A and B of Protein Databank (PDB) entry 3GBN6), the acyl-carrier protein 2 from M. tuberculosis (25%; Mt ACP2; PDB entry 2CGQ), and the Fc region of human IgG1 antibodies (13%; PDB entry 1L6X7). The structures of the scaffold proteins for binder design were taken from the PDB and their surfaces were redesigned for binding using the computational method mentioned above1. As a reference set of solved co-crystal structures we used the docking benchmark 3.08 comprising 120 protein complexes with experimentally determined dissociation constants9 ranging from 10-5 to 10-14M. These sets of natural and computationally designed complexes were provided to participants in CAPRI, noting in each case whether a complex was designed or natural. At the beginning of the experiment 9 designed proteins had not been experimentally tested for binding and these served as unmarked blind cases.
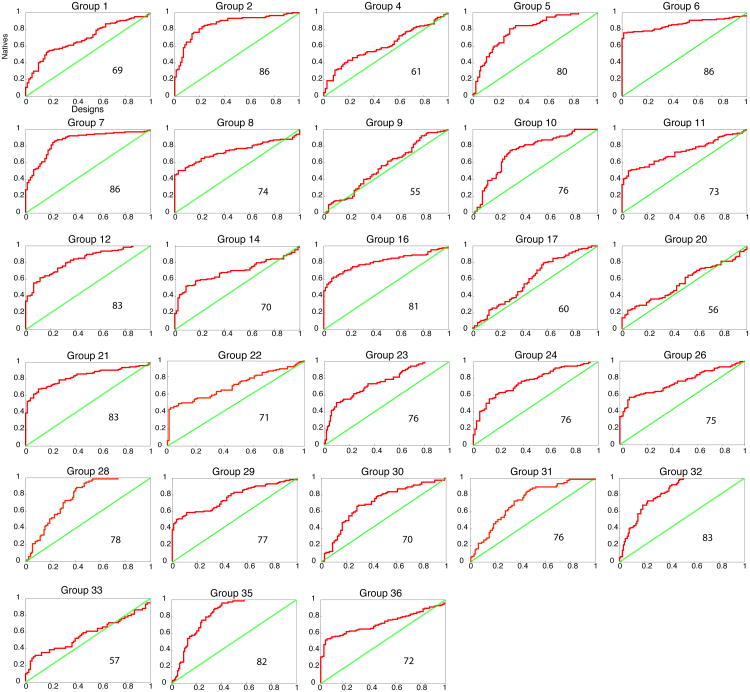
Each participating group (Table 1) was asked to provide a method for ranking the complexes according to their binding energy (all of the values provided by participants are available as Supplemental Data). To get at the underlying physical chemistry of binding, groups were asked not to train their methods on the data, i.e., the information on whether a complex was designed or natural could not be used in training the parameters used in the evaluation strategy. Otherwise, the groups were free to choose which metrics or combinations of metrics to use. Figure 2 shows a Receiver-operator Characteristic (ROC) curve for each participating group, plotting the true-positive rate vs. the false-positive rate. The Area Under the Curve (AUC, in percent units) is marked in each panel. The participating groups were additionally asked to categorize each complex according to the following criteria: the two partners (i) bind, (ii) are likely to bind, (iii) are likely not to bind, (iv) do not bind, and (v) unknown (Figure S1), and were free to choose thresholds to maximize discrimination.
Table 1.
List of participating groups and a brief explanation of the methods. A complete description of each method is provided in the Methods section.
| Group number1 | Affiliation2 | van der Waals packing3 | solvation3 | Pair terms3 | electrostatics3 | others3 | Use of prior knowledge4 |
|---|---|---|---|---|---|---|---|
| 1 | 3 | 1 | Electrostatic interaction free energy, calculated on the transient complex, by solving the Poisson-Boltzmann equation | a | |||
| 2 | 4,5 | NA | NA | NA | NA | SVM | b |
| 5 | 6 | 1 | a | ||||
| 6 | 7 | 0.1 | 0.4 | 0.16 | c | ||
| 7 | 8 | - | - | ATTRACT score of the minimized complex (0.33) in RT units | - | Rank of minimized complex relative to docking solutions from systematic search (0.33); deviation of complex from nearest minimum (0.33) | a |
| 8 | 9 | 0.18 | Sequence conservation score (0.52) | a | |||
| Sidechain entropy (0.13) | |||||||
| 9 | 10-13 | NA | NA | NA | NA | Genetic algorithms | a |
| 10 | 14,15 | DiffColl (1.0) | a | ||||
| The difference in the increase in degree collectivity between chains A and B | |||||||
| 11 | 16 | 0.41 (vdW attractive) | 0.42 | 0.13 - 0.21 (four independent weights for short/long/attractive/repulsive; average is 0.16) | c | ||
| 12 | 17 | -0.5 | 0.5 | a | |||
| 14 | 18 | 0.056 | Sum of weights for 18 terms of DeLisi-Zhang atomic solvation (0.563) | Dfire (0.369) | 0.013 | Sum of linear fitting weights for DeLisi-Zhang atomic solvation (4.101), for pair solvation and hydrogen bond (-1.167), and for many-body graph (-3.712) | a |
| 16 | 19 | a | |||||
| 17 | 20 | - | - | - | - | Relative sequence entropy score comparing the degree of conservation of the interface core versus the rim (1.0) | a |
| 20 | 21 | 0 | 0 | 1 | 0 | 0 | a |
| 21 | 22 | NA | NA | NA | NA | Interface descriptors: polar solvent accessible surface area buried at the interface is smaller in designs | c |
| 22 | 23 | 0.2 | 0 | 0.2 | 0.2 | 0.2 | b |
| 23 | 24-27 | 0.09 | 0.28 | 0.44 | SCRsurf | a | |
| -0.19 | a | ||||||
| 24 | 28,29 | NA | NA | NA | NA | Interface intra- and intermolecular energies scaled to differentiated total energy, and scaled surface area buried at the interface | a |
| 26 | 30 | 0 | 0 | 1 | 0 | 0 | a |
| 28 | 31 | 0.3 | 0.26 (Lazaridis-karplus solvation+Buried surface area) | 0.24 | Hydrogen bonding | a | |
| 0.2 | |||||||
| 29 | 32 | 0.25 | 0.25 | 0.25 | Internal energy | a | |
| 30 | 33 | NA | NA | NA | NA | Interface area per residue of the complex | c |
| 31 | 34 | 0.75 | 0.05 | 0.2 | a | ||
| 32 | 35 | N/A | N/A | N/A | N/A | Frequency and geometric similarity of interaction patterns of interfacial residues to the native (classical) ones. | a |
| 33 | 36 | vdw attractive (0.49) | 0.01 | 0.35 | short range attractive (0.06) and | hydrogen bonding | a |
| short range repulsive (0.07) | |||||||
| 35 | 37 | NA | NA | NA | NA | binding energy (dG) per surface area (PSA) (0.25), hydrogen-bond energy per dG (0.25), cavity score (0.25), unsatisfied hydrogen bonds PSA (0.25) | a |
| 36 | 38 | NA | NA | ITScore/PP | NA | NA | a |
The group number refers to the numbers in the Text and Figures.
The affiliation number in the Author Affiliation section.
Weights on the major score terms used by the discriminators. The weights in the Table are reported after normalizing the sum of all weights used by each group to 1.0.
Extent to which prior knowledge was used: a) none b) score was trained on Rosetta models provided in the past, but not on the design benchmark c) different discrimination models or parameters were tested and the best performing one was selected
Figure 2.
Ability of different methods to discriminate between native and designed complexes. Receiver-operator Characteristic (ROC) curves are shown for each group, with the true- and false-positive classification on the y- and x-axes, respectively. The steeper the ascent of the curve and the larger the Area Under the Curve (AUC) the better the discrimination between natural and designed complexes. The green diagonal represents the expected output of random prediction. Percent AUC is noted within each plot. Groups 2 and 22 trained their metrics, in part, on Rosetta models published in the past, but not on the current set of designs (see Methods for more details).
The methods used by participating groups span a wide spectrum. Many groups computed binding energies, typically dominated by electrostatics, solvation, and knowledge-based pair terms (Groups 1, 5, 6, 11, 12, 14, 20, 23, 26, 28, 29, 31, 33, and 36); Groups 1 and 6 used continuum solvation methods to compute binding energies, similar to widely used MM-PBSA approaches for computing binding afinities10. Others utilized features such as hydrogen-bonding patterns and buried surface area (Groups 16, 21, 23, 24, 30, 32, 35). Groups 2 and 22 used machine learning to determine which features discriminate previously published Rosetta models from natural complexes. Groups 8 and 17 used the low sequence conservation at the designed interface as a discriminator. Group 10 analyzed low-frequency dynamics; and Group 7 tested the low-resolution compatibility of the surfaces compared to randomly docked decoys of the same partners,
Discrimination between the designed interfaces and some, but not all, categories of natural ones
Many different metrics provide useful posteriori discriminators between designed and naturally occurring complexes (Fig. S1), with several groups achieving AUC values above 85% (Fig. 2). However, the ROC curves also point out that even well-performing metrics suffer from poor discrimination between designs and many native complexes. That is, many of the best discriminators rank a large fraction of the natural complexes as better binders than the designed complexes, but still rank many designed and natural ones equally. Consequently, many of the native complexes were predicted as unlikely to bind or as not binding by most groups. These results suggest that the designs share some features with a substantial fraction of the natural complexes but not with all.
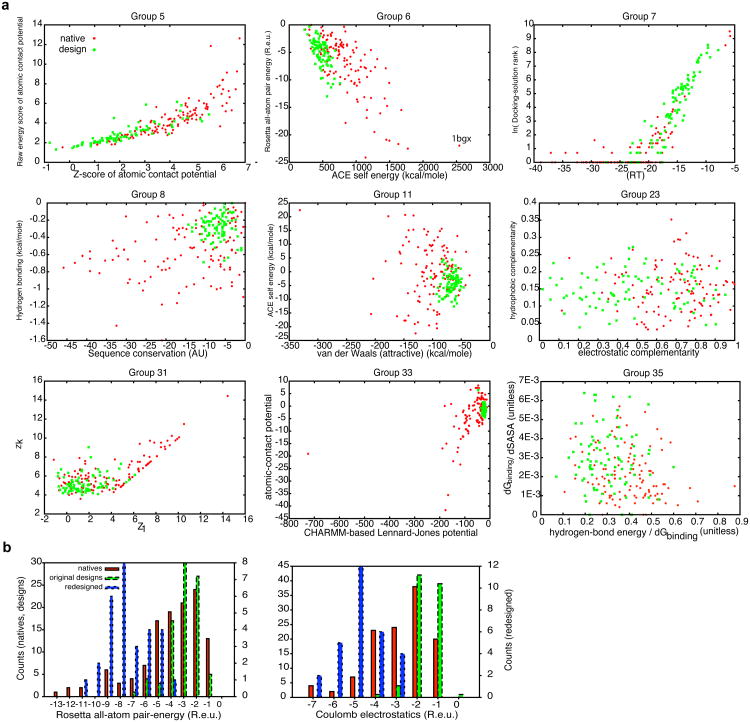
To get a more detailed view of the individual features that contribute most to discrimination, we compared the distributions for designed and natural interfaces of the two most heavily weighted terms given by several participating groups (Fig. 3A). As with the full metrics (Figs 2 & S1), the individual-score values for natural complexes span and exceed the range of designed complexes, and hence no single or indeed pair of scores unambiguously discriminates designed from natural complexes. Nevertheless, the designed complexes typically stand out as having on average less optimal values than a majority of the natural complexes in terms of their van der Waals contacts, solvation self energy, and electrostatic complementarity. To understand the commonalities between designed and natural complexes that were predicted not to bind, we analyzed in detail the results from Group 6, one of the best-performing participants (Fig. 2). We found that those natural interfaces that scored more favorably than designs according to the two-metric analysis (Fig. 3A) were typically larger and comprised many saltbridge or backbone-mediated interactions (see per-group two-metric analysis in Supplemental Data). By contrast, the natural interfaces that were predicted not to bind were smaller, more hydrophobic, and contained few if any charges and paired backbone atoms. The de novo designed interfaces share many of the same features as the latter category of smaller, more hydrophobic interfaces, explaining why many metrics showed natural complexes to span the range of values for the designs but did not clearly discriminate the two groups (Figs. 2 & 3A). Many of these natural hydrophobic protein complexes bind quite strongly, implying that even the best-performing metrics do not fully reflect binding thermodynamics. This is highlighted by the fact that the natural complex best separated from the designs (predicted most strongly to be a binder) was a structure, which after its publication was deemed by several studies to be likely incorrect11, and was recommended for retraction by the University of Alabama (PDB entry: 1BGX12). In retrospect, the bias towards hydrophobic interfaces was a failing of our design benchmark set. We remedied this failing in two ways (below): by adding more polar interfaces to the design set and by contrasting the designs with the most apolar natural interfaces in the docking dataset.
Figure 3.
Individual features that partially discriminate native and designed complexes. (A) Comparison of native and designs using the two most heavily weighted terms in the scoring function for each group. The points represent individual natives or designs, and the axes represent the most heavily weighted scoring terms. The scatter plots provide insight into some of the discriminatory power of the methods. While the phase-plane occupied by designs and natives overlap, in these cases, the designs occupy a small fraction of the plane with many of the natives having more favorable values. The results from Groups 11 and 33 suggest that the van der Waals contacts in designed interfaces are weaker than in natives. Likewise, Groups 6 and 11 suggest that solvation self energy (ACE) and electrostatics (the dominant contribution to Rosetta pair energy) are more optimized in natives. See individual groups' methods for more details. (B) Modification of the design protocol yields distributions of interface pairwise and Coulomb-electrostatic energies similar to those in natural complexes. Natural complexes (natives) and designs generated with (redesigns) and without (designs) an increased pairwise attractive term (weight=0.98) and Coulomb electrostatic interaction with a distance-dependent dielectric (weight=1.0). The distributions were calculated using pairwise attractive term and electrostatic interaction of 0.49 and 0.25, respectively, for all complexes. These designs have many flaws as potential binders, but can serve as decoys with more native-like distributions of electrostatic interactions.
Reducing the polarity discrepancy between natural and designed interfaces identifies methods that discriminate designs based on functional site rigidity
To address the problem of unequal polarities in designed and natural interfaces, we redesigned the set of 87 designed complexes, increasing the contributions from residue pairwise-interaction probabilities and Coulomb electrostatics to the energy function used by RosettaDesign, and selected 29 designs with high buried surface area and computed binding energies. In these redesigned interfaces, the distributions of contributions to binding from electrostatic and pairwise-interaction probabilities are comparable to those of natural interfaces (Fig. 3B). While these new redesigned complexes have many flaws (sidechain packing is not ideal and their interfaces contain many unsatisfied hydrogen-bond donors and acceptors), the addition of interfaces with higher charge complementarity reduces the polarity discrepancy between designed and natural interfaces in our set and makes the benchmark more representative of the physical-chemical diversity of natural interfaces. We have added these new, more polar complexes to the benchmark set (Supplemental Data). The improved benchmark set should provide an even better test of current understanding of binding physical chemistry than the original set.
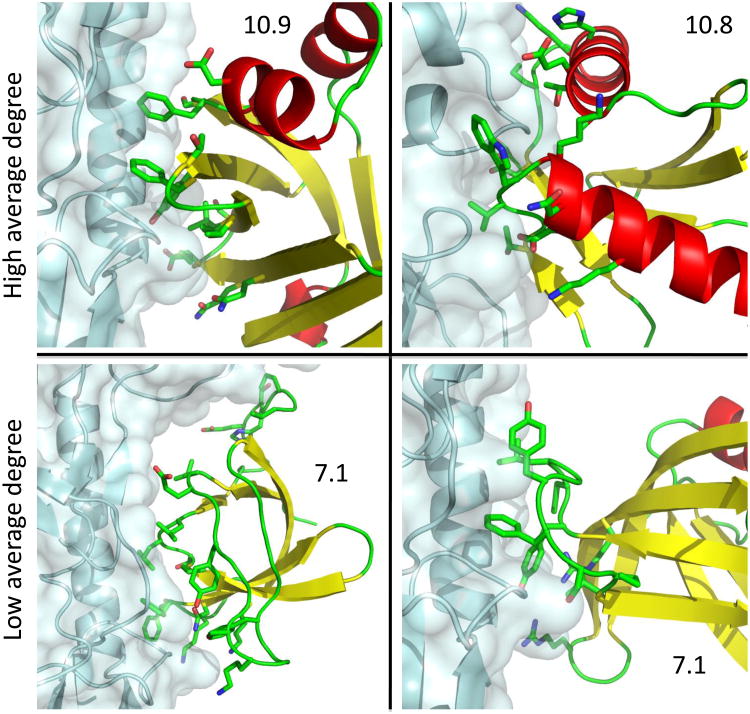
To isolate metrics that discriminate the designs from a set of apolar natural interfaces, we selected 25 natural interfaces with the lowest electrostatic desolvation penalty according to the Rosetta all-atom energy (Table S2). As expected, the AUC of many of the metrics deteriorated in this analysis compared to the results of Figure 2, while a few methods performed as well on this stricter test as in the one shown in Figure 2 (Table S3). Group 7 (AUC=81% in this analysis) used low-resolution docking and favored those complexes where close-to-native conformations had lower interaction energies than far-from-native ones. An analysis of the worst and best-performing designs according to this method showed that it penalized designs with poor low-resolution shape complementarity, and conversely favored designs with intricate ‘knobs-into-holes’ features, which allow more residue-to-residue interactions. Group 10 (AUC=79% in this analysis) used a single feature based on the compatibility of the low-frequency vibrational modes of the partner proteins. Interfaces where the vibrational modes of the two partners were incompatible were penalized. An analysis of the worst-performing designs according to this method showed that it penalized designs where the binding surface was positioned on loops or secondary-structural elements that were poorly embedded in the designed monomer, and conversely favored interfaces that integrated the designed surface through many interactions in the host monomer. Group 10 found that a simpler related metric based on the average degree of connectivity of interfacial residues on the designed monomer (see methods) performed more poorly than the analysis of vibrational modes, but was also discriminatory. Indeed, in following up on the Group 10 results we found that most designed proteins with an average degree of less than 8.5 residue neighbors at the interface (∼15% of designs in the set) utilize loops or secondary structural elements that are poorly anchored to the designed protein and, retrospectively, are unlikely to form the modeled surfaces in experiment (Fig. 4). That such a high fraction of designs employ backbones that are poorly anchored in the designed monomer is unsurprising given that binding to a target surface is typically hindered by other surfaces on the target molecule; designed surfaces that are less embedded in their host monomers suffer less from such hindrance. We have implemented this degree of connectivity metric in the Rosetta software and expect it to improve the likelihood of obtaining active designed binders in future.
Figure 4.
Average number of neighbors (average degree) of interface residues within the designed monomer discriminates some designed complexes from native complexes. Surfaces with low average degree (bottom) tend to comprise segments, including unstructured regions, which are poorly embedded in the host monomer. By contrast, surfaces with high average degree (top) comprise secondary-structural elements and short loops that are better structurally connected to the host monomer. Following sequence design poorly connected surfaces might have altered conformations from those seen in the wild-type protein structure, providing some explanation for the failure of these designs to experimentally bind their targets. Average degree is marked on each panel. Clockwise from top-left, the panels represent designs 47, 59, 78, and 77 (coordinates are available in the online supplement). The target proteins are rendered in cyan. The backbones of the designed monomers are colored according to secondary structure (red – helix; yellow – strand; green – loop). Designed interfacial residues are shown in sticks with carbon, oxygen, and nitrogen, colored in green, red, and blue, respectively. Molecular representations were produced with PyMol20.
Failure to identify an experimentally validated designed binder as such
Of the 87 designed interfaces provided to participants for ranking, 9 designs had not been tested for binding at the start of the experiment and thus serve as a blind test of the ranking methods. Of these 9 one has been experimentally confirmed to bind its HA target surface (herein numbered design 45 or HB80 in ref. 1). In vitro selection of design 45 variants for higher affinity identified four point substitutions at the periphery of the interface that together produced an experimentally determined dissociation constant of 38nM, rivaling many of the affinities in the docking benchmark of naturally occurring binders8. Despite this high affinity, none of the groups predicted that design 45 binds, and a majority predicted it is unlikely to bind or that it would not bind (Fig. S2). Design 45 has a small nonpolar interface, which as noted above confounds discrimination of binders from non binders by most of the methods reported here. The failure with design 45 and the general difficulty in distinguishing the designs from non-polar natural interfaces suggest that considerable work remains in refining models of protein-interface thermodynamics.
Discussion
Defining the structural and energetic determinants of high-affinity binding is crucial for our mechanistic understanding of protein-interaction networks and the ability to intervene in physiologically important systems. Our analysis provides a snapshot of current understanding of binding energetics. While certain features emerge as discriminators between designs and a majority of the natural protein complexes in our dataset, all of the metrics misclassify some natural complexes as non-binders. In many areas of computational biology, ranging from sequence alignment13 to function annotation14, the availability of comprehensive benchmarks has provided strong impetus to method development and a powerful means of gauging progress. The benchmark provided here, the first to contain complexes that are predicted to associate but have been experimentally determined not to interact, provides a valuable orthogonal axis for evaluating both the relative and absolute performance of alternative approaches.
The design discrimination test is complementary to traditional docking tests. In this test, large-scale sampling of rigid-body or backbone freedom is not needed, allowing more direct focus on the energy function. On the other hand, it must be kept in mind that the failure of a computational design to experimentally bind its target could be related not only to overestimation of the computed binding energy due to energy function inaccuracies, but also imperfect design at the monomeric protein level: the design may not actually fold to the target structure. The high likelihood of designed sidechains to adopt binding-incompatible conformations in the unbound state has been suggested to play a role in the failure of design calculations to produce active binders15. Here, we find that changes to backbone structure in designed surfaces might play an equally significant role in compromising designs. Indeed, in the design of hemagglutinin binders, the two active designs used largely helical and conformationally restricted surfaces1. Our conclusion that surfaces that are not well anchored are poor choices for design can be easily used to eliminate such surfaces from design.
The 28 participating groups found many differences between the designed and natural complexes. In particular several metrics employing electrostatics and solvation show promise as discriminators; perhaps unsurprisingly, given that the three surfaces targeted in the design set were largely hydrophobic, whereas natural interfaces span the range of hydrophobicity and charge. On the other hand, most all-atom metrics fail to discriminate native and designed hydrophobic interfaces, even though most of the designs do not bind. This result underscores the importance of developing improved forcefields for protein interfaces that are able to discriminate binders from non-binders in all categories. One result of the community wide testing is that our original benchmark set could be “tricked” because of its too strong focus on nonpolar interfaces. We have now supplemented the benchmark with more polar and charged interfaces to remedy this deficiency and by suggesting a subset of 25 apolar natural interfaces for comparison to designs; we look forward to the improved metrics that will be developed to solve the discrimination problem posed by this more inclusive benchmark.
Solving the discrimination problem by all-atom methods may require explicit treatment of the various conformational-entropy penalties of binding, such as sidechain and backbone freezing15; 16. Additional aspects such as water molecules at the interface, and the likelihood that the designed protein adopts its target conformation may also need to be addressed. The availability of a comprehensive dataset should enable the development of improved energy functions, yielding a more complete understanding and formulation of the energetic contributions to binding free energy and increasing the reliability of tools for predicting and engineering protein interactions.
Materials & Methods
Experimental materials and methods and the computational methods used in discrimination are provided in the online supplement.
Computational methods
Preparation of input files
Designed and natural complexes were subjected to the same computational protocol consisting of full sidechain repacking and refinement of the rigid-body and sidechain conformations using the local-refine mode of RosettaDock17. All calculations were conducted in the Rosetta all-atom forcefield (score12), which is dominated by van der Waals, hydrogen bonding, and solvation terms5. A RosettaScript for complex-structure refinement is available in the online supplement. Refined structures were provided to the participants and are available in the online supplement.
Computed binding energy and buried-surface area calculations
The binding energy and buried-surface area (Fig. 1; Table S1) were computed within the Rosetta software suite. For the natural complexes, the biologically relevant interface was extracted from information provided with the docking benchmark18. Binding-energy calculations (using score12) were computed by subtracting the energy in the unbound complex from the energy in the bound complex, in each state allowing for repacking of interface sidechains. Binding energies were averaged over three repeats for numerical stability. A RosettaScript for computing the binding energies and buried surface areas is available in the online supplement.
Receiver-operator Characteristic (ROC) and the Area Under the Curve (AUC)
The raw scores from each group were numerically sorted from high to low propensity to bind, irrespective of the type of complex (natural or designed). To plot the ROC, for each natural complex in the sorted list, a step was taken along the y-axis, and conversely, for each designed complex, a step was taken along the x-axis. Step sizes were normalized such that the total lengths of the x- and y-axes were 1.0. The AUC was computed by summing the area added under the curve for each x-axis increment. Scripts for computing the AUC and plotting the ROC are available in the online supplement.
Degree of connectivity at the interface
For each interface residue on designed monomers and all interface residues on natural binders we calculate the number of residue neighbors on the host monomer within 8Å of the interfacial residue (ignoring the partner protein). We find that below 8.5 residue neighbors designed surfaces are poorly anchored in their host monomers (examples in Figure 4). Residues within 8Å of the partner protein were considered to be interfacial. This metric is implemented in RosettaScripts19 (see Supplemental Data).
Redesign for improved electrostatics
The 87 designed complexes served as starting structures for three iterations of sidechain design of scaffold interface residues followed by minimization of rigid-body, backbone, and sidechain degrees of freedom. During design and minimization, the Rosetta all-atom forcefield was augmented with a Coulomb electrostatic-interaction term with a distance-dependent dielectric (weight=1.0) and pair potential (weight=0.98, compared to 0.49 in the default all-atom forcefield). The 29 designs burying the highest surface areas were selected.
Pairwise and electrostatic contributions to binding (Fig. 3B) were these energetic components of binding-energy calculations (see above), and were computed assuming weights of 0.49 for the pairwise potential and 0.25 for Coulomb electrostatics. A RosettaScript for the design trajectory is available as Supplemental Data.
Source code
The Rosetta software suite is available free of charge to academic users at http://www.rosettacommons.org. Scripts used in analyzing the data and producing the graphics are provided in the online supplement.
Supplementary Material
Acknowledgments
The authors thank Sameer Velankar and Marc Lensink for their help in coordinating this experiment and Raik Grunberg for many helpful suggestions on a draft. SJF was supported by a long-term fellowship from the Human Frontier Science Program. SJW is Canada Research Chair Tier 1, funded by the Canadian Institutes for Health Research. Research in the Baker lab was supported by the Howard Hughes Medical Institute, the Defense Advanced Research Projects Agency, the NIH Yeast Resource Center, and the Defense Threat Reduction Agency.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–68. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 3.Janin J, Henrick K, Moult J, Eyck LT, Sternberg MJ, Vajda S, Vakser I, Wodak SJ. CAPRI: a Critical Assessment of PRedicted Interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 4.Karanicolas J, Corn JE, Chen I, Joachimiak LA, Dym O, Peck SH, Albeck S, Unger T, Hu W, Liu G, Delbecq S, G TM, C PS, Liu DR, Baker D. A de novo protein binding pair by computational design and directed evolution. Mol Cell. 2011;42:250–60. doi: 10.1016/j.molcel.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–82. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 6.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–51. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Idusogie EE, Presta LG, Gazzano-Santoro H, Totpal K, Wong PY, Ultsch M, Meng YG, Mulkerrin MG. Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc. J Immunol. 2000;164:4178–84. doi: 10.4049/jimmunol.164.8.4178. [DOI] [PubMed] [Google Scholar]
- 8.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein-protein docking benchmark version 3.0. Proteins. 2008;73:705–9. doi: 10.1002/prot.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kastritis PL, Moal IH, Hwang H, Weng Z, Bates PA, Bonvin AM, Janin J. A structure-based benchmark for protein-protein binding affinity. Protein Sci. 2011;20:482–91. doi: 10.1002/pro.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilson MK, Zhou HX. Calculation of protein-ligand binding affinities. Annu Rev Biophys Biomol Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 11.Sheffler W, Baker D. RosettaHoles: rapid assessment of protein core packing for structure prediction, refinement, design, and validation. Protein Sci. 2009;18:229–39. doi: 10.1002/pro.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murali R, Sharkey DJ, Daiss JL, Murthy HM. Crystal structure of Taq DNA polymerase in complex with an inhibitory Fab: the Fab is directed against an intermediate in the helix-coil dynamics of the enzyme. Proc Natl Acad Sci U S A. 1998;95:12562–7. doi: 10.1073/pnas.95.21.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson JD, Plewniak F, Poch O. BAliBASE: a benchmark alignment database for the evaluation of multiple alignment programs. Bioinformatics. 1999;15:87–8. doi: 10.1093/bioinformatics/15.1.87. [DOI] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleishman SJ, Khare SD, Koga N, Baker D. Restricted sidechain plasticity in the structures of native proteins and complexes. Protein Sci. 2011;20:753–757. doi: 10.1002/pro.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunberg R, Nilges M, Leckner J. Flexibility and conformational entropy in protein-protein binding. Structure. 2006;14:683–93. doi: 10.1016/j.str.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–99. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 18.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–7. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 19.Fleishman SJ, Leaver-Fay A, Corn JE, Strauch EM, Khare SD, Koga N, Ashworth J, Murphy PM, Richter F, Lemmon G, Meiler J, Baker D. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS One. 2011 doi: 10.1371/journal.pone.0020161. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeLano WL. The PyMol molecular graphics systems. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.