SUMMARY
The mammalian brain is composed of thousands of interacting neural cell types. Systematic approaches to establish the molecular identity of functional populations of neurons would advance our understanding of neural mechanisms controlling behavior. Here, we show that ribosomal protein S6, a structural component of the ribosome, becomes phosphorylated in neurons activated by a wide range of stimuli. We show that these phosphorylated ribosomes can be captured from mouse brain homogenates, thereby enriching directly for the mRNAs expressed in discrete subpopulations of activated cells. We use this approach to identify neurons in the hypothalamus regulated by changes in salt balance or food availability. We show that galanin neurons are activated by fasting and that prodynorphin neurons restrain food intake during scheduled feeding. These studies identify elements of the neural circuit that controls food intake and illustrate how the activity-dependent capture of cell-type-specific transcripts can elucidate the functional organization of a complex tissue.
INTRODUCTION
A basic goal of neuroscience is to link the activity of specific neuronal cell types to the various functions of the brain. This task is complicated by the extraordinary cellular diversity of the mammalian central nervous system (CNS) (Lichtman and Denk, 2011; Masland, 2004; Nelson et al., 2006; Stevens, 1998) and the fact that most neurons cannot be identified based solely on their morphology or location (Isogai et al., 2011; Siegert et al., 2009). Comprehensive analyses of gene expression in the nervous system, such as the GENSAT project and the Allen Brain Atlas, have revealed extensive heterogeneity in gene expression across brain regions (Gong et al., 2003; Lein et al., 2007), but there are significant gaps in our understanding of how this molecular diversity is linked to function.
The ability to profile the genes uniquely expressed in neurons that respond to a stimulus would facilitate the systematic molecular identification of the cell types that control behavior. The molecular identification of these cells would also enable their manipulation in vivo by using technologies that make it possible to activate or inhibit neurons with light (Yizhar et al., 2011), generate transcriptional profiles from neurons by using tagged ribosomes (Heiman et al., 2008; Sanz et al., 2009), or label and record from neurons by using fluorescent reporters (Gong et al., 2003). These tools achieve their selectivity by targeting protein expression using a promoter from a cell-type-specific marker gene, but in many cases, the marker genes that identify a functional population of neurons are unknown (Zhang et al., 2007).
Immediate early genes such as c-fos have been widely used to visualize the neurons that respond to numerous stimuli (Morgan and Curran, 1991). However, despite its utility in marking neurons that have been biochemically activated, c-Fos staining does not reveal the genetic identity of the labeled cells. Characterizing the coexpression of an activation marker such as c-Fos with even a limited set of candidate genes requires processing large numbers of histologic sections (Isogai et al., 2011). For this reason, systematic methods are needed to profile gene expression from discrete subpopulations of activated neurons in the brain.
Here, we show that phosphorylation of the ribosome can be used as a molecular tag to retrieve RNA selectively from activated neurons. This enables the unbiased discovery of the genes that are uniquely expressed in a functional population of neurons. By quantifying in parallel the enrichment of many such markers, it is possible to assess the activation or inhibition of numerous cell types in a complex tissue, revealing the coordinated regulation of ensembles of neurons in response to an external stimulus. We use this approach to identify cellular components of the neural circuit that controls feeding in the hypothalamus.
RESULTS
Ribosome Phosphorylation Often Correlates with Neural Activity
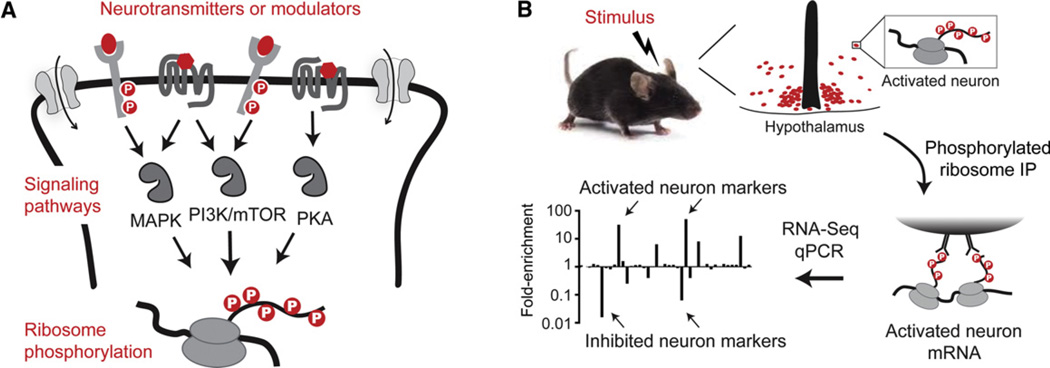
Immediate early genes such as c-fos are widely used to visualize activated neurons in the mouse brain (Morgan and Curran, 1991), but c-Fos staining does not reveal the molecular identity of the labeled cells. We thus set out to develop a method for generating expression profiles from activated neurons. We noted that many stimuli that trigger c-Fos expression in activated neurons also induce phosphorylation of ribosomal protein S6 (Cao et al., 2008; Valjent et al., 2011; Villanueva et al., 2009; Zeng et al., 2009). S6 is a structural component of the ribosome that is phosphorylated downstream of PI3-K/mTOR, MAPK, and PKA signaling (Figure 1A) (Meyuhas, 2008; Valjent et al., 2011). These same pathways regulate the transcription of activity-dependent genes such as c-fos (Flavell and Greenberg, 2008). We reasoned that, because S6 phosphorylation introduces a tag on ribosomes that reside in biochemically activated neurons, it might be possible to immunoprecipitate these phosphorylated ribosomes from mouse brain homogenates and thereby enrich for messenger RNA (mRNAs) expressed in the activated cells (Figure 1B). By comparing the abundance of each transcript in the pS6 immunoprecipitate to its abundance in the tissue as a whole, it would thus be possible to rank in an unbiased way the genes that are uniquely expressed in a population of neurons that respond to a stimulus.
Figure 1. Phosphorylated Ribosome Profiling.
(A) Neurotransmitters and neuromodulators activate a core set of signaling pathways in neurons. Rps6 is a common target of these pathways and is phos-phorylated on five C-terminal residues.
(B) Asparse subpopulationofneuronsis activated in response a stimulus (red cells). Activated neurons show enhanced pS6, and thus, capture of phosphorylated ribosomes enriches for the mRNA expressed in the activated cells. Quantifying the enrichment (IP/input) of a large panel of cell-type-specific marker genes reveals the genes uniquely expressed in the neurons that were activated.
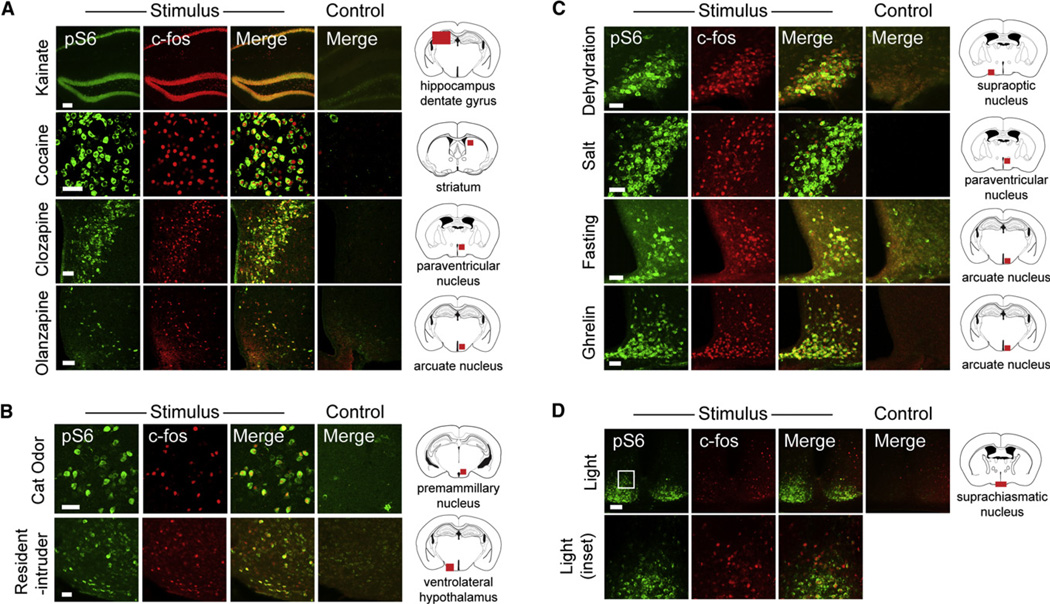
To confirm that S6 was phosphorylated in cells expressing c-Fos, we exposed mice to a diverse panel of stimuli and then performed double immunohistochemistry for c-Fos and pS6 in brain slices (Figure 2). We found that treatment of mice with drugs such as cocaine (a stimulant), kainate (a convulsant), and clozapine and olanzapine (antipsychotics) all induced colocalization of pS6 and c-Fos in a variety of brain regions (Figure 2A and Figure S1 available online). Exposure of male mice to an intruder induced an overlapping pattern of c-Fos and pS6 expression in brain regions that are known to mediate aggression (Lin et al., 2011), such as the ventrolateral hypothalamus and periaqueductal gray (Figures 2B and S1). We found that a cat odorant, which signals to rodents the presence of a predator, induced c-Fos and pS6 in the dorsal premammillary nucleus, a region knowntomediate fear and defensive responses (Dielenberg et al., 2001) (Figure 2B). A wide variety of nutritional stimuli, including fasting, dehydration, salt challenge, and ghrelin treatment, also resulted in extensive colocalization of c-Fos and pS6 in regions of the hypothalamus that are known to regulate water and food intake (Figures 2C and S1). In some cases, we observed that one of these markers labeled a broader population of activated neurons than the other; for example, light induced strong pS6 but only scattered c-Fos within the suprachiasmatic nucleus (Figure 2D), a region that regulates circadian rhythms and receives input from the retina (Cao et al., 2008). However, in general, we found that a wide range of stimuli induced expression of c-Fos and pS6 in largely overlapping neural populations throughout the brain.
Figure 2. Colocalization between c-Fos and pS6 following Diverse Stimuli.
(A) Response to the drugs kainate (pS6 235/236), cocaine (pS6 244), clozapine (pS6 235/236), and olanzapine (pS6 235/236).
(B) Response to stimuli that induce defensive behavior or aggression, including introduction of an intruder (pS6 244) or exposure to a worn cat collar (pS6 244).
(C) Response to nutritional stimuli, including dehydration (pS6 244), salt challenge (pS6 235/236), overnight fast (pS6 244), and ghrelin (pS6 235/236).
(D) Response inthe suprachiasmatic nucleus (SCN) following light stimulation at the end of the dark phase (pS6 235/236). Inset region is shown in the second row.
All scale bars, 50 mm except kainate (100 µm). See also Figure S1.
Selective Capture of Phosphorylated Ribosomes
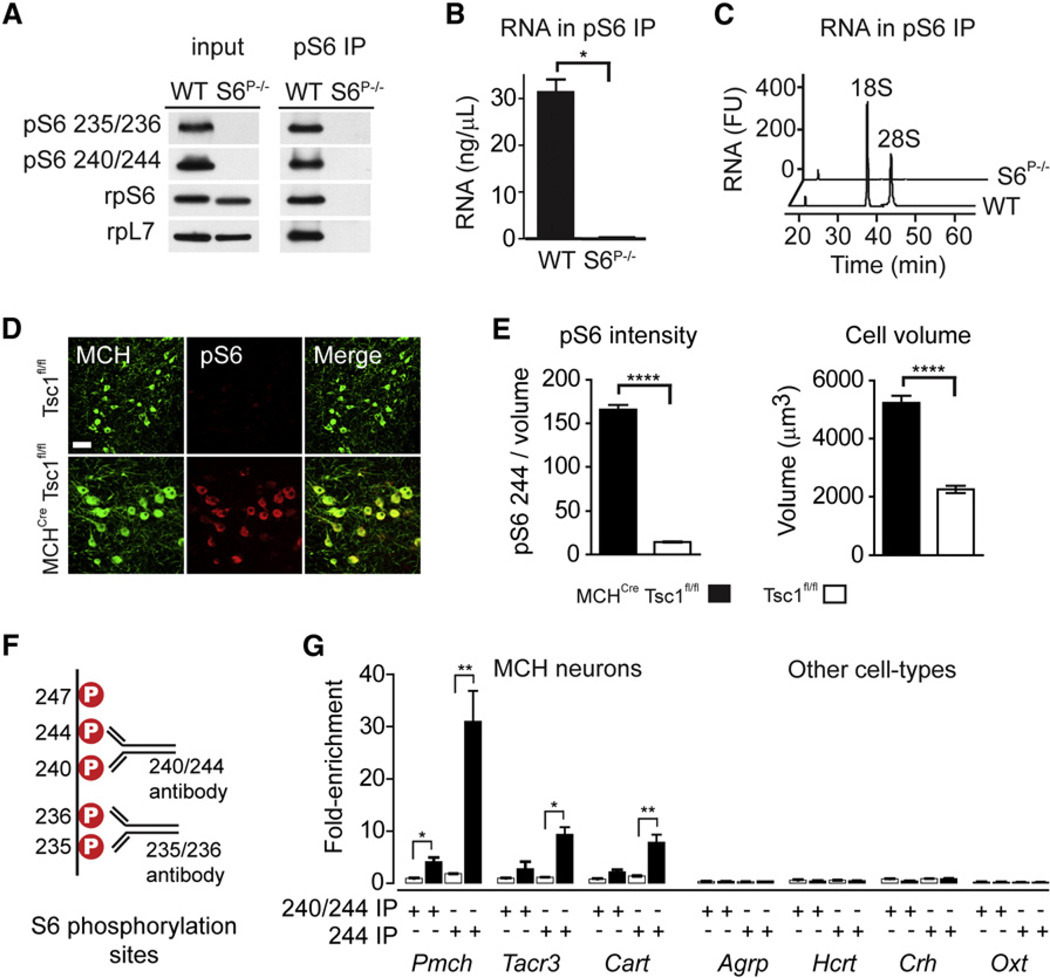
We next set out to confirm that we could selectively isolate phosphorylated ribosomes and their associated mRNA. We prepared lysates from wild-type mouse embryonic fibroblasts (MEFs) as well as knockin MEFs in which each of the five serine phosphorylation sites on S6 was mutated to alanine (Ser235, 236, 240, 244, and 247; S6S5A) (Ruvinsky et al., 2005). Antibodies that recognize pS6 240/244 efficiently immunoprecipitated ribosomes from lysates of wild-type MEFs, but not from S6S5A cells (Figure 3A). Approximately 100-fold more RNA was isolated in pS6 immunoprecipitates from wild-type MEFs compared to S6S5A controls (Figures 3B and 3C), confirming that phosphorylated ribosomes can be captured with high selectivity.
Figure 3. Selective Capture of Phosphorylated Ribosomes In Vitro and In Vivo.
(A) Western blot for ribosomal proteins from wild-type or S6S5A MEFs. Input lysate (left) and the pS6 240/244 immunoprecipitate (right) are shown.
(B) RNA yield from pS6 240/244 immunoprecipitates from wild-type and S6S5A MEFs.
(C) Bioanalyzer analysis of immunoprecipitated RNA from wild-type and S6S5A MEFs. 18S and 28S ribosomal RNA are labeled. FU, fluorescence units.
(D) Colocalization of MCH and pS6 in the hypo-thalamus of Tsc1fl/fl and MCHCre Tsc1fl/fl mice. Scale bar, 50 mm.
(E) Left: pS6 244 immunostaining density in MCH neurons following Tsc1 deletion. Right: volume of MCH neurons following Tsc1 deletion.
(F) Five major S6 phosphorylation sites and the diphospho-motifs recognized by commonly used pS6 antibodies.
(G) Enrichment of a panel of cell-type-specific genes following immunoprecipitation with antibodies recognizing pS6 240/244 or only pS6 244. Black bars represent immunoprecipitates from hypothalamic homogenates of MCHCre Tsc1fl/fl mice, whereas white bars represent Tsc1fl/fl controls. The enrichment of cell-type-specific genes in pS6 immunoprecipitates was determined by Taqman.
*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 using two-tailed unpaired t test. All error bars are mean ±SEM. See also Figure S2.
To confirm that we could enrich for mRNA from a single neuronal cell type in vivo, we generated mice in which the gene encoding Tsc1 was selectively deleted in melanin-concentrating hormone (MCH) neurons of the lateral hypothalamus (MCHCre Tsc1fl/fl). Deletion of Tsc1 activates the mTORC1 pathway, resulting in constitutive S6 phosphorylation (Figures 3D and 3E) and, consistent with prior reports, increased cell size (Figure 3E) (Meikle et al., 2007). We prepared tissue homogenates from hypothalami of these mice, immunoprecipitated phosphorylated ribosomes, and analyzed the purified RNA. We found that commercially available antibodies that recognize either pS6 235/236 or 240/244 could enrich for Pmch mRNA by ~4-fold (Figures 3F and 3G). Because Tsc1 deletion results in uniform and complete S6 phosphorylation in the targeted cells (Meikle et al., 2007), this 4-fold enrichment represented an upper limit on the RNA enrichment we could achieve, and at this level of enrichment, it was challenging to identify markers for cell types that underwent graded or heterogeneous activation in response to a physiologic stimulus (Z.A.K. and J.M.F., unpublished data). We therefore explored ways to capture RNA from activated neurons more selectively.
Phosphorylation of S6 is believed to occur sequentially (in the order 236,235,240,244, and 247), such that the most C-terminal sites (244 and 247) are phosphorylated at much lower stoichiometry than the N-terminal sites at baseline (Figure 3F) (Meyuhas, 2008). We reasoned that phosphorylation of these C-terminal sites should therefore exhibit a wider dynamic range in response to changes in neural activity and that an antibody recognizing only one of the C-terminal sites might enable greater enrichment of cell-type-specific transcripts. Following extensive optimization, we discovered that a polyclonal antibody targeting pS6 240/244 could be made more selective by preincubation with a phosphopeptide containing the pS6 240 phosphorylation site, thereby yielding antibodies that recognize only phosphorylation at 244 (hereafter referred to as pS6 244 antibodies) (Figure 3F). Immunoprecipitation of phosphorylated ribosomes by using pS6 244 antibodies resulted in more than 30-fold enrichment of Pmch transcripts from MCHCre Tsc1fl/fl mice, but not Tsc1fl/fl controls (Figure 3G). Importantly, we also observed robust enrichment (8- to 10-fold) for genes coexpressed in only a subset of MCH neurons, such as Cart and Tacr3 (Croizier et al., 2010), but observed no enrichment for genes expressed in a set of different hypothalamic cell types, such as the neuropeptides Hcrt, Oxt, Agrp, and Crh (Figure 3G). Consistent with this quantitative PCR (qPCR) data, brain slices stained by using pS6 244 antibodies showed enhanced contrast between pS6-positive and -negative neurons compared to slices stained with commercial antibodies that recognize a broader set of phosphorylation sites (Figure S2). Thus, using this optimized approach, we were able to achieve highly selective enrichment of the transcripts expressed in neurons with induction of pS6 in vivo.
Molecular Identification of Hypothalamic Neurons Activated by Salt
We next asked whether we could identify neurons activated by a well-characterized physiologic stimulus. Plasma osmolarity is controlled by a hypothalamic system that includes vasopressin and oxytocin neurons, and the levels of these peptides are known to increase in response to salt loading. We therefore challenged mice with a concentrated salt solution and stained brain sections for pS6 244.
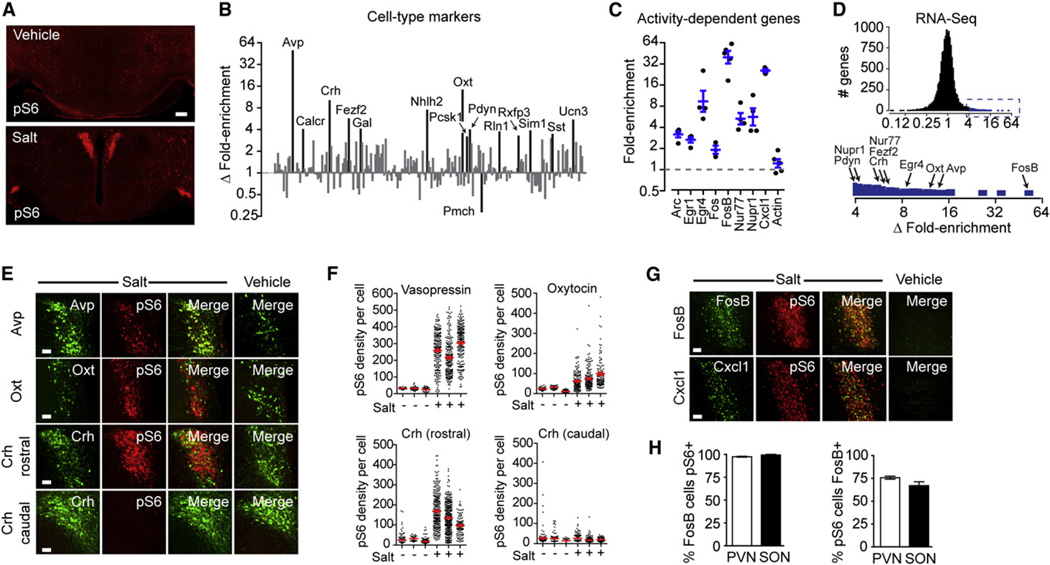
Salt challenge induced a dramatic increase in pS6 in regions of the hypothalamus that mediate osmoregulation, including the paraventricular (PVN) and supraoptic nuclei (SON) and median eminence (Figure 4A). We immunoprecipitated phosphorylated ribosomes from hypothalamic homogenates of salt-challenged and control animals and analyzed the enriched mRNAs. To enable the rapid and sensitive quantification of low abundance transcripts, we designed a custom array of 225 Taqman probes comprised of marker genes that show anatomically restricted expression within the hypothalamus, including neuropeptides, receptors, and transcription factors (Table S1). The expression data for these genes are plotted as the log of the differential enrichment of each gene in response to the stimulus (Figure 4B). Similar results were obtained by using RNA sequencing and microarrays (Figures 4D and S3 and Table S3).
Figure 4. Identification of Neurons Activated by Salt Challenge.
(A) Hypothalamic staining for pS6 244 from mice given an injection of vehicle (PBS) or 2 M NaCl.
(B) Differential enrichment of cell-type-specific genes in pS6 immunoprecipitates determined by Taqman. Data are expressed as the ratio of fold enrichment (IP/input) for salt-treated animals divided by the fold enrichment (IP/input) for controls and plotted on a log scale. Genes with a fold enrichment > 3.0 and p < 0.05 are labeled.
(C) Fold enrichment (IP/input) for a panel of activity-dependent genes following osmotic stimulation. Actin is shown for reference.
(D) RNA-seq analysis of differential enrichment of cell-type-specific genes in pS6 immunoprecipitates. Inset, magnification of region (blue) showing >4-fold enrichment. Genes labeled in (B) are highlighted.
(E) Colocalization between Avp, Oxt, and Crh with pS6 244 in salt-treated and control animals. Crh neurons were analyzed as two separate populations in the rostral and caudal PVN.
(F) pS6 intensity within individual Avp, Oxt, and Crh neurons from salt-treated and control animals.
(G) Colocalization between FosB, Cxcl1, and pS6 in salt-treated and control animals.
(H) Percentage of FosB-positive cells in the PVN and SON that are also pS6 positive.
(I) Percentage of pS6-positive cells in the PVN and SON that are also FosB positive.
All scale bars, 50 mm except (A) (200 mm). All error bars are mean ±SEM. See also Figure S3.
We found that markers for the major neural populations that respond to salt challenge were among the most highly enriched genes in pS6 immunoprecipitates. These include vasopressin (Avp; 49-fold enriched), oxytocin (Oxt; 14-fold), and corticotro-phin-releasing hormone (Crh; 10-fold) (Figure 4B and Table S2). The degree of enrichment of these marker genes correlated with the quantitative induction of pS6 in the corresponding cells as assayed by immunohistochemistry (Figures 4E and 4F). We likewise detected specific enrichment at a lower level for genes that partially overlap in expression with Avp and Oxt, such as the neuropeptides galanin (Gal; 4.0-fold) and prodynorphin (Pdyn; 3.9-fold) and the PVN-specific transcription factors Nhlh2 (7.4-fold), Fezf2 (5.6-fold), and Sim1 (3.8-fold) (Figure 4B) (Gai et al., 1990; Sherman et al., 1986). These data show that pS6 immunoprecipitation can enrich for transcripts that identify activated cell types and, further, that the fold enrichment of these genes reflects their selective expression in the activated cells.
Some of the genes enriched in pS6 immunoprecipitates identify neural populations not previously known to be activated by salt challenge. For example, we detected specific enrichment for relaxin-1 (Rln1; 3.7-fold), a neuropeptide that stimulates water intake (Thornton and Fitzsimons, 1995) and activates vasopressin and oxytocin neurons (Sunn et al., 2002) but that had not been characterized in the hypothalamus due to its low expression level. Other enriched neuropeptides include urocortin 3 (Ucn3; 5.4-fold), which is related to Crh and is expressed in a small population of neurons in the perifornical region, and somatostatin (Sst; 3.4-fold), which is known to promote vasopressin release (Brown et al., 1988).
In addition to these cell-type markers, we also detected enrichment for biochemical markers that are known to be induced in activated neurons (Figure 4C). The most highly enriched activity-dependent gene was Fosb (43-fold), and immunostaining revealed essentially complete colocalization between FosB and pS6 in the PVN and SON (Figures 4G and 4H). We likewise detected enrichment for Cxcl1 (26-fold), a chemokine that is not expressed in the hypothalamus at baseline but is selectively induced in the PVN by salt (Figure 3G) (Koike et al., 1997). These results were confirmed by microarray analysis, which identified Avp, Oxt,Fosb, and Cxcl1as the four most highly enriched genes in the genome in pS6 immunoprecipitates from salt-challenged animals relative to controls (Table S3). A similar pattern of marker gene enrichment was observed by RNA sequencing (Figure 4C and Table S3). In contrast, immunoprecipitation of total ribosomes from salt-challenged animals enriched for none of these genes (Table S2). Thus, by capturing phosphorylated ribosomes and by analyzing the associated mRNA, we are able to systematically identify genetic markers for neurons that are activated by a stimulus, revealing the coordinated response of numerous intermingled cell types to a physiologic signal.
The Hypothalamic Response to Fasting
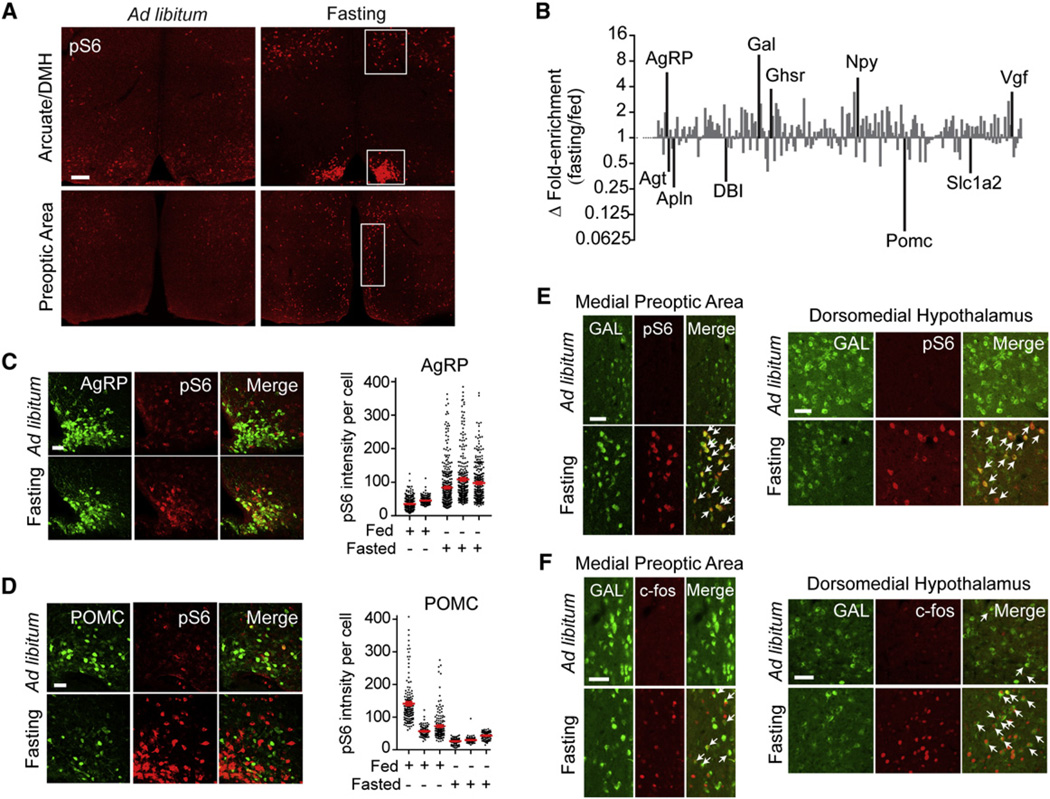
A different set of neurons in the hypothalamus regulate food intake and the response to food restriction. To identify components of this system, we exposed mice to a series of nutritional perturbations, beginning with fasting. Mice were fasted overnight and sacrificed at the end of the dark phase, and the extent of ribosome phosphorylation was assayed by immunostaining. We found that fasting induced strong pS6 in the arcuate nucleus of the hypothalamus as well as in the dorsomedial hypothalamus (DMH) and scattered cells of the medial preoptic area (MPA) and PVN (Figures 5A and S4). To identify fasting-regulated neurons in each of these regions, we immunoprecipitated phosphorylated ribosomes from hypothalamic homogenates of fasted and fed animals and analyzed the enrichment of cell-type-specific RNAs.
Figure 5. Identification of Neurons Activated by Fasting.
(A) Hypothalamic staining for pS6 244 from fasted and fed mice. Top panels show the arcuate nucleus and DMH, and bottom panels show the preoptic area (highlighted).
(B) Relative enrichment of cell-type-specific genes in pS6 immunoprecipitates from fasted and fed animals. Data are expressed as the ratio of fold enrichment (IP/input) for fasted animals divided by the fold enrichment (IP/input) for fed controls and are plotted on a log scale. Genes with fold enrichment >2.5 and p < 0.05 are labeled.
(C) Left: colocalization between AgRP and pS6 244 in fed and fasted mice. Right: pS6 intensity in AgRP neurons.
(D) Left: colocalization between POMC and pS6 in fed and fasted mice. Right: pS6 intensity in POMC neurons.
(E) Colocalization between Gal and pS6 in fed and fasted mice in the MPA and DMH.
(F) Colocalization between Gal and c-Fos in fed and fasted mice in the MPA and DMH.
All scale bars, 50 mm except (A) (100 mm). All error bars are mean ±SEM. See also Figure S4.
Markers for many cell types that are known to regulate feeding were differentially enriched in pS6 immunoprecipitates. Thus, two of the most enriched transcripts in response to fasting were Agrp and Npy (Figure 5B). These two neuropeptides are coexpressed in critical neurons of the arcuate nucleus that promote food intake (Elmquist et al., 2005), and we confirmed that fasting induces a selective increase in pS6 in these cells (Villanueva et al., 2009) (Figure 5C). We also observed enrichment for genes such as the ghrelin receptor (Ghsr), which is expressed in most AgRP/NPY neurons (Willesen et al., 1999), and the neuropeptide Vgf, which is induced in AgRP neurons following fasting (Hahm et al., 2002).
Galanin Neurons Are a Distinct Population of Fasting-Activated Cells
The neuropeptide galanin was one of the most strongly enriched genes in pS6 immunoprecipitates from fasted animals (Figure 5B). Galanin has been shown to stimulate feeding when injected directly into the hypothalamus (Parker and Bloom, 2012), but the regulation of galanin neurons by changes in nutritional state has not been described (Schwartz et al., 1993). We found that fasting induced a marked increase in pS6 in a specific subset of galanin neurons located in the DMH and MPA (Figure 5E). Galanin neurons in these two regions also expressed c-Fos after an overnight fast (Figure 5F), confirming that they are activated by food restriction. We further characterized the neurochemical identity of galanin neurons in the DMH and found that the majority were positive for GAD67, indicating that they produce the inhibitory neurotransmitter GABA, but did not express the leptin receptor, indicating that they do not directly sense changes in plasma leptin (Figure S4). Thus, galanin neurons in the DMH and MPA represent a population of fasting-activated cells in the hypothalamus with a localization and regulation distinct from AgRP neurons.
Markers for Inhibited Cells Are Depleted from pS6 Immunoprecipitates
As all neurons have a basal level of ribosome phosphorylation, we expected that neural inhibition might result in a decrease in pS6, which would be detected as the depletion of transcripts from pS6 immunoprecipitates. Consistent with this, we found that the neuropeptide Pomc was the most depleted transcript in response to fasting (Figure 5D). Pomc is expressed in a key population of neurons in the Arc that inhibit food intake, and Pomc expression is downregulated during food deprivation (Elmquist et al., 2005), whereas leptin increases c-Fos in Pomc neurons as well as the firing rate of these cells (Cowley et al., 2001). Although fasting increases the level of pS6 in the Arc overall (largely as a result of AgRP neuron activation, Figure 5A), we showed by quantitative imaging that fasting decreases the density of pS6 specifically within Pomc-expressing cells (Figure 5D). Thus, the depletion of specific transcripts from pS6 immunoprecipitates can reveal the identity of inhibited neurons. It is important to emphasize that the depletion we observe for Pomc is not the result of a change in its expression level, as we analyze only the ratio of RNA in the immunoprecipitate versus the tissue as a whole (immunoprecipitate [IP]/input). Rather, we enrich or deplete for RNA from neurons based on whether the state of activation of that neuron has changed. This ability to detect inhibition by ribosome profiling contrasts with c-Fos staining, which has a limited ability to detect downregulation due to the low level of c-Fos expression in most cells at baseline.
In addition to Pomc, we observed depletion following fasting of several additional neuropeptides that have been reported to inhibit feeding (Figure 5B), including apelin (Apln, which is coexpressed with Pomc), angiotensin (Agt), and diazopam-binding inhibitor (Dbi), suggesting that each of these peptides may reside in a population of fasting-inhibited cells (Porter and Potratz, 2004; Reaux-Le Goazigo et al., 2011; de Mateos-Verchere et al., 2001).
Scheduled Feeding Synchronizes Ribosome Phosphorylation with Food Availability
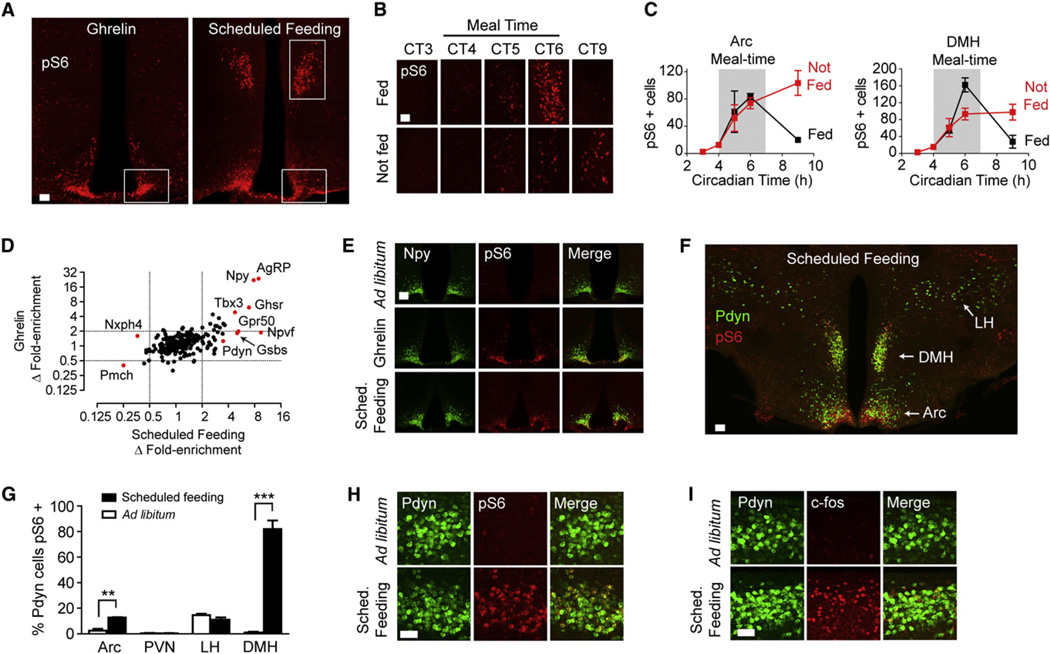
Although fasting can reveal the response to chronic energy deficit, most human feeding takes place intermittently at regular times in the day, and the timing of meals is associated with numerous biochemical and behavioral responses. Similarly, rodents allowed daily access to food only during a scheduled window are known to synchronize their metabolism and activity to the time of food availability (Mistlberger, 2011). This behavioral adaptation is known as food-anticipatory activity (FAA) and is associated with the activation of neurons in multiple hypothalamic regions, including prominently the DMH and Arc. Despite extensive investigation into the mechanism of FAA, the identity of the activated cell types and their specific roles, in particular those in the DMH, are largely unknown. Thus, we sought to identify neurons with a specialized function associated with scheduled feeding. Unlike fasting, scheduled feeding also allows for more precise synchronization of behavior, enabling a more refined analysis of temporal changes in cell activation.
We restricted the access of mice to food to a 3 hr window in the middle of the light phase, which resulted in the emergence of robust FAA within 10 days (Movies S1 and S2). We performed pS6 staining of brain slices from these mice at several time points to establish the dynamics of ribosome phosphorylation in the hypothalamus. We found that scheduled feeding induced intense pS6 staining in the DMH and Arc (Figure 6A) that peaked within the meal window and declined to baseline thereafter (Figures 6B and 6C). This DMH staining was enriched in the compact part of the DMH, a region that does not show a change in ribosome phosphorylation after a single overnight fast (Figure 5A).Once the mice were entrained, this pattern of S6 phosphorylation no longer depended on the presence of food because brain sections from mice that were acclimated to scheduled feeding but that were not fed on the day of the experiment showed a similar pattern of pS6 (although with lower intensity in the DMH; Figures 6B and 6C). This suggests the existence of unidentified neural populations that are regulated in part by a circadian signal entrained by food availability.
Figure 6. Identification of Neurons Activated by Ghrelin and Scheduled Feeding.
(A) Hypothalamic staining for pS6 244 in response to ghrelin (intraperitoneal injection, 1 hr) or scheduled feeding (2 hr following food presentation).
(B) pS6 staining in the DMH in mice acclimated to a protocol of scheduled feeding between circadian time (CT) 4–7. Mice were either fed (top) or not fed (bottom) on the day of the experiment.
(C) Number of pS6-positive cells in the DMH (left) and Arc (right) in mice on a scheduled feeding protocol. Black, fed on the day of the experiment. Red, not fed.
(D) Differential enrichment of cell-type-specific transcripts in pS6 IPs from mice that were given ghrelin (y axis) or subjected to scheduled feeding (x axis). Data are expressed as the ratio of fold enrichment (IP/input) from ghrelin or scheduled feeding animals relative to the fold enrichment of their controls and are plotted on a log scale. Key genes are labeled.
(E) Colocalization between NPY and pS6 in ad libitum, ghrelin-treated, and scheduled feeding mice.
(F) Expression of Pdyn in the hypothalamus and its colocalization with pS6 in mice subjected to scheduled feeding and sacrificed at CT6. The Arc, DMH, and LH are labeled for reference.
(G) Colocalization between Pdyn and pS6 in various hypothalamic nuclei of mice fed ad libitum or subjected to scheduled feeding and sacrificed at CT6.
(H) Colocalization between Pdyn and pS6 in the DMH of ad libitum and scheduled feeding.
(I) Colocalization between Pdyn and c-fos at CT6 in mice subjected to scheduled feeding.
Scale bars, 50 mm except (A) and (F) (100 µm). All error bars are mean ±SEM. See also Figure S5 and Movies S1, S2, and S3.
Scheduled Feeding Activates AgRP Neurons and Inhibits MCH Neurons
To identify neurons activated during scheduled feeding, we immunoprecipitated phosphorylated ribosomes from the hypothalamus of animals sacrificed 2 hr after food presentation and analyzed the enriched mRNAs. To provide a comparison data set, we also performed ribosome profiling from mice that received an injection of the hormone ghrelin. Levels of plasma ghrelin increase prior to meals, and this increase has been hypothesized to promote scheduled feeding (LeSauter et al., 2009; Mistlberger, 2011; Verhagen et al., 2011). We found that ghrelin induced strong pS6 in the Arc but had little effect on pS6 in the DMH (Figure 6A). Thus, we compared these two profiles in order to segregate enriched cell-type markers according to their potential anatomic location (i.e., DMH versus Arc) and function.
Ghrelin and scheduled feeding both induced strong enrichment of Agrp (24- and 8.9-fold), Npy (22- and 7.8-fold), and Ghsr (6.1- and 6.9-fold), and we confirmed extensive colocalization between pS6 and AgRP/NPY neurons of the Arc under both conditions (Figure 6E). The activation of AgRP/NPY neurons is consistent with the voracious eating displayed by animals acclimated to scheduled feeding following food presentation (Movie S3) and suggests that ghrelin and scheduled feeding activate a common set of neural targets in the Arc.
In contrast to the enrichment of Agrp and Npy transcripts, we noticed that Pmch was consistently depleted from pS6 immuno-precipitates after scheduled feeding (Figure 6D). We performed double immunostaining for pS6 and MCH in brain slices from these animals, which revealed a selective decrease in pS6 localized to MCH neurons from mice subjected to scheduled feeding relative to ad libitum controls (Figure S5). This decrease in ribosome phosphorylation was specific to MCH neurons, as neighboring pS6-positive cells were observed in the lateral hypothalamus in the same sections. Thus, MCH neurons appear to be selectively inhibited during scheduled feeding. Interestingly, we also observed depletion of transcripts for a second neuropeptide, Nphx4, which is expressed in the lateral hypothalamus in a pattern that resembles Pmch (Figure 6D). As deletion of both Pmch and its receptor (Mch1r) has been reported to induce hyperactivity activity in mice (Zhou et al., 2005), inhibition of these neurons may be related to the locomotor phenotype observed during scheduled feeding.
Molecular Identification of Activated Neurons in the DMH
We focused our attention on identifying activated neurons in the DMH because our understanding of the function and identity of the cell types in this region that regulate feeding is limited. We identified four transcripts—Npvf, Pdyn, Gpr50, and Gsbs—that were enriched in pS6 immunoprecipitates from mice subjected to scheduled feeding relative to ghrelin treatment (Figure 6D). Analysis of in situ hybridization data from the Allen Brain Atlas confirmed that these transcripts show localized expression in the DMH (Figure S5). Among these, the neuropeptide Npvf has previously been shown to colocalize with c-Fos in a sparse population of DMH cells activated during FAA (Acosta-Galvan et al., 2011), and the G-protein-coupled receptor Gpr50 is known to be regulated by leptin and nutritional state (Ivanova et al., 2008) but has not previously been linked to scheduled feeding.
We further characterized the neurons in the DMH that express Pdyn, a neuropeptide that has complex effects on mood, nociception, and reward but that has not previously been linked to scheduled feeding. Immunostaining revealed extensive colocal-ization between Pdyn and pS6 across the entire rostrocaudal axis of the DMH; 82% of Pdyn neurons in the DMH were pS6 positive in mice subjected to scheduled feeding compared to just 1% in ad libitum controls (Figures 6F–6H). We observed a smaller increase in colocalization between pS6 and Pdyn in the Arc and observed no change in the level of pS6 in Pdyn neurons in the PVN or lateral hypothalamus (Figures 6F and 6G). Thus, these data suggest that the Pdyn neurons in the DMH represent a functionally distinct population with a specialized role in feeding. Immunostaining for c-Fos also revealed extensive colocalization with Pdyn neurons in the DMH during scheduled, but not ad libitum, feeding (Figure 6I), confirming that Pdyn neurons are biochemically activated when mice are exposed to this protocol. c-Fos expression was also observed in other cells in the DMH, indicating the existence of additional populations of activated neurons in this region.
Prodynorphin Restrains Bouts of Intense Feeding
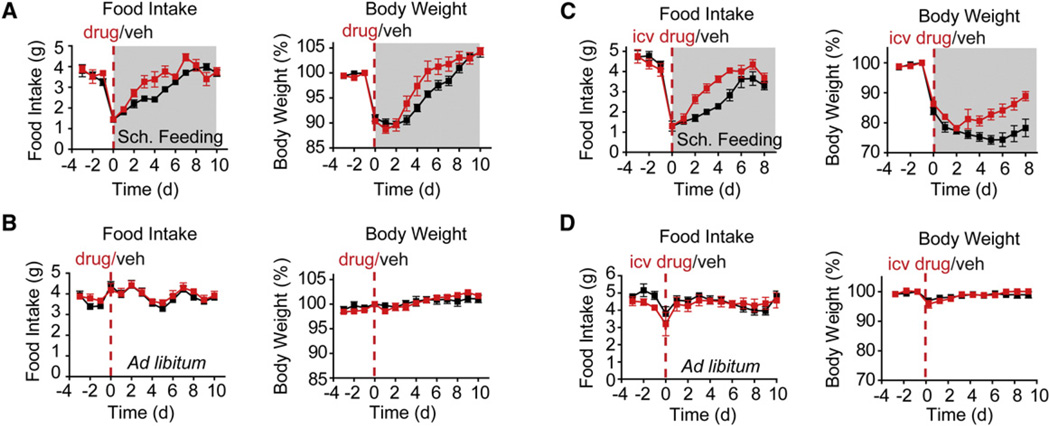
We hypothesized that Pdyn might play a role in meal termination following bouts of intense feeding. This hypothesis was based on the observation that pS6 induction in Pdyn neurons is evident only late in the meal window (Figures 6B and 6C), requires food presentation for full expression (Figures 6B and 6C), and is not observed in response to orexigenic signals such as fasting or ghrelin (Figures 5A and 6A). Pdyn signals by activating the k-opioid receptor (KOR) and potent, highly selective KOR antagonists have been developed (Bruchas et al., 2007; Carroll et al., 2004). We therefore assessed the function of Pdyn during scheduled feeding by using pharmacological inhibitors of KOR. We administered an intraperitoneal injection of either a selective KOR antagonist (JDTic) or vehicle to mice and then assigned animals to two groups—one exposed to the scheduled feeding paradigm and the other fed ad libitum. Because KOR antagonists have a characteristic long duration of action in vivo (up to 3 weeks), only a single dose was required (Bruchas et al., 2007; Carroll et al., 2004).
Vehicle-treated animals initially consumed less food per day and lost weight after shifting to scheduled feeding, after which their weight gradually recovered over the course of 7 days (Figure 7A; black). Mice that were treated with JDTic showed a similar decrease in food intake and body weight at first, but relative to the control group, their food intake increased more rapidly, and they showed a more rapid regain of body weight (Figure 7A). In contrast, JDTic had no impact on food intake or body weight in ad-libitum-fed animals (Figure 7B), indicating that the increased feeding induced by the drug is only evident under conditions in which the Pdyn neurons are activated.
Figure 7. KOR Signaling Restrains Food Intake during Scheduled Feeding.
Mice were treated with KOR inhibitors (red) or vehicle (black) by central or peripheral injection, and their food intake and body weight were recorded during ad libitum or scheduled feeding (gray).
(A) Mice givenanintraperitoneal injection ofthe KOR antagonistJDTic (red) orvehicle (black) and switched from adlibitum toscheduled feedingon day 0.p=0.01 for the difference in cumulative food intake on days 2–7. p = 0.055 for the body weight difference for days 4–7.
(B) Mice given an intraperitoneal injection of JDTic (red) or vehicle (black) and maintained on an ad libitum diet.
(C) Mice given an icv injection ofthe KOR antagonist norbinaltorphimine (red)orvehicle (black). Mice were switched from adlibitum toscheduled feedingonday 0. p < 0.01 for the difference in food intake for days 2–5 by t test. p < 0.02 for difference in body weight for days 5–8.
(D) Mice given an icv injection of the norbinaltorphimine (red) or vehicle (black) and fed ad libitum.
All error bars are mean ±SEM; p values calculated by two-tailed unpaired t test.
To test whether this effect was mediated by central KOR signaling, we delivered a second, structurally unrelated KOR antagonist (norbinaltorphimine) to mice by intracerebroventricular (icv) injection and then exposed these animals to a scheduled feeding protocol. Norbinaltorphimine-treated animals consumed more than 50% more food during scheduled feeding than vehicle-treated controls (Figure 7C). Drug-treated animals likewise gained weight much faster than controls (Figure 7C). Remarkably, this effect was specific to the scheduled feeding paradigm, as icv norbinaltorphimine had no effect on food intake or body weight in ad-libitum-fed animals (Figure 7D). Taken together, these data show that Pdyn neurons in the DMH are selectively activated during scheduled feeding and that central KOR signaling downstream of Pdyn acts to limit food intake following the intense feeding that accompanies this paradigm. As Pdyn neurons are also found in other brain regions, it is possible that Pdyn produced outside the DMH also contributes to the effects we observe. Understanding how Pdyn signaling is able to selectively regulate episodic feeding will require further characterization of the Pdyn cells in the DMH and their relation to other elements of the circuitry that controls food intake.
DISCUSSION
A vast array of experiments has sought to establish the functional importance of discrete neurons in controlling behavior (Lichtman and Denk, 2011). However, these efforts are often limited by a lack of molecular information about the relevant cell types. In 2001, Francis Crick and Christ of Koch predicted that the development of techniques “based on the molecular identification and manipulation of discrete and identifiable subpopulations” of neurons would enable the elucidation of the functional anatomy of the CNS (Koch and Crick, 2001). With the development of optogenetics and related methods, the means for manipulating cells are now available. In contrast, there has been less progress toward the development of approaches for the molecular identification of functional populations of neurons, and for many neural functions, the molecular identity of the relevant cell types remains unknown (Lin et al., 2011; Wu et al., 2012; Zhang et al., 2007). This problem of linking cell types to function has persisted despite increasingly sophisticated analyses of the molecular heterogeneity of the brain as a whole (Gong et al., 2003; Lein et al., 2007).
We report here a conceptually distinct way to map the functional organization of gene expression in the brain. This approach takes advantage of the fact that marker genes can be used to identify specific cell types within an anatomic region such as the hypothalamus (Siegert et al., 2009). We have shown that it is possible to capture RNA from cells in proportion to their activity, quantify the enrichment of these cell-type-specific marker genes, and then use this information to assay in parallel the functional state of a large number of intermingled cell types. A key advantage of this approach is that it enables the use of powerful tools of molecular biology, such as qPCR or RNA sequencing, to make a measurement of cellular activity that would otherwise require analysis of large numbers of samples by histology. As a result, it is possible to identify in an unbiased way the specific genes that are most uniquely expressed in a coregulated population of neurons in the brain. Once identified, such genes can serve as markers that enable the functional interrogation of those cells by using optogenetics or other approaches.
In this paper, we demonstrated that phosphorylation of ribosomal protein S6 can be used as a tag to enable the capture of mRNA from activated cells. This is possible because the same signaling pathways that trigger S6 phosphorylation are themselves often correlated with neural activity (Flavell and Greenberg, 2008; Meyuhas, 2008; Valjent et al., 2011). As the phosphorylation sites on S6 are evolutionarily conserved (Meyuhas, 2008), this approach, in principle, can be used to study a range of species, including those that are not amenable to genetic modification. Moreover, as S6 phosphorylation is controlled by extracellular stimuli in all cells (Meyuhas, 2008), this strategy could also reveal the regulation of nonneuronal cell types that reside in other complex tissues besides the brain, such as the immune system, lung, intestine, kidney, and others. We have validated the fidelity of this approach by identifying many neurons known to be activated or inhibited in response to well-characterized stimuli such as salt challenge and fasting. In addition to recapitulating the known components of these systems, we have also identified markers for activated neurons that have been overlooked, such as Gal neurons during fasting and Pdyn neurons during scheduled feeding. As many functional populations of neurons have been visualized by c-Fos staining, but not molecularly characterized (Dielenberg et al., 2001; Lin et al., 2011; Wu et al., 2012), phosphorylated ribosome profiling provides a general way to identify these cells. Once marker genes for these cell types have been identified, techniques such as BacTRAP or Ribotag can be used to genetically deliver tagged ribosomes to these cells, enabling deep profiling of their transcriptomes (Heiman et al., 2008; Sanz et al., 2009).
Although our data suggest that this approach will find broad application in neuroscience, it is important to emphasize that pS6, like other surrogates for neural activity such as c-Fos, measures only one dimension of “neural activation” and therefore will not retrieve markers for all neurons that are activated in all contexts. c-Fos and pS6 are induced by the biochemical activation of neurons, and therefore, these markers may be most responsive to stimuli that modulate neurons such as neuro-peptides and biogenic amines. Although we have shown that many stimuli that activate neurons induce pS6, it is not fully understood to what extent the induction of pS6 is correlated with changes in firing rate itself.
Different stimuli induce pS6 with varying efficiency, and for this reason, it is important to optimize the stimulus protocol to maximize the fold enrichment of the relevant neural markers (see Extended Experimental Procedures for further discussion). For the stimuli we have explored, we find that pS6 is induced with kinetics that range from tens of minutes to 2 hr. This is comparable to the expression of many immediate early genes but is somewhat slower than c-fos transcription, which is often complete within 20 min of stimulation. In addition, we have focused in these experiments primarily on the hypothalamus, but brain regions that have a high level of pS6 at baseline may be less amenable to this approach.
Several large-scale efforts are currently underway to map the functional organization of the mammalian brain (Alivisatos et al., 2012; Gong et al., 2003; Koch and Reid, 2012). These projects are being supported by efforts to develop new imaging technologies that can probe the complex anatomy of this tissue (Lichtman and Denk, 2011). The approach we describe here represents a complementary way to link structure to function in the nervous system. Unlike current efforts, our approach suggests a way, in principle, to simultaneously measure the activity of every cell type within a region of the brain, a goal that is not addressed by any existing technology. Although we have focused here on the use of ribosome phosphorylation to identify activated neurons, it should also be possible to capture RNA in response to other signals that reflect the functional state of a cell. For example, many proteins dynamically associate with polysomes in response to extracellular stimuli, and these proteins could also function as ribosome tags that enable enrichment of RNA from cells that received a specific signal. Alternatively, it might be possible to engineer ribosomes that are modified in response to the expression of immediate early genes, such as c-fos, or to use mass spectrometry to identify posttranslational modifications of the ribosome that correlate with specific stimuli. The combination of such approaches may eventually enable the use of RNA sequencing to measure the functional state of a complex tissue, such as the brain, along multiple dimensions and at the resolution of molecularly defined cell types.
EXPERIMENTAL PROCEDURES
Animal Treatment
Animal experiments were approved by the Rockefeller University IACUC. Male C57Bl/6J mice were maintained on a 12 hr light-dark schedule and were 9–13 weeks old at the time of sacrifice. All animals were sacrificed between CT4 and CT6 except as noted. For osmotic stimulation experiments, animals were given an intraperitoneal injection of 2 M NaCl (350 µl), water was removed from the cage, and mice were sacrificed 120 min later. For ghrelin experiments, animals were given an intraperitoneal injection of ghrelin (66 µg), food was removed from the cage, and mice were sacrificed 70 min later. For scheduled feeding, mice were allowed access to food between CT4 and CT7 each day for a minimum of 10 days, fed on the day of the experiment, and then sacrificed at CT6. For fasting experiments, animals were transferred to a new cage without food at CT7 and then sacrificed at CT23 the following morning. Control mice for fasting experiments were fed ad libitum and sacrificed at the same time. For drug treatments, mice were given an intraperitoneal injection of the following dose and then sacrificed at the indicated time: cocaine (30 mg/kg, 60 min), kainate (12.5 mg/kg, 120 min), olanzapine (20 mg/kg, 120 min), clozapine (10 mg/kg, 45 min). For cat odor experiments, a domestic cat was fitted with a fabric collar (Safe Cat) for 3 weeks; the collar was removed, mice were exposed to the collar for 60 min, and then they were sacrificed by perfusion. For the resident-intruder test, a male mouse was single caged for at least 2 weeks, a foreign male was introduced into the cage, the animals were monitored for attacks, and the resident was perfused after 60 min. For dehydration experiments, water was removed from the cage, and mice were perfused 24 hr later.
Ribosome Immunoprecipitations
Mice were sacrificed by cervical dislocation. Hypothalami were rapidly dissected on ice, pooled (10–20 per experiment), homogenized, and clarified by centrifugation. Ribosomes were immunoprecipitated by using polyclonal antibodies against pS6 240/244 combined with a pS6 240-containing peptide. RNA from the tissue as a whole (input) and the immunoprecipitate were then purified and analyzed. Detailed procedures are described in Extended Experimental Procedures.
Immunohistochemistry
Free-floating sections were stained, mounted, and then imaged by using a Zeiss LSM 810 confocal microscope. Image analysis and quantification were performed by using Imaris software. Additional details are described in Extended Experimental Procedures.
Treatment with KOR Antagonists during Scheduled Feeding
JDTic was delivered by intraperitoneal injection (10 mg/kg). Norbinaltorphimine (5 ml of a 1 mg/ml solution) was delivered via Hamilton syringe into the lateral ventricle by using the coordinates L/M 1.0 mm from Bregma, A/P −0.4 mm from Bregma, and 2.5 mm beneath the cortex.
Taqman Array Analysis
Primers and internally quenched probes were synthesized to detect 225 genes that mark discrete populations of hypothalamic neurons. Complementary DNA (cDNA) was prepared by using the QuantiTect Reverse Transcription Kit, and the abundance of these genes in the pS6 IP and input RNA was quantified by using Taqman Gene Expression Master Mix. Transcript abundance was normalized to rpL27. Differential fold enrichment values were calculated by dividing the average fold enrichment in the stimulus samples by the average fold enrichment in the controls. The log-transformed fold enrichment values for each gene were analyzed by calculating a p value (unpaired two-tailed t test) and a q value to estimate the false discovery rate at different thresholds of significance. See also Tables S1 and S2.
Microarray and RNA-Seq Analysis
For microarray analysis, amplified cDNA was prepared by using the Ovation RNA Amplification System V2 and was hybridized to MouseRef-8 v2 BeadChips (Illumina). For RNA sequencing (RNA-seq) analysis, cDNA was prepared by using the SMARTer Ultralow Input RNA for Illumina Sequencing Kit and then sequenced by using an Illumina HiSeq 2000. See also Table S3.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by funding from the JPB Foundation (J.M.F.) and NIH grant DK083531 (Z.A.K.). The authors have filed a patent related to this work. Dr. Friedman is a Founder of Envoy Therapeutics and holds equity in it and receives a retainer from the company. Envoy holds an option to license this technology from the Rockefeller University.
Footnotes
ACCESSION NUMBERS
The GEO accession number for the microarray and RNA-seq data reported in this paper is GSE40995.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, five figures, three tables, and three movies and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.10.039.
REFERENCES
- Acosta-Galvan G, Yi CX, van der Vliet J, Jhamandas JH, Panula P, An-geles-Castellanos M, Del Carmen Basualdo M, Escobar C, Buijs RM. Interaction between hypothalamic dorsomedial nucleus and the suprachiasmatic nucleus determines intensity of food anticipatory behavior. Proc. Natl. Acad. Sci. USA. 2011;108:5813–5818. doi: 10.1073/pnas.1015551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alivisatos AP, Chun M, Church GM, Greenspan RJ, Roukes ML, Yuste R. The brain activity map project and the challenge of functional connectomics. Neuron. 2012;74:970–974. doi: 10.1016/j.neuron.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MR, Mortrud M, Crum R, Sawchenko P. Role of somatostatin in the regulation of vasopressin secretion. Brain. Res. 1988;452:212–218. doi: 10.1016/0006-8993(88)90025-x. [DOI] [PubMed] [Google Scholar]
- Bruchas MR, Yang T, Schreiber S, Defino M, Kwan SC, Li S, Chavkin C. Long-acting kappa opioid antagonists disrupt receptor signaling and produce noncompetitive effects by activating c-Jun N-terminal kinase. J. Biol. Chem. 2007;282:29803–29811. doi: 10.1074/jbc.M705540200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Lee B, Cho HY, Saklayen S, Obrietan K. Photic regulation of the mTOR signaling pathway in the suprachiasmatic circadian clock. Mol. Cell. Neurosci. 2008;38:312–324. doi: 10.1016/j.mcn.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll I, Thomas JB, Dykstra LA, Granger AL, Allen RM, Howard JL, Pollard GT, Aceto MD, Harris LS. Pharmacological properties of JDTic: a novel kappa-opioid receptor antagonist. Eur. J. Pharmacol. 2004;501:111–119. doi: 10.1016/j.ejphar.2004.08.028. [DOI] [PubMed] [Google Scholar]
- Cowley MA, Smart JL, Rubinstein M, Cerda´n MG, Diano S, Horvath TL, Cone RD, Low MJ. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature. 2001;411:480–484. doi: 10.1038/35078085. [DOI] [PubMed] [Google Scholar]
- Croizier S, Franchi-Bernard G, Colard C, Poncet F, La Roche A, Ri-sold PY. A comparative analysis shows morphofunctional differences between the rat and mouse melanin-concentrating hormone systems. PLoS ONE. 2010;5:e15471. doi: 10.1371/journal.pone.0015471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mateos-Verchere JG, Leprince J, Tonon MC, Vaudry H, Costentin J. The octadecaneuropeptide [diazepam-binding inhibitor (33–50)] exerts potent anorexigenic effects in rodents. Eur. J. Pharmacol. 2001;414:225–231. doi: 10.1016/s0014-2999(01)00771-3. [DOI] [PubMed] [Google Scholar]
- Dielenberg RA, Hunt GE, McGregor IS. “When a rat smells a cat”: the distribution of Fos immunoreactivity in rat brain following exposure to a predatory odor. Neuroscience. 2001;104:1085–1097. doi: 10.1016/s0306-4522(01)00150-6. [DOI] [PubMed] [Google Scholar]
- Elmquist JK, Coppari R, Balthasar N, Ichinose M, Lowell BB. Identifying hypothalamic pathways controlling food intake, body weight, and glucose homeostasis. J. Comp. Neurol. 2005;493:63–71. doi: 10.1002/cne.20786. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai WP, Geffen LB, Blessing WW. Galanin immunoreactive neurons in the human hypothalamus: colocalization with vasopressin-containing neurons. J. Comp. Neurol. 1990;298:265–280. doi: 10.1002/cne.902980302. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Hahm S, Fekete C, Mizuno TM, Windsor J, Yan H, Boozer CN, Lee C, Elmquist JK, Lechan RM, Mobbs CV, et al. VGF is required for obesity induced by diet, gold thioglucose treatment, and agouti and is differentially regulated in pro-opiomelanocortin- and neuropeptide Y-containing arcuate neurons in response to fasting. J. Neurosci. 2002;22:6929–6938. doi: 10.1523/JNEUROSCI.22-16-06929.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suá rez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y, Si S, Pont-Lezica L, Tan T, Kapoor V, Murthy VN, Dulac C. Molecular organization of vomeronasal chemoreception. Nature. 2011;478:241–245. doi: 10.1038/nature10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova EA, Bechtold DA, Dupré SM, Brennand J, Barrett P, Luckman SM, Loudon AS. Altered metabolism in the melatonin-related receptor (GPR50) knockout mouse. Am. J. Physiol. Endocrinol. Metab. 2008;294:E176–E182. doi: 10.1152/ajpendo.00199.2007. [DOI] [PubMed] [Google Scholar]
- Koch C, Crick F. Neural basis of consciousness. In: Smelser NJ, Baltes PB, editors. International Encyclopedia of the Social & Behavioral Sciences. New York: Elsevier; 2001. pp. 2600–2604. [Google Scholar]
- Koch C, Reid RC. Neuroscience: observatories of the mind. Nature. 2012;483:397–398. doi: 10.1038/483397a. [DOI] [PubMed] [Google Scholar]
- Koike K, Sakamoto Y, Kiyama H, Masuhara K, Miyake A, Inoue M. Cytokine-induced neutrophil chemoattractant gene expression in the rat hypothalamus by osmotic stimulation. Brain Res. Mol. Brain. Res. 1997;52:326–329. doi: 10.1016/s0169-328x(97)00300-8. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- LeSauter J, Hoque N, Weintraub M, Pfaff DW, Silver R. Stomach ghrelin-secreting cells as food-entrainable circadian clocks. Proc. Natl. Acad. Sci. USA. 2009;106:13582–13587. doi: 10.1073/pnas.0906426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Denk W. The big and the small: challenges of imaging the brain’s circuits. Science. 2011;334:618–623. doi: 10.1126/science.1209168. [DOI] [PubMed] [Google Scholar]
- Lin D, Boyle MP, Dollar P, Lee H, Lein ES, Perona P, Anderson DJ. Functional identification of an aggression locusin the mouse hypothalamus. Nature. 2011;470:221–226. doi: 10.1038/nature09736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. Neuronal cell types. Curr. Biol. 2004;14:R497–R500. doi: 10.1016/j.cub.2004.06.035. [DOI] [PubMed] [Google Scholar]
- Meikle L, Talos DM, Onda H, Pollizzi K, Rotenberg A, Sahin M, Jensen FE, Kwiatkowski DJ. A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J. Neurosci. 2007;27:5546–5558. doi: 10.1523/JNEUROSCI.5540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O. Physiological roles of ribosomal protein S6: one of its kind. Int. Rev. Cell Mol. Biol. 2008;268:1–37. doi: 10.1016/S1937-6448(08)00801-0. [DOI] [PubMed] [Google Scholar]
- Mistlberger RE. Neurobiology of food anticipatory circadian rhythms. Physiol. Behav. 2011;104:535–545. doi: 10.1016/j.physbeh.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Morgan JI, Curran T. Stimulus-transcription coupling in the nervous system: involvement of the inducible proto-oncogenes fos and jun. Annu. Rev. Neurosci. 1991;14:421–451. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- Nelson SB, Sugino K, Hempel CM. The problem of neuronal cell types: a physiological genomics approach. Trends. Neurosci. 2006;29:339–345. doi: 10.1016/j.tins.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Parker JA, Bloom SR. Hypothalamic neuropeptides and the regulation of appetite. Neuropharmacology. 2012;63:18–30. doi: 10.1016/j.neuropharm.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Porter JP, Potratz KR. Effect of intracerebroventricular angiotensin II on body weight and food intake in adult rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;287:R422–R428. doi: 10.1152/ajpregu.00537.2003. [DOI] [PubMed] [Google Scholar]
- Reaux-Le Goazigo A, Bodineau L, De Mota N, Jeandel L, Chartrel N, Knauf C, Raad C, Valet P, Llorens-Cortes C. Apelin and the proopiomelanocortin system: a new regulatory pathway of hypothalamic α-MSH release. Am. J. Physiol. Endocrinol. Metab. 2011;301:E955–E966. doi: 10.1152/ajpendo.00090.2011. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. USA. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MW, Sipols AJ, Grubin CE, Baskin DG. Differential effect of fasting on hypothalamic expression of genes encoding neuropeptide Y, galanin, and glutamic acid decarboxylase. Brain Res. Bull. 1993;31:361–367. doi: 10.1016/0361-9230(93)90228-4. [DOI] [PubMed] [Google Scholar]
- Sherman TG, Civelli O, Douglass J, Herbert E, Watson SJ. Coordinate expression of hypothalamic pro-dynorphin and pro-vasopressin mRNAs with osmotic stimulation. Neuroendocrinology. 1986;44:222–228. doi: 10.1159/000124649. [DOI] [PubMed] [Google Scholar]
- Siegert S, Scherf BG, Del Punta K, Didkovsky N, Heintz N, Roska B. Genetic address book for retinal cell types. Nat. Neurosci. 2009;12:1197–1204. doi: 10.1038/nn.2370. [DOI] [PubMed] [Google Scholar]
- Stevens CF. Neuronal diversity: too many cell types for comfort? Curr. Biol. 1998;8:R708–R710. doi: 10.1016/s0960-9822(98)70454-3. [DOI] [PubMed] [Google Scholar]
- Sunn N, Egli M, Burazin TC, Burns P, Colvill L, Davern P, Denton DA, Oldfield BJ, Weisinger RS, Rauch M, et al. Circulating relaxin acts on subfornical organ neurons to stimulate water drinking in the rat. Proc. Natl. Acad. Sci. USA. 2002;99:1701–1706. doi: 10.1073/pnas.022647699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton SM, Fitzsimons JT. The effects of centrally administered porcine relaxin on drinking behaviour in male and female rats. J. Neuroendocrinol. 1995;7:165–169. doi: 10.1111/j.1365-2826.1995.tb00743.x. [DOI] [PubMed] [Google Scholar]
- Valjent E, Bertran-Gonzalez J, Bowling H, Lopez S, Santini E, Mata-males M, Bonito-Oliva A, Herve D, Hoeffer C, Klann E, et al. Halo-peridol regulates the state of phosphorylation of ribosomal protein S6 via activation of PKA and phosphorylation of DARPP-32. Neuropsychopharmacology. 2011;36:2561–2570. doi: 10.1038/npp.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen LA, Egecioglu E, Luijendijk MC, Hillebrand JJ, Adan RA, Dickson SL. Acute and chronic suppression of the central ghrelin signaling system reveals a role in food anticipatory activity. Eur. Neuropsychopharmacol. 2011;21:384–392. doi: 10.1016/j.euroneuro.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Villanueva EC, Mü nzberg H, Cota D, Leshan RL, Kopp K, Ishida-Taka-hashi R, Jones JC, Fingar DC, Seeley RJ, Myers MG., Jr Complex regulation of mammalian target of rapamycin complex 1 in the basomedial hypothalamus by leptin and nutritional status. Endocrinology. 2009;150:4541–4551. doi: 10.1210/en.2009-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willesen MG, Kristensen P, Rømer J. Co-localization of growth hormone secretagogue receptor and NPY mRNA in the arcuate nucleus of the rat. Neuroendocrinology. 1999;70:306–316. doi: 10.1159/000054491. [DOI] [PubMed] [Google Scholar]
- Wu Q, Clark MS, Palmiter RD. Deciphering a neuronal circuit that mediates appetite. Nature. 2012;483:594–597. doi: 10.1038/nature10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Zeng LH, Rensing NR, Wong M. The mammalian target of ra-pamycin signaling pathway mediates epileptogenesis in a model of temporal lobe epilepsy. J. Neurosci. 2009;29:6964–6972. doi: 10.1523/JNEUROSCI.0066-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Aravanis AM, Adamantidis A, de Lecea L, Deisseroth K. Circuit-breakers: optical technologies for probing neural signals and systems. Nat. Rev. Neurosci. 2007;8:577–581. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
- Zhou D, Shen Z, Strack AM, Marsh DJ, Shearman LP. Enhanced running wheel activity of both Mch1r- and Pmch-deficient mice. Regul. Pept. 2005;124:53–63. doi: 10.1016/j.regpep.2004.06.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.