Abstract
Autophagy is an important membrane transport pathway that is conserved among eukaryotic cells. Although first described as an intracellular catabolic pathway used to break down self components, autophagy has been found to play an important role in the elimination of intracellular pathogens. A variety of host mechanisms exist for recognizing and targeting intracellular bacteria to autophagosomes. Several intracellular bacteria have evolved ways to manipulate, inhibit, or avoid autophagy in order to survive in the cell. Thus, the autophagy pathway can be viewed as an evolutionarily conserved host response to infection.
Keywords: autophagy, xenophagy, intracellular bacteria
Cellular autophagy
Macroautophagy (hereafter autophagy) is an intracellular degradative process that is highly conserved from yeast to mammals. It involves the formation of double membrane vesicles that sequester cytosolic cargo and transport them to the lysosome for degradation. A basal level of autophagy occurs in most cell types in order to maintain cellular homeostasis through the breakdown of proteins and organelles. This is important for cell survival, especially during periods of limited nutrients [1, 2]. In this process, an isolation membrane forms and expands, which requires the activity of a complex of the autophagy proteins Atg5, Atg12, and Atg16 [3]. This protein complex also promotes the lipidation of Atg8, which is needed for the eventual complete maturation of the autophagosome. Once the isolation membrane expands around a cytoplasmic target and fuses upon itself to form the double membrane autophagosome [4]. These autophagosomes then traffic along the endocytic pathway to eventually fuse with lysosomes so that the cargo can be degraded [5]. In addition to breaking down self components, autophagy has been demonstrated to be an important cell-autonomous defense mechanism against intracellular bacteria [6]. As a result, bacteria have evolved a variety of mechanisms to manipulate autophagy to counteract this immune response. This highlights the importance of autophagy as an ancient, cell- autonomous, innate immune response.
Autophagy as a defense mechanism against intracellular bacteria
The process of autophagic elimination of intracellular pathogens has been termed xenophagy [7]. In this process, intracellular bacteria are targeted to autophagosomal membranes and engulfed into the autophagosome, then delivered to the lysosome for degradation. In this manner, autophagy is able to restrict the intracellular replication of certain bacteria as in the case of group A streptococcus (GAS). GAS is typically an extracellular bacterium, but when internalized into endosomes these bacteria are able to invade the cytosol of the host cell by secreting the pore-forming toxin streptolysin O [8]. The cytosolic GAS are then captured into large membrane-bound organelles that stain positive for LC3 (a mammalian homologue of Atg8), which is a protein attached to autophagosome membranes that plays an important role in formation of the compartments. Eventually, approximately 80% of the intracellular GAS are contained within autophagosomes. In contrast, autophagydeficient cells contained no GAS within autophagosomes. The small GTPases Rab5 and Rab7 are involved throughout these steps of bacterial invasion, endosome maturation, and autophagosome formation [9, 10]. In examining the fate of these bacteria, it was observed that these GAS-containing autophagosomes eventually fuse with lysosomes. By examining bacterial viability in wild type (WT) versus autophagy-deficient host cells, it was demonstrated that autophagy contributes to the killing of most intracellular GAS and prevents GAS replication [8]. Upon entry into the host cytosol the bacteria Salmonella enterica [11], Listeria monocytogenes [12, 13] and Francisella tularensis [14] have also been shown to be targeted by autophagy.
Autophagy has been demonstrated to be an important defense mechanism used to limit infection by Mycobacterium tuberculosis. Earlier work demonstrated that stimulation of autophagy in macrophages resulted in more effective killing of Mycobacterium in vitro [15]. More recent data suggests that autophagy is important in controlling infection in vivo as well. A conditional knockout of Atg5 that eliminated the autophagy pathway in monocytes resulted in mice that were killed faster than WT mice by Mycobacterium and displayed more severe tissue necrosis and lung pathology [16]. These phenotypes appear to be attributed to not only higher bacterial loads in the animals, but also a greater pro-inflammatory response by the autophagy-deficient macrophages. Thus, autophagy in vivo is important in both bacterial clearance as well as prevention of host-inflicted tissue destruction. These studies highlight the importance of autophagy as a cell autonomous innate immune defense mechanism to intracellular bacteria.
Mechanisms for bacterial targeting to autophagosomes
In order for xenophagy to be an effective defense mechanism against intracellular bacteria, the cell has several mechanisms to recognize and target these bacteria to autophagosomes, one of which is ubiquitination. One of the mechanisms that contribute to ubiquitination of bacteria involves the E3 ligase LRSAM1, which localizes to the bacteria through its LRR domain [17]. It then promotes ubiquitination in a process that requires its RING domain. Targeting of the ubiquitinated bacteria to autophagosomes then involves adaptor proteins that are able to interact with both LC3 [18–20] and ubiquitin [21]. These adapter proteins link ubiquitinated cargo to forming autophagosomes. The host protein p62 along with other SLRs (sequestosome 1/p62-like receptors) have been reported to be important in the targeting of intracellular bacteria to autophagosomes, and can be considered as a specialized category of pattern recognition receptors (PRRs).
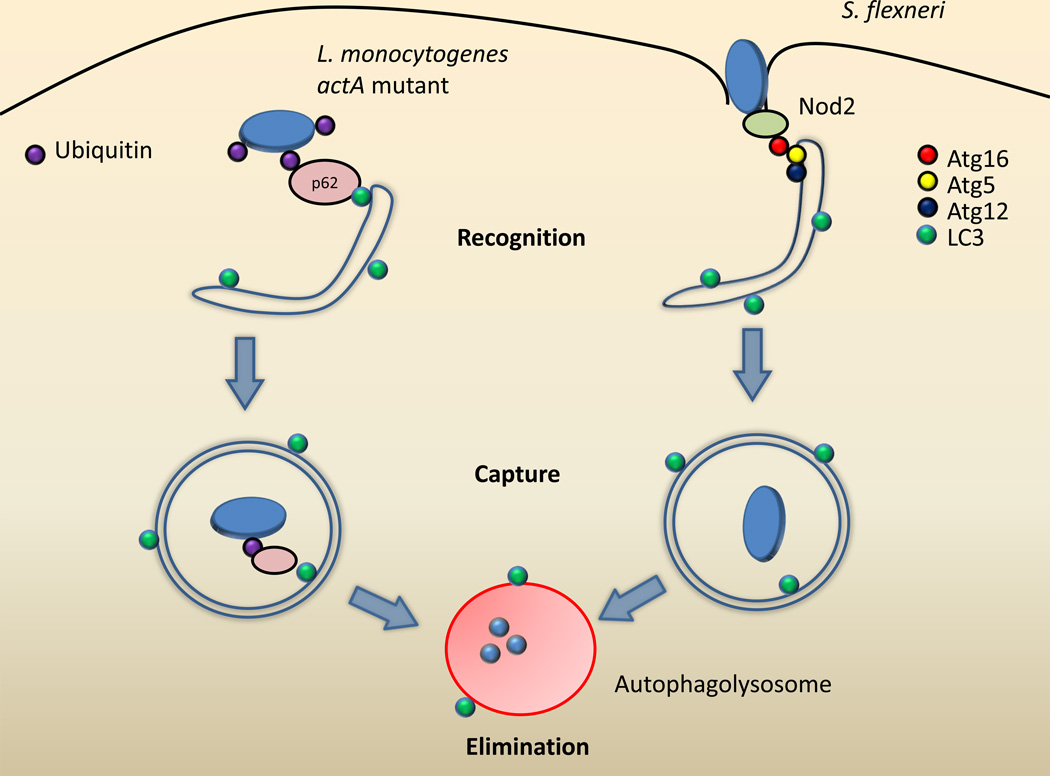
The protein p62 has been demonstrated to be important for the clearance of Listeria and Salmonella species by autophagy [22, 23]. Listeria is a Gram-positive bacterium that escapes from its phagosome into the cytosol using the pore-forming toxin listeriolysin O, where it is able to replicate proficiently [24]. Once in the cytosol, Listeria uses a surface protein called ActA to recruit the host Arp2/3 complex and polymerize actin filaments for motility inside of the cell [25]. However, actA mutants of Listeria that are unable to recruit the Arp2/3 complex are targeted for host ubiquitination, which results in the binding of p62 [22] (Figure 1). LC3 recruitment follows and these mutant bacteria are then eliminated by autophagy as their replication is reduced in WT mouse embryonic fibroblasts (MEF) compared with autophagy-deficient MEFs [22].
Figure 1. Mechanisms for recognition of intracellular bacteria by autophagy.
Listeria monocytogenes actA mutants (left) that are unable to bind to the host proteins Arp2/3 and VASP become ubiquitinated. This recruits p62 to the ubiquitinated Listeria through its ubiquitin-binding domain. The p62 also binds to LC3 on the surface of forming autophagosome membranes, thus targeting Listeria to be captured in the autophagosome where it will be eliminated after lysosomal fusion. Shigella flexneri (right) can be recognized immediately upon entry into the cell by Nod2 localized on the cytoplasmic side of the plasma membrane. Nod2 then recruits Atg16 to this site, targeting Shigella to the forming autophagosome.
NDP52 and optineurin (OPTN) are two other SLRs that have been demonstrated to assist in autophagic targeting of bacteria [26, 27]. Both these SLRs appear to work in conjunction with p62 in the clearance of ubiquitinated Salmonella [26, 27]. Why multiple adaptors exist for targeting Salmonella to autophagy is unclear. Perhaps bacteria have evolved mechanisms to inhibit the function of one or more of these adaptors and the cell has evolved these redundant SLRs in response. Alternatively, these different adaptors may have distinct functions based on their differential localization to Salmonella. Whereas OPTN and NDP52 seem to co-localize to the same areas on Salmonella [27], it was observed that NDP52 and p62 are recruited to different micro-domains of Salmonella and do not co-localize with each other [28]. This suggests a differential spatial ubiquitination on the surface of Salmonella, perhaps indicating that p62 and NDP52 have different targets and serve non-redundant functions. This is evidenced by the observation that NDP52 is distinct from p62 in its ability to directly interact with LC3C (other autophagy studies commonly use LC3B), which is critical in control of Salmonella by autophagy [29]. In addition, OPTN is unique in that it requires phosphorylation by TANK binding kinase 1 for optimal clearance of Salmonella [27]. This may suggest that several distinct targeting steps are required for targeting of bacteria to autophagosomes to ensure that the appropriate cargo is eliminated by autophagy.
Direct recognition by the autophagic machinery of bacteria is another mechanism that cells can use to target bacteria to autophagosomes. Shigella flexneri is a Gram-negative bacterium that also escapes from the phagosome into the cytosol of the host cell. During Shigella infection, Atg5 is able to recognize and bind to the Shigella protein IcsA [30]. This appears to facilitate the recruitment of Shigella to expanding phagophores through the protein Tecpr1 which binds to Atg5 and Atg18 bound to phosphatidylinositol 3-phosphate [PI(3)P] on phagophore membranes [31]. As such, Shigella replicates more efficiently in Atg5-deficient [30] and Tecpr1-deficient [31] cells compared to WT cells. Furthermore, Shigella targeted to these autophagosomes are trapped in septin cage-like structures, restricting their mobility to prevent escape [32].
A form of non-canonical autophagy has also been described in macrophages [33]. This process involves the direct recruitment of LC3 to the phagosome by a process that requires phagocytosis of cargo displaying TLR (Toll-like receptor) ligands. Phagosomes that recruit LC3 show enhanced rates of acidification and fusion with lysosomes [33]. Thus, this non-canonical autophagy pathway rapidly transports bacteria to lysosomes, and provides further evidence that autophagy has been co-opted by the innate immune system to defend against bacteria.
Finally, bacteria also appear to be targeted for autophagy through signaling pathways mediated by classical PRRs. Nod1 and Nod2 are members of the nucleotide-binding oligomerization domain (NOD)-like receptor (NLR) family of cytosolic PRRs [34], and detect bacterial entry into the cytosol by sensing fragments of peptidoglycan [35]. During Shigella infection, Nod1 and Nod2 are recruited to the plasma membrane at the site of bacterial entry [36]. Nod1 and Nod2 then recruit Atg16L1 to this site (Figure 1). Signaling by this complex then promotes the capture of the bacterium in an autophagosome after entry. Furthermore, stimulation of Nod 1 or Nod2 results in higher levels of autophagy [36, 37]. Thus, it is clear that a variety of cellular mechanisms have evolved to detect and target invading bacteria for autophagic destruction, highlighting the importance of autophagy as an innate immune defense against intracellular bacteria.
Bacterial evasion of autophagy
As it has been demonstrated that autophagy is capable of eliminating intracellular bacteria, successful intracellular bacterial pathogens must have evolved mechanisms to avoid autophagic degradation. There have been several reported examples of bacteria that manage to escape autophagy. The common theme among these different bacteria is a strategy of avoiding autophagic detection mechanisms outlined in the previous section.
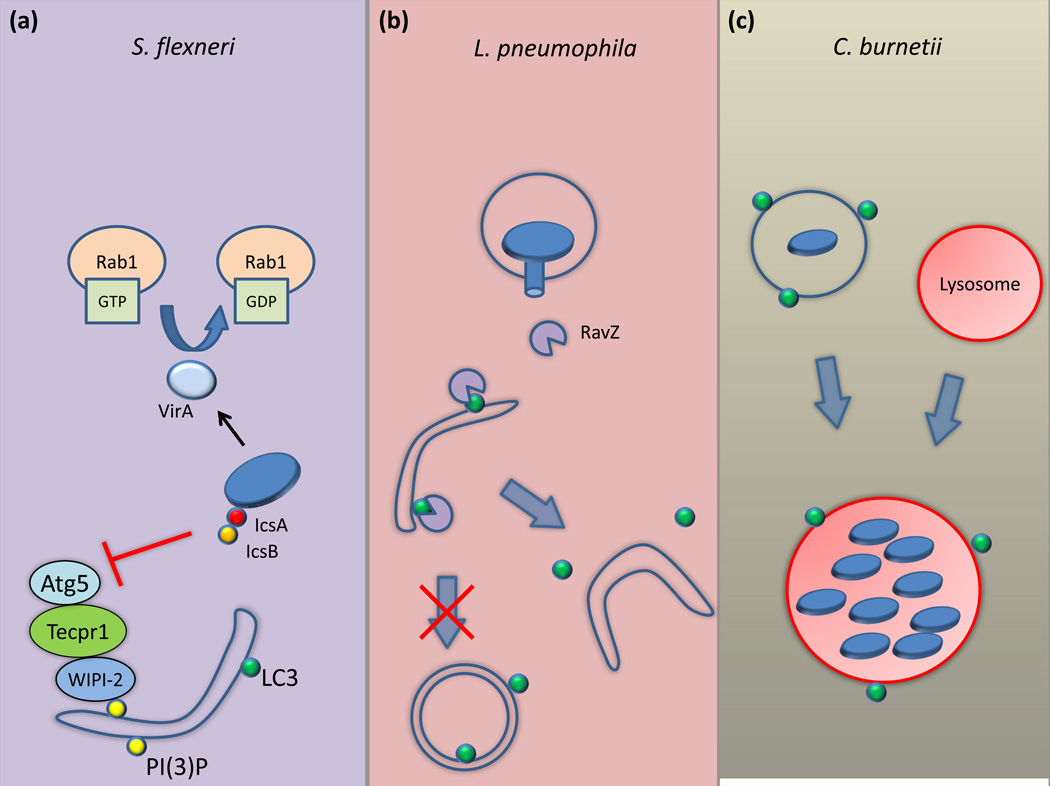
As mentioned previously, Atg5 and Tecpr1 are instrumental in the targeting of Shigella to autophagosomes by binding to the Shigella effector protein IcsA, a protein required for actin-based motility. However, Shigella manages to avoid this recognition by secreting the effector protein IcsB that competitively binds to IcsA, reducing Atg5 binding and Tecpr1 recruitment [30, 31] (Figure 2a). This results in the inability of the cell to target Shigella to autophagosomes. As such, icsB mutants traffic to LC3-positive compartments and do not replicate as well as WT Shigella. Shigella encodes another effector protein, VirA, which is important for autophagy evasion. VirA functions as GTPase activating protein activity that inactivates the host protein Rab1 [38] (Figure 2a). Rab1 function has been linked to autophagosome formation [39], which might explain how inactivation of Rab1 by VirA results in less recruitment of LC3-positive membranes to Shigella [38]. Thus, Shigella has multiple mechanisms for autophagy evasion.
Figure 2. Bacterial manipulation of autophagy.
A) For the Shigella that manage to evade Nod2 detection, it secretes the effector IcsB that binds to the surface protein IcsA. This prevents the binding of Atg5 to IcsA, which would normally target the Shigella to autophagosomes. Shigella also secretes the protein VirA to inactivate Rab1, which disrupts autophagosome formation. B) Legionella pneumophila secretes the effector protein RavZ, which irreversibly cleaves LC3 off of pre-autophagosomal membranes. This prevents the formation of mature autophagosomes. C) Coxiella burnetii replicates in a low-pH LC3-positive vacuole, suggesting that it may exploit the autophagic pathway to create its replicative vacuole.
Listeria can be targeted for autophagic elimination through ubiquitination and recruitment of the adaptor protein p62 [22]. However, most intracellular Listeria avoid this targeting system using an intracellular motility process stimulated by the bacterial ActA protein. ActA directly recruits the host Arp2/3 complex which is important for bacterial motility. Without this ability to recruit these host proteins, Listeria becomes ubiquitinated, after which p62 and recruitment of LC3-positive membranes follows. Alternatively, Listeria has another virulence factor InlK that it also uses to avoid autophagy. InlK is a surface protein that recruits the major vault protein (MVP) to the surface of Listeria [40]. This recruitment of MVP also impedes the recruitment of p62 to Listeria by an unknown mechanism. Interestingly, InlK is poorly expressed in vitro, but highly expressed in vivosuggesting that it is important for the infection process. These multiple independent mechanisms that Listeria and Shigella use for autophagy evasion suggests that this autophagy-bacteria interaction is a constantly evolving arms-race. Over time, the cell has evolved a variety of different mechanisms to recognize and target bacteria for autophagic degradation. Successful intracellular bacteria have in turn evolved multiple mechanisms to evade autophagy to remain one step ahead of the host cell.
Bacterial inhibition of autophagy
Although a variety of bacterial mechanisms used for autophagy avoidance have been reported in the past decade, it was only recently where a mechanism for autophagy inhibition was demonstrated that involved the direct manipulation of the core machinery used for autophagosome formation. It was shown that infection of mammalian cells by the intracellular bacterium Legionella pneumophila results in a global block in host autophagy. Genetic analysis revealed that Legionella encodes a single effector protein called RavZ that is necessary and sufficient to disrupt the autophagy pathway [41]. After RavZ is delivered into mammalian cells by the Legionella Dot/Icm type IV secretion system, this effector targets pre-autophagosomal membranes and functions as a cysteine protease that specifically cleaves the C-terminal region of lipid-conjugated Atg8 proteins such as LC3 (Figure 2b). This cleavage occurs between the conserved C-terminal glycine and an adjacent aromatic amino acid found in all Atg8 family members. Cleavage results in an Atg8 product that is resistant to re-conjugation to membranes as it lacks the reactive C-terminal glycine. Without Atg8 present on the surface of pre-autophagosomal membranes, these structures are unable to mature and develop into autophagosomes. Thus, Legionella is able to inhibit host autophagy through direct interaction with the host autophagic machinery. Interestingly, Legionella lacking RavZ do not have an intracellular growth defect despite being unable to inhibit autophagy [41], which suggests these bacteria evade being targeted by functional autophagy pathway, which would be similar to what is observed for Shigella and Listeria.
Bacterial subversion of autophagy
In contrast to the bacteria that try to avoid autophagic elimination, certain bacteria embrace autophagy and exploit it to support creation of the specialize vacuole in which they replicate. Coxiella burnetii is an intracellular bacterium that traffics along the endosome-lysosome pathway, replicating in a low-pH environment [42]. The vacuole containing Coxiella begins to accumulate LC3 after uptake [43, 44]. Maintaining LC3 on the vacuole containing Coxiella appears to require bacterial protein synthesis, as treatment of cells with the antibiotic chloramphenicol prevents LC3 association [45] (Figure 2c). Because Coxiella requires a type IV secretion system to create the specialized lysosome-like vacuole that supports bacterial replication [46, 47], it is likely that a secreted protein factor is stimulating the recruitment of host vesicles containing LC3 or perhaps LC3 directly to the vacuole. It is possible that LC3 on the Coxiella-containing vacuole (CCV) may be involved in the fusion of membranes that contribute to the generation of the CCV, similar to what has been observed for phagosome maturation stimulated by non-conventional autophagy [33]. Indeed, development of the mature CCV is hampered in cells when LC3-association is blocked [45], which suggests that membrane transport via the autophagy pathway is critical for the biogenesis of the vacuole that supports replication of Coxiella. Thus, the ability of Coxiella seems to exploit autophagy to facilitate biogenesis of a lysosome-like organelle.
Brucella abortus is another intracellular bacterium that uses the autophagic machinery to create its replicative vacuole. Brucella is similar to Coxiella in that the Brucella containing vacuole (BCV) also traffics along the late endosome-lysosome pathway [48]. This is required for induction of a type IV secretion system in Brucella called VirB [49]. The BCV is converted into a vacuole with autophagic features late in the life cycle and this requires the autophagy-initiation proteins ULK-1, Beclin-1, and ATG14L [50]. This process is needed to complete the intracellular life cycle and promote Brucella cell-to-cell spread. This provides another example for how a bacterium that tolerates a low pH environment endosomal environment has evolved the ability to co-opt autophagy for host pathogenesis.
Concluding remarks
From these studies it is clear that autophagy can be an effective cell-autonomous immune response to eliminate intracellular bacteria. However, certain bacteria have evolved mechanisms to avoid autophagic elimination, and in some cases co-opt autophagy for their own replication. Emerging research suggests that the induction of autophagy as a treatment for certain infectious diseases can be an effective strategy both in vitro [51] and in vivo [52]. However, only a few pathogenic bacteria have been examined in detail, and it is unclear at the moment if this can be applied broadly to a variety of different bacteria. Understanding the relationship between specific bacteria and autophagy is important in determining which bacteria would be a suitable target for autophagic elimination. Furthermore, it is known that a variety of different stimuli can trigger anti-bacterial autophagy, such as IFN-γ treatment [53] or TLR stimulation [54, 55]. It will be interesting to determine whether certain methods for autophagy activation are more effective at bacterial elimination than others.
Now that it has been discovered that bacteria have the ability to directly interfere with autophagy, it is important to try and unveil novel mechanisms for autophagy inhibition. Considering the variety of mechanisms that enable autophagy to respond to pathogens, it is likely that pathogens have multiple strategies for disrupting host autophagy that remain undiscovered. Understanding these mechanisms would allow for determining an effective approach for autophagy induction that would counteract bacterial evasion and inhibition of autophagy. Along these lines, it is also important to understand the role of different SLRs in xenophagy and determine if they are redundant or if they serve different functions. Could SLRs that would not be triggered by a certain pathogen under normal infection condition be induced pharmacologically during infection to promote xenophagy and host protection? This would be especially useful in cases where the bacteria have evolved mechanisms to persist intracellularly and sterilization is difficult to achieve using standard antibiotic treatments. In summary, there are more questions than answers at this moment in the field of autophagy-mediated clearance of bacterial pathogens (Box 1), which makes this an exciting area for future investigation.
Box 1. Outstanding questions.
What other mechanisms for bacterial evasion, inhibition, and subversion exist?
Does autophagy contribute to progression of bacterial diseases in humans?
Can induction of autophagy be used to treat a wide variety of bacterial pathogens?
Would certain methods of autophagy induction be more effective than others at eliminating bacteria?
Do different SLRs serve synergistic or redundant functions?
Highlights.
Autophagy is an important immune response used to eliminate intracellular bacteria.
A variety of cellular mechanisms exist to target bacteria to autophagosomes.
Bacteria have evolved different mechanisms to avoid, inhibit or subvert autophagy.
Autophagic induction is being pursued as an approach to treat bacterial infections.
Acknowledgements
This work was supported by National Institutes of Health Grants AI041699 and AI048770.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for selfeating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanada T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 4.Juhasz G, Neufeld TP. Autophagy: a forty-year search for a missing membrane source. PLoS Biol. 2006;4:e36. doi: 10.1371/journal.pbio.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn WA., Jr Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J Cell Biol. 1990;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Nakagawa I, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 9.Sakurai A, et al. Specific behavior of intracellular Streptococcus pyogenes that has undergone autophagic degradation is associated with bacterial streptolysin O and host small G proteins Rab5 and Rab7. J Biol Chem. 2010;285:22666–22675. doi: 10.1074/jbc.M109.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi H, et al. An initial step of GAS-containing autophagosome-like vacuoles formation requires Rab7. PLoS Pathog. 2009;5:e1000670. doi: 10.1371/journal.ppat.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birmingham CL, et al. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 12.Py BF, et al. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy. 2007;3:117–125. doi: 10.4161/auto.3618. [DOI] [PubMed] [Google Scholar]
- 13.Rich KA, et al. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol. 2003;5:455–468. doi: 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- 14.Checroun C, et al. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 16.Castillo EF, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci U S A. 2012;109:E3168–E3176. doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huett A, et al. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella Typhimurium. Cell Host Microbe. 2012;12:778–790. doi: 10.1016/j.chom.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 19.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 21.Geetha T, Wooten MW. Structure and functional properties of the ubiquitin binding protein p62. FEBS Lett. 2002;512:19–24. doi: 10.1016/s0014-5793(02)02286-x. [DOI] [PubMed] [Google Scholar]
- 22.Yoshikawa Y, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 23.Zheng YT, et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 24.Portnoy DA, et al. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haglund CM, Welch MD. Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. J Cell Biol. 2011;195:7–17. doi: 10.1083/jcb.201103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thurston TL, et al. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 27.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cemma M, et al. The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy. 2011;7:341–345. doi: 10.4161/auto.7.3.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Muhlinen N, et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogawa M, et al. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa M, et al. A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe. 2011;9:376–389. doi: 10.1016/j.chom.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 32.Mostowy S, et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe. 2010;8:433–444. doi: 10.1016/j.chom.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 33.Sanjuan MA, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 34.Elinav E, et al. Regulation of the antimicrobial response by NLR proteins. Immunity. 2011;34:665–679. doi: 10.1016/j.immuni.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Rubino SJ, et al. Nod-like receptors in the control of intestinal inflammation. Curr Opin Immunol. 2012;24:398–404. doi: 10.1016/j.coi.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 36.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 37.Cooney R, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 38.Dong N, et al. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell. 2012;150:1029–1041. doi: 10.1016/j.cell.2012.06.050. [DOI] [PubMed] [Google Scholar]
- 39.Zoppino FC, et al. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 40.Dortet L, et al. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog. 2011;7:e1002168. doi: 10.1371/journal.ppat.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choy A, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heinzen RA, et al. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun. 1996;64:796–809. doi: 10.1128/iai.64.3.796-809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beron W, et al. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect Immun. 2002;70:5816–5821. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gutierrez MG, et al. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol. 2005;7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 45.Romano PS, et al. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell Microbiol. 2007;9:891–909. doi: 10.1111/j.1462-5822.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 46.Beare PA, et al. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. MBio. 2011;2:e00175–e00111. doi: 10.1128/mBio.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carey KL, et al. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog. 2011;7:e1002056. doi: 10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Starr T, et al. Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic. 2008;9:678–694. doi: 10.1111/j.1600-0854.2008.00718.x. [DOI] [PubMed] [Google Scholar]
- 49.Boschiroli ML, et al. The Brucella suis virB operon is induced intracellularly in macrophages. Proc Natl Acad Sci U S A. 2002;99:1544–1549. doi: 10.1073/pnas.032514299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Starr T, et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11:33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Campbell GR, Spector SA. Autophagy induction by vitamin D inhibits both Mycobacterium tuberculosis and human immunodeficiency virus type 1. Autophagy. 2012;8:1523–1525. doi: 10.4161/auto.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shoji-Kawata S, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deretic V, et al. Autophagy in immunity against Mycobacterium tuberculosis: a model system to dissect immunological roles of autophagy. Curr Top Microbiol Immunol. 2009;335:169–188. doi: 10.1007/978-3-642-00302-8_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delgado MA, et al. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y, et al. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]