Abstract
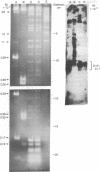
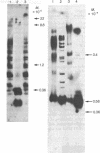
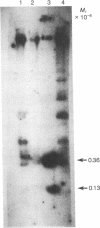
Several continuous lymphoid cell lines have been established from tumors induced by Herpesvirus saimiri. At least a portion of the viral DNA in the marmoset lymphoid cell line 1670, which does not produce detectable virus, is present as covalently closed circular episomal DNA. The use of restriction endonuclease digestion, transfer to nitrocellulose filters, and hybridization of the virus-specific DNA has produced strong evidence that viral DNA sequences present in total 1670 cell DNA and in isolated episomes are extensively methylated. The restriction endonuclease Hpa II has the same recognition sequence as Msp I but, unlike Msp I, fails to cleave when the C of the C-G dinucleotide is methylated. Viral DNA sequences of 1670 cells are refractory to cleavage by Hpa II but not Msp I; greater than 80% of the Hpa II cleavage sites appear to be methylated. Similarly, viral DNA sequences of 1670 cells are refractory to cleavage by Sma I (C-C-C-G-G-G) and Sac II (C-C-G-C-G-G) but not Sac I, Pvu II, or Pst I, which lack the dinucleotide C-G in their recognition sequences. Methylation of mammalian DNA has been previously found exclusively at C residues in the dinucleotide C-G. H. saimiri DNA sequences of another nonproducer cell line, 70N2, also appeared to be extensively methylated, but analysis of total cell DNA extracted from three virus-producing lymphoid lines revealed no evidence of methylation of viral DNA sequences. It remains to be seen if methylation of viral DNA plays a role in the lack of complete expression of H. saimiri genome information in nonproducing lymphoid cell lines.
Keywords: intracellular virus DNA, restriction endonucleases, lymphoma, virus-producing and nonproducing cells
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bornkamm G. W., Delius H., Fleckenstein B., Werner F. J., Mulder C. Structure of Herpesvirus saimiri genomes: arrangement of heavy and light sequences in the M genome. J Virol. 1976 Jul;19(1):154–161. doi: 10.1128/jvi.19.1.154-161.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman J. K., Price P., Pedrinan L., Acs G. Correlation between hypomethylation of DNA and expression of globin genes in Friend erythroleukemia cells. Eur J Biochem. 1977 Nov 15;81(1):53–61. doi: 10.1111/j.1432-1033.1977.tb11926.x. [DOI] [PubMed] [Google Scholar]
- Fleckenstein B., Müller I., Werner J. The presence of Herpesvirus Saimiri genomes in virus-transformed cells. Int J Cancer. 1977 Apr 15;19(4):546–554. doi: 10.1002/ijc.2910190416. [DOI] [PubMed] [Google Scholar]
- Gautier F., Bünemann H., Grotjahn L. Analysis of calf-thymus satellite DNA: evidence for specific methylation of cytosine in C-G sequences. Eur J Biochem. 1977 Oct 17;80(1):175–183. doi: 10.1111/j.1432-1033.1977.tb11869.x. [DOI] [PubMed] [Google Scholar]
- Gunthert U., Schweiger M., Stupp M., Doerfler W. DNA methylation in adenovirus, adenovirus-transformed cells, and host cells. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3923–3927. doi: 10.1073/pnas.73.11.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey J. K., Brackmann K. H., Green M. R., Green M. Preparation and characterization of highly radioactive in vitro labeled adenovirus DNA and DNA restriction fragments. Biochemistry. 1977 Oct 4;16(20):4478–4483. doi: 10.1021/bi00639a023. [DOI] [PubMed] [Google Scholar]
- Marczynska B., Falk L., Wolfe L., Deinhardt F. Transplantation and cytogenetic studies of Herpesvirus saimiri-induced disease in Marmoset monkeys. J Natl Cancer Inst. 1973 Feb;50(2):331–337. doi: 10.1093/jnci/50.2.331. [DOI] [PubMed] [Google Scholar]
- Miller G. The oncogenicity of Epstein-Barr virus. J Infect Dis. 1974 Aug;130(2):187–205. doi: 10.1093/infdis/130.2.187. [DOI] [PubMed] [Google Scholar]
- Munns T. W., Podratz K. C., Katzman P. A. A method for determination of the methylated constituents of transfer ribonucleic acid. Biochemistry. 1974 Oct 8;13(21):4409–4416. doi: 10.1021/bi00718a026. [DOI] [PubMed] [Google Scholar]
- Roy P. H., Weissbach A. DNA methylase from HeLa cell nuclei. Nucleic Acids Res. 1975 Oct;2(10):1669–1684. doi: 10.1093/nar/2.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Biswal N. Studies on the in vivo methylation of replicating herpes simplex virus type 1 DNA. Virology. 1977 Oct 15;82(2):265–274. doi: 10.1016/0042-6822(77)90002-2. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutter D., Westphal M., Doerfler W. Patterns of integration of viral DNA sequences in the genomes of adenovirus type 12-transformed hamster cells. Cell. 1978 Jul;14(3):569–585. doi: 10.1016/0092-8674(78)90243-x. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F. J., Bornkamm G. W., Fleckenstein B. Episomal viral DNA in a Herpesvirus saimiri-transformed lymphoid cell line. J Virol. 1977 Jun;22(3):794–803. doi: 10.1128/jvi.22.3.794-803.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de-Thé G., Geser A., Day N. E., Tukei P. M., Williams E. H., Beri D. P., Smith P. G., Dean A. G., Bronkamm G. W., Feorino P. Epidemiological evidence for causal relationship between Epstein-Barr virus and Burkitt's lymphoma from Ugandan prospective study. Nature. 1978 Aug 24;274(5673):756–761. doi: 10.1038/274756a0. [DOI] [PubMed] [Google Scholar]