Abstract
The promises of cardiac stem cell therapy have yet to be fully realized, in part because of poor survival and engraftment efficacy of implanted cells. Cells die after implantation owing to ischemia, inflammation, immune response, as well as mis-injection or implantation into fibrotic tissue. Imaging tools can help implant cells in areas of the heart most receptive to stem cell therapy and monitor the efficacy of treatment by reporting the viability, location, and number of implanted stem cells. We describe a multimodal, silica-based nanoparticle that can be used for cell sorting (fluorescence), real-time guided cell implantation ultrasound, and high-resolution, long-term monitoring by magnetic resonance imaging (MRI). The nanoparticle agent increased the ultrasound and MRI contrast of labeled human mesenchymal stem cells (hMSCs) 700 and 200% versus unlabeled cells, respectively, and allowed cell imaging in animal models for 13 days after implantation. The agent had no significant impact on hMSC cell metabolic activity, proliferation, or pluripotency, and it increased the production of many paracrine factors implicated in cardiac repair. Electron microscopy and ultrasound imaging suggest that the mechanism of action is in vivo aggregation of the 300-nm silica nanoparticles into larger silica frameworks that amplify the ultrasound backscatter. The detection limit in cardiac tissue was 250,000 hMSCs via MRI and 70,000 via ultrasound. This ultrasound-guided cell delivery and multimodal optical/ultrasound/MRI intra-cardiac cell-tracking platform could improve cell therapy in the clinic by minimizing misdelivery or implantation into fibrotic tissue.
INTRODUCTION
Morbidity and mortality owing to ischemic heart disease continues to be a major clinical challenge in cardiovascular medicine. The therapeutic role of stem cells, including human mesenchymal stem cells (hMSCs) in cardiovascular disease (CVD) such as myocardial infarction, has recently been detailed in a number of animal studies and human clinical trials (1–3). hMSCs have been implicated in a variety of repair mechanisms including the recruitment of endogenous cardiac stem cells, differentiation into important cardiomyocytes and vascular cells, and the release of therapeutic paracrine factors (cytokines, growth factors, and chemokines) that enhance angiogenesis, reduce inflammation, and encourage proliferation of endogenous progenitor cells (4–7). Although there are several clinical trials (NCT 01392625 and NCT 00587990) in progress or completed to study stem cell therapy (SCT) for CVD patients, safe, consistent, and effective results have yet to be demonstrated (1, 3, 5).
Specific limitations to SCT include cell death owing to ischemia, anoikis, or immune response, contamination by undifferentiated cells, and cell delivery into fibrotic tissue. In one of the first human examples of cell therapy, dendritic cells were mis-injected in 50% of melanoma patients (8, 9). In that study, the injection needle was positioned under ultrasound guidance by an experienced surgeon, but real-time cell imaging was not performed, and the poor injection rates were not identified until post-procedure magnetic resonance imaging (MRI) studies 2 days after injection. This work was in the lymph nodes, which is a more straightforward region to inject than in the cardiac tissue (8).
Imaging can improve the efficacy of SCT by ensuring that a sufficient number of cells are implanted in the areas of the heart most receptive to regeneration. Ultrasound is a promising tool for SCT because of its high resolution, low cost, and high depth penetration. Unlike positron emission tomography (PET) and MRI, ultrasound can facilitate the real-time guidance of stem cell implantation. Ultrasound is especially promising for cardiac applications because of the ease and broad clinical acceptance of echocardiography. Although the catheter position is easily monitored via angiography and ultrasound, proper catheter position does not ensure sufficient delivery and immobilization of cells at the desired location, and thus, the development of contrast agents to highlight the transplanted cells is a critical goal (8, 10).
Ultrasound for cell tracking is challenged by a lack of effective imaging agents (11). Although microbubbles have been used for vascular applications, their large size and composition prevent intracellular labeling, which is critical for cell implantation (12). Microbubbles also fail to produce contrast beyond 30 min, which is too short for a typical cell-tracking study that requires imaging for many days. To address this limitation, we studied recent reports detailing submicron ultrasound contrast agents and hypothesized that they could be tailored to include both fluorescent and MR reporters and used for SCT (13, 14). Silica nanoparticles (SiNPs) were particularly attractive because of their high impedance mismatch and use in clinical trials (15, 16). This approach also removes any concerns regarding gas release into the vasculature that may occur with microbubbles (17).
Here, we describe an alternative to MRI-based stem cell imaging via SiNPs that can be detected by fluorescent, MR (Gd3+-doped), and ultrasound imaging. After systemic characterization of their toxicity, cellular distribution, and stability, we used them to image hMSCs via ultrasound in animals after intracardiac implant. These probes offer an intense and stable signal that can be detected in real time. This approach offers a convenient and facile method to monitor cellular implantation in living subjects and monitor their progression throughout an organ of interest.
RESULTS
Multimodal SiNP characterization
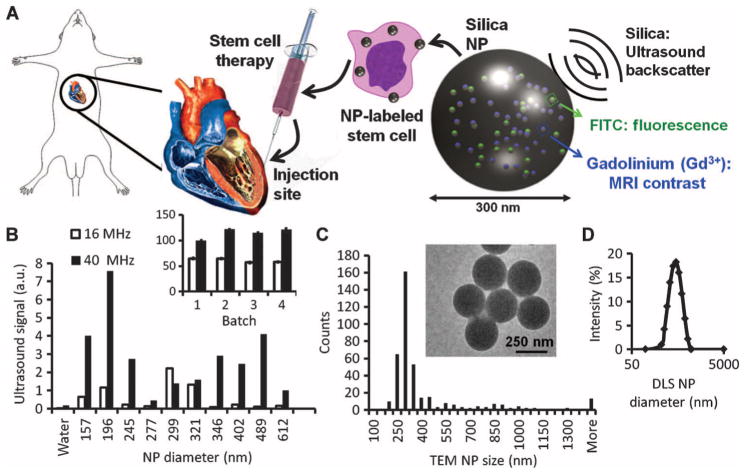
The SiNPs were developed to have fluorescence, ultrasound, and MR signal (Fig. 1A). Different sizes of SiNPs (Fig. 1A) were made by changing the volume of water added during synthesis (18). Ultrasound contrast of the different batches was then evaluated. We selected ~300-nm particles because they had an intense signal at both 40 and 16 MHz (Fig. 1B). Yields of 250 ± 30 mg of NPs per batch were typical. There were no changes to excitation and emission features of fluorescein isothiocyanate (FITC). We characterized the physical size of the product with transmission electron microscopy (TEM) and dynamic light scattering (DLS) (Fig. 1C). TEM imaging of 45 different fields of view (FOVs), each with more than 20 SiNPs, indicated a mode size of 300 nm and a mean size of 403.8 ± 11.4 nm (SEM). More than 80% of the SiNPs were between 200 and 500 nm by TEM. The hydrodynamic radius was also determined by DLS, and the Z average value was 600 nm with a polydispersity index (PDI) of 0.144. The batch-to-batch variation in size of the product for these four different lots was 25% by TEM. The ζ potential was −6.5 ± 6.2 mV in water and −29.0 ± 16.0 mV in 50 mM saline.
Fig. 1.
SiNPs are a multimodal contrast agent for SCT. (A) Cardiac SCT uses hMSCs loaded ex vivo with nanoparticles, which consist of silica (SiO2) framework that backscatters ultrasound (black waves) and stabilizes Gd3+ and FITC fluorophores. (B) The ultrasound signals for 0.5 mg of different-sized SiNPs in an agarose phantom are shown at 40 and 16 MHz. Inset, batch-to-batch variability in ultrasound signal of 5 mg of SiNPs. (C) The size distribution within the 299-nm batch size selected from (B) had a mode size of 300 nm via TEM. Inset, TEM image of SiNPs. (D) DLS of the 299-nm batch.
We studied the ultrasound, fluorescence, and MRI signaling capacity of the SiNPs. The SiNPs were diluted from 10 and 0 mg/ml in an agar phantom and analyzed at 16- and 40-MHz ultrasound with 100% power, gain of 20 dB, and dynamic range of 50 dB. The limit of detection (LOD; also known as sensitivity) at 40 MHz was 11.5 μg/ml (4.0 × 108 SiNPs/ml) and 18.5 μg/ml (6.4 × 108 SiNPs/ml) at 16 MHz. The relative SD (RSD) in ultrasound intensity, as measured at replicate samples (n = 4) of 1.25 mg/ml, was 3.0 and 2.4% at 40 and 16 MHz, respectively (intrabatch RSD). The variation (RSD) between batches was 11.1% at 40 MHz and 5.5% at 16 MHz. For fluorescence imaging, the LOD was 10.6 μg/ml.
The T1-based MR signal of the SiNPs was evaluated with tissue phantoms and spin echo imaging. The LOD was 29.0 μg/ml. Concentrations above 10 mg/ml showed a decreased signal because of water exclusion. Finally, the relaxivity of the SiNPs was measured via decreasing concentrations of material and found to be 6.0 × 106 mM−1 s−1 at 1.0 T with batch-to-batch signal variation of 1.04%. The relaxivity per Gd3+ was 4.1 mM−1 s−1. The amount of Gd3+ per SiNP was determined by inductively coupled plasma (ICP) analysis to be 147 × 104 ± 3.06 × 104 (SD) for nine replicate samples. ICP analysis indicated that 3.22 and 4.75% of the Gd3+ became dissociated from the SiNP at 2 and at 24 hours of incubation in serum at 37°C, respectively. Incubation in water caused 1.80% dissociation. The RSD of these measurements was <5%.
Cell labeling
hMSCs were incubated for 7.5 hours with SiNPs (0 to 1.0 mg/ml) to measure endocytosis (fig. S1A). The incubation time was evaluated in a separate experiment using SiNPs (0.5 mg/ml) between 1 and 18 hours (fig. S1B). Positive (fluorescent) hMSCs ranged from 59.0 to 79.6% of the total population, with the highest percentages at 0.5 mg/ml. We also saw limited gains in fluorescent signal between 6 and 18 hours. Thus, SiNPs (0.5 mg/ml) for 6 hours were used for all subsequent labeling protocols. ICP analysis of the cells indicated 6540 ± 620 SiNPs per cell or 92.4 μm3 of material. The hMSCs had a volume of 2.3 × 105 μm3, and the SiNPs occupied 0.5% of the cell volume at an intracellular concentration of 1.0 mg/ml. To measure the temporal stability of SiNP loading, we replated 25,000 loaded hMSCs in each well of a 12-well plate. The fluorescent signal was measured daily and suggests that, by 48 hours, the cell intensity is reduced by 50%, which corresponds to the doubling time of 2 days (fig. S1C).
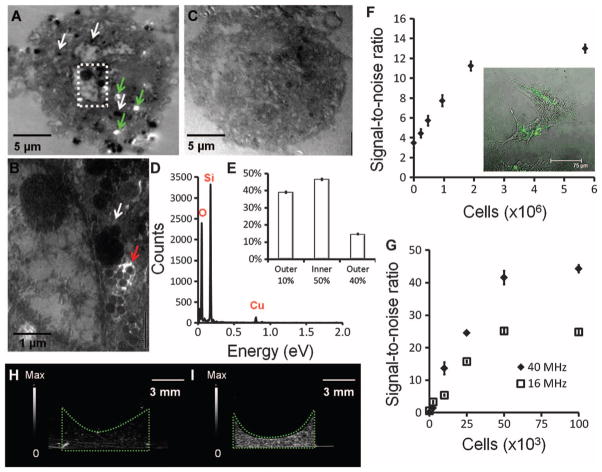
TEM images of SiNP-labeled hMSCs show dark clusters that correspond to SiNPs (Fig. 2A). Individual SiNPs could be seen at higher magnification (Fig. 2B). Control hMSCs that were not exposed to SiNPs did not have these darkened areas (Fig. 2C). These areas were confirmed to be SiNPs via energy-dispersive spectroscopy (Fig. 2D). The TEM images of 40 unique 500-nm sections indicated that 16.6 ± 3.5 (SEM) SiNP clusters were located per slice with a mode SiNP size of 1100 nm and a mean size of 1400 ± 500 nm (SD). Extrapolation to the total cell volume suggests 850 SiNPs of these large clusters per cell.
Fig. 2.
Imaging hMSCs loaded with SiNPs. hMSCs were fixed, stained, sectioned, and placed on copper grids for TEM imaging. (A and B) SiNP-loaded cell. White arrows point to SiNPs distributed throughout the cell. White areas indicate ripping of the epoxy resin support matrix that occurs because of SiNPs during microtoming (green arrows). Area in box is subjected to higher magnification in (B) and shows individual SiNPs (blue arrow) and SiNP clusters (white arrow). (C) Control, unloaded cell. (D) The dark areas inside the hMSCs were confirmed to be SiNPs by energy-dispersive spectroscopy, which showed the characteristic Si peak at 1740 eV. (E) Distribution of SiNP location in hMSCs throughout 45 cells. Data are means ± SEM. (F) Ultrasound signal versus noise as a function of cell number loaded into the agarose phantom. Data are averages ± SD (n = 5 replicate measurements at each concentration for one batch and performed for all batches). (G) T1-weighted spin echo MR signal was measured as a function of cell number in the agarose phantom. (H and I) Echogenicity of 50,000 hMSCs without SiNPs (H) and 50,000 SiNP-loaded hMSCs (I) imaged at 40 MHz. The green dotted line highlights the cell pellet in an agarose phantom.
TEM also determined the location of the SiNPs. The location was assigned to either the center 50% of the cell area, the outer 40% of the cell area, or the periphery (outer 10%) of the cell. SiNPs were present on both the periphery and interior of the cell (Fig. 2E). Confocal microscopy confirmed that SiNPs are located throughout the hMSC interior (Fig. 2F, inset). The hMSCs were placed in an agarose phantom and imaged with ultrasound (Fig. 2G). Labeled cells had 3.0- and 2.4-fold higher ultrasound B-mode signal at 40 and 16 MHz, respectively (Fig. 2, H and I). When different cell counts were imaged, the LOD at 16 MHz was 1430 hMSCs and 635 cells at 40 MHz (Fig. 2G). Cells were also imaged with T1 spin echo MR imaging (Fig. 2F) for a LOD of 338,000 hMSCs. The T1 signal intensity of 9.5 × 105 SiNP-loaded hMSCs was twofold higher than that of an equivalent number of unloaded cells.
We found that size plays an important role in the generation of contrast. The SiNPs were allowed to settle overnight and separate into the larger, irregular sedimented particles (1% of total) and the remaining supernatant (fig. S3A). The sediment yielded ultrasound signal in phantoms 36-fold higher than that of the smaller supernatant (fig. S3B). However, when these two fractions were used to label cells, there was no statistical difference between the two populations (P = 0.38, two-tailed t test), with a sevenfold increase in signal above unlabeled cells at 40 MHz (fig. S3C). This suggests that even the <300-nm SiNPs can aggregate inside a cell to produce larger aggregates that modulate backscatter.
To confirm that the SiNPs were compatible with clinical ultrasound imaging systems, we scanned SiNPs alone, SiNP-loaded hMSCs, and an agarose phantom blank using a preclinical and a clinical scanner. The LOD of contrast agent with the clinical scanner was SiNPs (17 μg/ml) and 14,000 SiNP-labeled hMSCs. The signal-to-background ratio of these cells versus the agarose/saline vehicle was 9.2 and 10.9 for the 40- and 16-MHz pre-clinical transducers, respectively. On the clinical system, the values were 8.3 and 6.7 for the 5- and 10-MHz transducers, respectively, suggesting that this contrast agent is suitable for use in clinically deployed imaging systems.
Impact of SiNPs on hMSCs
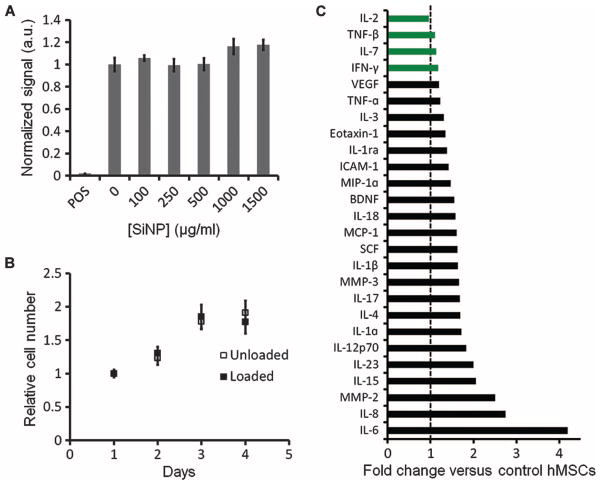
We next performed a series of experiments to measure the impact of SiNPs on hMSC metabolism, viability, proliferation, cytokine expression, and pluripotency. Metabolic activity was perturbed by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay and indicated that there was no significant (P > 0.05, two-tailed t test) difference between the cells with no SiNPs and those up to 1.5 mg/ml (Fig. 3A). Cell proliferation was also studied with MTT. There was no difference in the growth rate between the SiNP-loaded and unloaded cell populations (Fig. 3B). A cell migration assay demonstrated that unloaded and SiNP-loaded cells both migrated to an area depleted of hMSCs within 24 hours (fig. S2). After 5 min of imaging with either 16- or 40-MHz ultrasound, we saw no morphological changes in either control (unloaded) or SiNP-loaded cells and saw no vacuole formation by calcein staining, suggesting that ultrasound imaging does not induce vacuole formation.
Fig. 3.
Influence of SiNPs on hMSC metabolism, proliferation, and cytokine secretion. (A) Triplicate wells of 20,000 cells were incubated with SiNPs or media only, and their metabolic activity was determined by MTT assay. “POS” indicated positive control of hMSCs with cytotoxic agent. (B) Growth rates of loaded and unloaded cells over time. (C) Cytokine expression levels in cell culture media from SiNP-loaded hMSCs are shown as fold change versus control (unloaded) hMSCs. Black bars indicate a statistically significant (P < 0.05) change in expression; green bars indicate nonsignificance (P > 0.05), two-tailed t test. Data in (A) and (B) are means ± SEM.
To determine whether hMSC metabolism caused leaching of Gd3+ from the SiNPs, we replated hMSCs loaded with SiNPs after loading and allowed them to grow for 24 hours. The media from the wells (n = 4) had a Gd3+ content of 7.7 ± 5.0 (SD) ng/liter, and control media without any hMSCs or SiNPs (n = 3) had a Gd3+ content of 7.4 ± 1.7 (SD) ng/liter. This shows no increase in Gd3+ content as a result of incubating SiNP-loaded hMSCs.
Secretome analysis by Luminex assay of SiNP-loaded and control hMSCs indicated that 26 of the 31 analyzed proteins were measurable in cell culture media. Cytokine concentrations from the SiNP-loaded cells were within onefold of control hMSC concentrations, except for interleukin-6 (IL-6), IL-8, and matrix metalloproteinase-2 (MMP-2) (Fig. 3C).
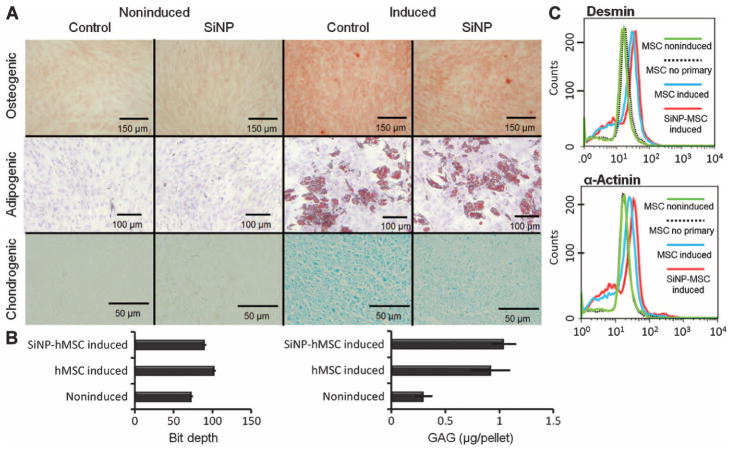
We next determined whether SiNPs affected the pluripotency of the loaded hMSCs. We were especially concerned that the SiNPs might transform hMSCs into osteogenic cells because silicon-based structures have previously been shown to induce osteogenic differentiation (19). SiNP-loaded cells differentiated into both osteogenic and adipogenic lineages similar to control (unloaded) hMSCs (Fig. 4A). Cell labeling with nanoparticles has been reported to inhibit chondrogenesis in hMSCs (20). Although the SiNP-loaded cells differentiated down the chondrocyte lineage, we noted a reduced staining intensity compared with unloaded controls (Fig. 4B). Nevertheless, the faint but consistent staining suggests that this pathway does remain intact. We further confirmed the transformation into chondrocytes by assaying for glycosaminoglycans (GAGs) with dimethylmethylene blue (DMMB) (Fig. 4B).
Fig. 4.
hMSC pluripotency is retained after labeling with SiNPs. (A) Cells loaded with SiNPs or unloaded cells were plated and cultured in either control media (noninduced) or differentiation media (induced). Osteogenic lineage was confirmed with Alizarin Red S staining. Adipogenesis (presence of lipid vacuoles) was confirmed with Oil Red O. Chondrogenesis was confirmed by Alcian blue staining. Images are representative of at least three replicates. (B) Chodrogenesis was quantified by measuring pixel density [in (A)] or amount of GAGs via DMMB assay. Data are means ± SD for three pellets. (C) Flow cytometry data of desmin and α-actinin expression in induced and noninduced hMSCs.
Finally, levels of α-actinin and desmin were determined by flow cytometry in cells treated and untreated with 5-azacytidine (21) (Fig. 4C). The α-actinin levels in SiNP-labeled hMSCs and control hMSCs treated with 5-azacytidine were 1.9- and 1.4-fold higher than nontreated hMSCs, respectively. The desmin levels in SiNP-labeled hMSCs and control hMSCs treated with 5-azacytidine were 2.1- and 1.9-fold higher than nontreated hMSCs, respectively. Control cells stained with secondary antibody only were identical to cells untreated with 5-azacytidine.
Tracking SiNPs in preclinical cardiac applications
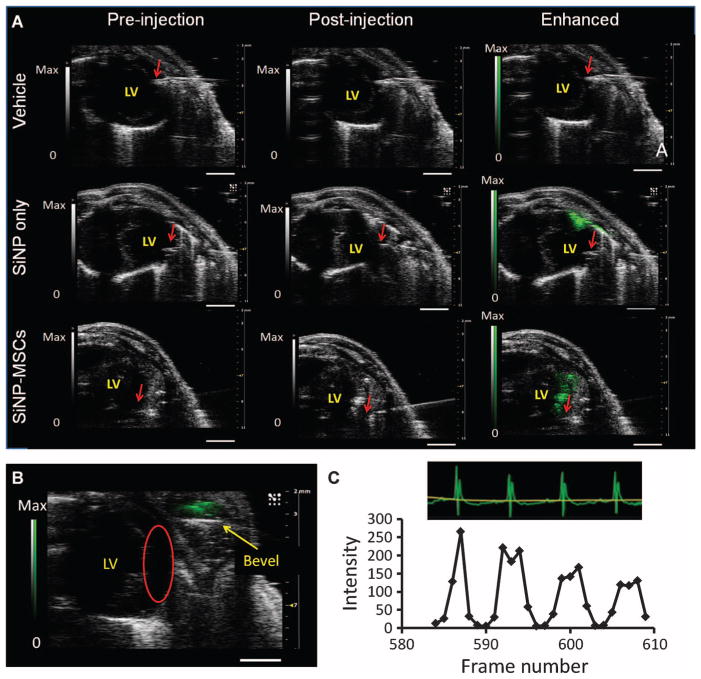
To evaluate the utility of these labeled cells in vivo, we imaged labeled cells with ultrasound and MR in the left ventricle (LV) wall of healthy nude mice and then confirmed this signal with histology. SiNP-labeled hMSCs were injected in a 1:1 saline/Matrigel mixture into the LV wall of healthy mice (n = 3 at different cell numbers). Control mice received the vehicle or SiNPs only (n = 3 each). All echocardiograms were in short-axis mode. Ultrasound was collected during injection, and MR images were collected before and 15 min after injection. In the vehicle controls, there was no increase in B-mode signal (Fig. 5A), indicating that there was no trauma during injection that could lead to increased ultrasound backscatter. SiNPs alone increased B-mode signal 3.8-fold at the site of injection compared with saline-injected controls (Fig. 5A). When 106 SiNP-loaded hMSCs were injected into the LV wall, B-mode signal increased 6.4-fold at the injection site (Fig. 5A). This was higher than SiNPs alone because of the signal from hMSCs and SiNPs and intracellular aggregation. Movie S1 shows real-time injection of labeled hMSCs into the LV. Cells are clearly seen entering at the 0:11 time point. The use of ultrasound imaging allowed immediate identification of any mis-injected cells (Fig. 5B). We also observed movement of the hMSC bolus in ultrasound relative to the LV (as determined by electrocardiogram) and observed precise integration between the two (Fig. 5C and movie S2).
Fig. 5.
Ultrasound imaging of hMSCs after intracardiac implantation in mice. (A) Short-axis views of the LV of nu/nu mice. Leftmost panels are after insertion of a needle catheter, but before injection of the vehicle, SiNPs (6 mg), or 106 SiNP-loaded hMSCs. The middle panel is immediately after injection of pay-load. The rightmost images are enhanced versions of the post-injection image after background subtraction and image analysis. The red arrow represents the bevel of the needle catheter. Images are representative of three mice per payload group. Scale bars, 2 mm. (B) Ultrasound can immediately identify mis-infection. Red circle, target site. (C) Subset of mouse electrocardiograms after injection of an SiNP-labeled hMSC bolus (500,000 hMSCs). This section was chosen because it was in between respiratory cycles. The signal of a region of interest (ROI) containing the hMSCs was measured as a function of time (frame rate number).
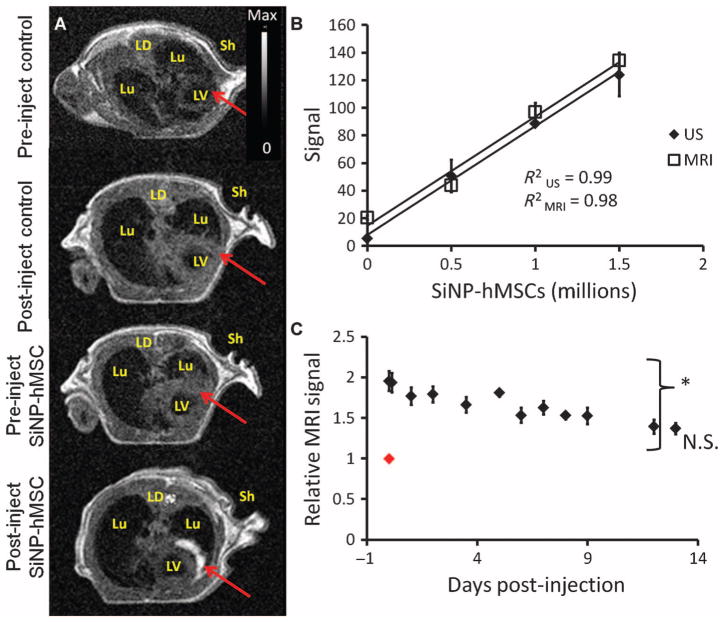
The hMSCs could also be imaged with MR after ultrasound-guided injection. Figure 6A shows axial sections of the murine chest cavity before and after injection for animals administered either vehicle control (n = 3) or SiNP-loaded hMSCs (n = 3). The labeled cells are clearly visible after injection, and the T1-weighted signal in the treated area increases 2.5 ± 0.18–fold versus preinjection images. To confirm that hMSCs were embedded in the wall of the LV, we measured the wall thickness before (2.25 ± 0.1 mm) and after (2.49 ± 0.4 mm) injection via T1 SE images and found that there was no significant difference between the two for any of the animals (P > 0.05, two-tailed t test). hMSCs outside the LV wall would have caused a much larger change.
Fig. 6.
MR imaging of hMSCs after ultrasound-guided intracardiac delivery. (A) TI SE axial sections of the chest cavity of nu/nu mice including pre- and post-injection images for the vehicle control and hMSC experiments. Lu, lung; Sh, left shoulder; LD, latissimus dorsi muscle. Red arrow indicates injection site before and after implant. (B) MRI and ultrasound (US) signal as function of the number of injected SiNP-loaded cells. (C) Animals injected (on day 0; red datum) with 106 hMSCs were monitored sequentially for 13 days after injection. Data in (C) are means relative to day 0 ± SEM (n = 3 animals per sample population). *P < 0.05 versus day 0, two-tailed t test.
Cell quantification in vivo was demonstrated by injecting increasing concentrations of SiNP-hMSCs into the LV wall (n = 3 animals per concentration). The animals were imaged with both ultrasound and MR. The intracardiac LOD was 250,000 hMSCs via MRI and 70,000 via ultrasound (Fig. 6B). The subjects injected with 106 cells were also monitored longitudinally via MRI. The MR signal stayed significantly elevated above baseline for 13 days (Fig. 6C).
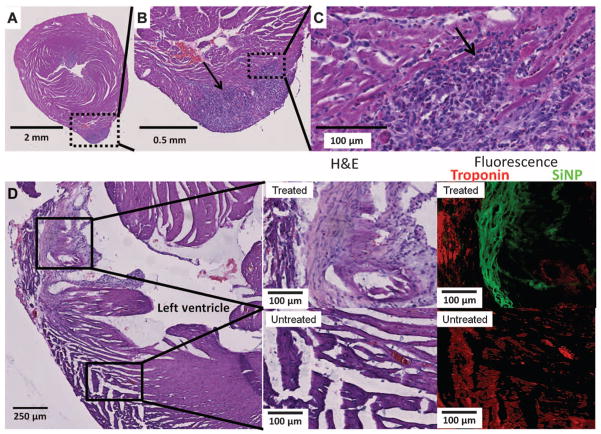
We confirmed that the signal seen by ultrasound and MRI correlates to hMSCs via histology. In one study, mouse hearts (n = 5) were excised 24 hours after injection of SiNP-hMSCs and imaged with hematoxylin and eosin (H&E) staining. Figure 7 (A to C) shows increasing magnification of the LV injection site. The darker purple area protruding from the ventricle wall in Fig. 7A is hMSCs as determined by morphology. In another set of mice (n = 3), the heart was removed 10 days after implant and imaged with H&E as well as fluorescence in both treated (Fig. 7D, red box) and untreated (Fig. 7D, blue box) areas of the heart. Although both treated and untreated areas were positive for troponins, only the treated area presents green fluorescence indicative of the hMSCs (Fig. 7D).
Fig. 7.
Histology validation of in vivo imaging. We used histology and immunofluorescence to image cardiac tissue after hMSC transplant. (A to C) H&E stains of cardiac tissue under increasing magnification. This sample was explanted 1 day after treatment with 5 × 105 hMSCs in the LV wall. Arrow points to dark purple hMSCs. The dashed box illustrates the area subjected to increased magnification in the following panel. (D) H&E stains of the LV and LV wall explanted 10 days after cell injection. Boxes indicate area treated with SiNP-hMSCs (top) and untreated area (bottom). Middle panels are increased magnification of treated and untreated areas. Rightmost panels are immunofluorescence from an adjacent slice from the same tissue sample. Red, troponin immunofluorescence; green, SiNP-hMSC fluorescence.
Four days after treatment, we desanguinated animals with no injection, sham injection (vehicle only), 500,000 hMSC injection, and 500,000 SiNP-hMSC injection. Serum was tested for liver enzymes, electrolytes, and cardiac enzymes (table S1). The liver enzyme alkaline phosphatase was significantly modulated for the SiNP-hMSC and hMSC groups relative to no injection animals and sham injection animals. However, these levels are still well within the normal reference range of rodents [alkaline phosphatase (20 to 275 U/liter)] (22, 23). The SiNP-hMSC group also showed increased total bilirubin and carbon dioxide levels and decreased anion gap relative to uninjected animals, but again within the reference range. All other values were statistically the same as untreated animals by two-tailed t test.
DISCUSSION
We used SiNPs to label hMSCs via a clathrin- and actin-dependent endocytosis (24) and then implanted the cells in living animals. The fluorescent mode was used during cell culture and histology to confirm the presence of SiNPs within hMSCs and heart tissue. Ultrasound was used during the implantation step and offered real-time guidance of the catheter and payload. The ultrasound signal was quantifiable, so we could obtain a cell count immediately upon implantation. Finally, the MR mode offered high-resolution imaging to confirm the location of hMSCs and longitudinally track cells. The SiNPs are highly echogenic and are compatible with clinical and preclinical ultrasound frequencies. We used ultrasound backscatter signal for image quantitation. This signal will change with different imaging systems and settings (dB scale, normalization, beamforming, saturation, etc.). Careful calibration will be needed when transitioning between imaging systems. The radio frequency signal could also be used for quantitative results. Higher frequencies are useful for high-resolution imaging, whereas lower frequencies offer a better depth of penetration (25). The temporal resolution achieved with this technique was 3.3 ms, which is 10,000-fold lower than PET/SPECT (single-photon emission computed tomography), which is a real-time, deep tissue, cell-tracking technique (26).
Previous work with ultrasound imaging in regenerative medicine has successfully used gold microcapsules (27) or perfluorcarbon droplets (28) as ultrasound contrast agents for imaging pancreatic islet transplantation in mice. Photoacoustic imaging has been used for surface imaging of hMSCs, but is limited by optical scatter and absorption by tissue (29, 30). Microbubbles are highly echogenic contrast agents but offer poor ultrasound signaling capabilities in cell-tracking applications because of their large size and short half-life (31). The SiNPs use backscatter mode to increase the impedance mismatch—increasing both the size and the number of nanoparticles increases signal (17). Although we could have used only larger SiNPs rather than rely on nanoparticles to aggregate, the micrometer-sized aggregates present several difficulties. First, they are only a tiny fraction (typically <1%) of the yield. As a byproduct of the reaction, it is impossible to control the size, morphology, porosity, and surface chemistry of these aggregates. Third, the membrane transport properties and cytotoxicity profiles of the larger aggregates are much more erratic because of the large PDI (0.8) within any given population. Thus, we used the smaller, monodisperse SiNPs for imaging and allowed the hMSC to enhance ultrasound signal.
hMSC action has been shown to be partially secretome-mediated (4, 7), and physiological preconditioning, genetic manipulation, molecular preconditioning, or pharmacological treatment can increase the expression of these beneficial agents by hMSCs (5). Secretome analysis showed a general increase in levels of cytokines with the addition of SiNPs. Many proteins broadly implicated in cardiac regeneration, including vascular endothelial growth factor (VEGF), MMP-2, stem cell factor (SCF), and monocyte chemotactic protein-1(MCP-1), were significantly up-regulated after SiNP labeling. Thus, the SiNPs facilitated cellular imaging while increasing the output of beneficial cytokines. Stem cell differentiation is another potential mechanism of action, and the differentiation capacity of labeled hMSCs is retained.
The particles contained free Gd ions for MR contrast, which is a drawback for clinical translation at this point. Although the toxicity of free Gd3+ is an important concern owing to cases of nephrogenic systemic fibrosis (32), less than 5% of Gd3+ becomes dissociated after 24 hours of incubation with mouse serum at physiological temperature. If 1 × 106 cells were implanted, the corresponding Gd dose would be 9.6 × 1015 Gd3+ (6540 SiNPs/hMSC; 1.47 × 106 Gd3+/SiNP) or 2.5 μg of gadolinium (assuming 100% dissociation of Gd3+ from SiNP). From the package insert of the clinically approved gadolinium-containing agent, Prohance, the recommended dose is 0.1 mmol/kg. For an 82-kg individual, this corresponds to 1.29 × 106 μg. However, this Prohance dose is chelated unlike our SiNPs, but the chelation has a dissociation half-life of 3 hours and circulation time of 1.5 hours, resulting in a dose of free Gd3+ of 4.6 × 105 μg at 1.5 hours (33). The Gd dose from the SiNPs is 184,000-fold lower than this dose. Patients with renal disease would be excluded from any study using SiNPs. Future work with these SiNPs and this contrast agent will use better chelation systems.
Another limitation of this work is that the contrast agent was diluted during successive cell generations (fig. S1C). The initial intra-cellular concentration was 1 mg/ml. Assuming even distribution of SiNPs between daughter cells, the cells will still have a concentration above the detection limit through generation 5 (30 μg/ml). Histology suggested that the ultrasound signal is indeed representative of hMSCs that have taken up SiNPs, but we cannot rule out uptake by macrophages, which is commonly reported (34). Fluorescence was generated from the SiNPs and not the hMSCs; thus, some free SiNPs resulting from dead hMSCs may have been endocytosed by macrophages. However, because the probe is small and intense, we could label only 5 to 10% of the cells and use them to estimate the entire cohort. This percentage would still be feasible in clinical applications because millions of cells will be used in humans (35), and the 10% that are labeled would still be within the detection range of the assay.
These studies were performed in mice and have smaller organs that are more readily accessible to high-frequency ultrasound imaging. Nevertheless, the SiNPs still had excellent signaling capabilities with clinical systems. Echocardiography is already deployed in many areas of cardiology, and transitioning to ultrasound-based imaging has a low clinical barrier. Ultrasound also facilitates more accurate intra-cardiac injection in addition or intracoronary delivery. Future work toward clinical translation should include biodistribution studies and tuning the silica biodegradation time.
Some researchers are developing MRI-compatible catheters for SCT applications (9). Although plausible, such an approach would not have the temporal resolution or cost-benefit of ultrasound. There is currently no alternative technique to allow real-time imaging of hMSC delivery with the temporal resolution and sensitivity of ultrasound. Our contrast agent allows one to differentiate between hMSCs and the surrounding tissue in vivo for multimodal (optical/MR/ultrasound) imaging in the clinic.
MATERIALS AND METHODS
Particle synthesis and characterization
SiNPs were synthesized through a modified Stober synthesis (18). An organosilane conjugate of FITC (Aldrich) was created by mixing 5.25 mg of FITC with 73 μl of (3-aminopropyl)-triethoxysilane (Alfa Aesar) in 1 ml of ethanol (Gold Shield) overnight, and confirmed by mass spectrometry. The next day, 25 ml of ethanol, 1.5 ml of 30% ammonium hydroxide (EMD), 12.5 mg of GdCl3, and 1.5 ml of distilled, deionized (DDI) water were added to a small Erlenmeyer flask with 100 μl of the silanized FITC product and mixed with a stir bar at 500 rpm. Two 1-ml aliquots of tetraethylorthosilicate (Acros) were added in rapid succession. The mixture turned from a translucent yellow color to opaque over ~2 hours, and the reaction was allowed to proceed overnight at room temperature.
The size of the resulting nanomaterial was monitored by DLS (Zetasizer-90; Malvern). After SiNP growth was completed, the particles were pelleted at 6000 rpm (4629 RCF) on an Eppendorf 5804 centrifuge for 10 min and washed thrice with ethanol and once with water. To remove any remaining free FITC or Gd3+, we dialyzed the particles overnight versus constantly refreshed water. Concentration values were determined by drying known volumes of the material overnight in a 140°C oven and measuring the resulting mass. The reproducibility of four batches was compared.
Cell culture and labeling
The hMSCs and growth media were purchased from Lonza and were used between passage 2 and 20. Cells were passaged when they reached 80% confluence with TripLE Express (Invitrogen) with about 3 to 7 days between each passage. Cells of the following passage number were used for the experiments: cell proliferation (2 to 6), MTT cell toxicity (4 to 8), secretome (3), cell differentiation (2 to 3), optimization of SiNP labeling (4 to 12), ex vivo phantoms (4 to 20), and in vivo implantation (3 to 8). Labeling with SiNPs was done without any exogenous transfection agents. SiNPs were added to media and allowed to incubate for 1 to 18 hours. The adherent cells were washed three times with phosphate-buffered saline (PBS) before removal from the flask. Cells were resuspended in PBS before flow cytometry analysis, ultrasound analysis, microscopy at 10×, or other analysis.
Cell proliferation was studied after labeling cells with SiNPs using MTT (Biotium). hMSCs (20,000) were incubated with SiNPs (0 to 1.5 mg/ml) for 6 hours, and their metabolic activity was then probed with the MTT assay. Cetyltrimethylammonium bromide is a cytotoxic agent and served as a positive control. MTT proliferation assays plated 5000 cells per well in each well of a 96-well plate with daily monitoring. Migration assays plated hMSCs at 5000 cells/cm2 in six-well plates. Twenty-four hours after plating, a section of the cells was removed by scraping with a pipette tip. That area was analyzed repeatedly with light microscopy. For perturbation of vacuole formation, we stained with 2 μM calcein for 30 min. For osteogenic, adipogenic, and chondrogenic differentiation, we used reagents and protocols from Lonza (see Supplementary Materials and Methods).
Cytokine expression and toxicity assays
Cells with and without SiNPs were plated at 20,000/cm2 in a 12-well plate and cultured for 2 days. The medium was then exchanged and allowed to stand for 24 hours. The medium from labeled and unlabeled cells was studied with a bead-based assay (Luminex) by a commercial operator (Rules-Based Medicine) (36). Serum electrolytes, liver function tests, and cardiac enzymes were measured in serum by the Veterinary Services Center at Stanford University.
Animal studies
Healthy female nu/nu mice age 6 to 16 weeks were used in this study with no cardiac injury. All animal work was conducted in accordance with the Administrative Panel on Laboratory Animal Care at Stanford University. Before imaging, mice were anesthetized with 2% isoflurane at 2 liters/min and confirmed with tail pinch. Intracardiac delivery of hMSCs was performed with a custom-made 27-gauge × 1-inch catheter. hMSCs were suspended in 50:50 pH 7.4 1× PBS/Matrigel 10 min before injection.
Histology
Cardiac tissue sections were removed and immediately placed in 10% buffered formalin for 2 days and then transferred to 30% sucrose in PBS. Sections were then placed in optimal cutting temperature media and frozen for 10 s in a bath of isopentane that was immersed in a bath of liquid nitrogen. Tissue sections (6 μm) were sliced and placed on positively charged slides. Immunofluorescence used a goat anti-rabbit biotinylated primary antibody specific to cardiac troponin T and Alexa Fluor 647–coated streptavidin. H&E sections were imaged with an automated histology slide reading tool (Nanozoomer; Hamamatsu).
Ultrasound imaging
Ultrasound imaging was performed with a preclinical Vevo2100 system with M550 and M250 transducers (VisualSonics) and a clinical scanner (iU22; Philips) with L12-5 and L17-5 transducers. The optimal settings were determined empirically and found to be 100% power, gain of 20 dB, and dynamic range of 50 dB. Video frame rate varied between 50 and 300 frames per second. At least five FOVs were collected for each sample.
MRI characterization and imaging
To measure the relaxivity of the SiNPs, we dissolved various concentrations of SiNPs from 0.1 to 2.0 mg/ml in DDI water. MRI imaging and T1/relaxivity measurements used an M2 system from Aspect Imaging that used a 1-T permanent magnet. After autoshimming and scouting, two-dimensional T1 spin echo (SE) multislice images were acquired with vertical frequency directions, FOVs of 100 mm, an echo time of 20 ms, repetition time of 500 ms, flip angle of 90°, a 512 × 512 matrix, one excitation, 20 μs of dwell time, and autogain calibration. Imaging after injection of contrast or cells retained the previous settings. Eleven to 13 slices were acquired with no gap between slices, and 1-mm thick slices. The φ and θ angle were set to zero for axial slices with no echo asymmetry. The T1 was determined by measuring the exponential time constant in a plot of peak intensity versus delay between pulses. The reciprocal of T1 was plotted as a function of concentration. The slope of this line measures relaxivity of the SiNPs.
Data analysis
Ultrasound images (RGB TIFF files or .avi videos) were analyzed with Vevo and ImageJ (version 1.44p) software by ROI analysis (37). The LOD was defined as signal-detectable three SDs above the mean signal of the blank. For the phantoms, signal was defined as the B-mode ultrasound contrast generated by the SiNP, and the background was defined as the B-mode ultrasound contrast generated by the agarose gel. Noise was defined as the SD in areas without contrast.
In animals, contrast mode in the ROI was used to determine increase in signal via subtraction of the preinjection image from the post-injection image and indicated areas of increased B-mode signal. Those pixels that had greater signal than the preinjection pixels were assigned to a green lookup table and then overlaid with the original image. Contrast mode videos used the average of at least 100 frames before injection of hMSCs as the baseline above which subsequent frames were compared. Subsequent pixels 20% above this average were coded green by the Vevo software. Maximum intensity projections sequentially added additional pixels throughout the cine loop. For LOD determinations, we used the mean intensity of the ROI for each animal subject after injection.
Data analysis of MR images used DICOM files and ImageJ. Distance measurements used MicroDICOM software. For the LOD calculations in MR images, ROIs were drawn around the injected cell bolus for those frames containing cells. Because the size of the implant was different between animals, we used the integrated density of the ROI rather than the mean. The integrated density values for those frames were summed and defined as signal. Because the imaging software scales each imaging session with a unique scaling factor and because of differences in receiver gain between studies, we normalized the value of signal by the mean intensity of latissimus dorsi muscle in the same slice as the cell bolus. That value was plotted as a function of injected cell number. For time course studies, we used the mean intensity of one DICOM slice per animal normalized by muscle intensity again with ImageJ.
Statistical analysis
Data means and SD were computed in Microsoft Excel 2007. SEM was calculated from the SD and n (number of independent trials). We used a two-tailed Student’s t test for significance. Analysis of the secretome used a two-tailed Student’s t test with 49 degrees of freedom (t = 1.960). We assumed that the coefficient of variation applied to both SiNP and control samples. The final P values were generated with the “T.DIST” command in MS Excel.
Supplementary Material
Acknowledgments
We appreciate the support of T. Lobovkina with confocal microscopy; P. Kempen with TEM sample preparation; A. Marshall with scanning TEM; S. Lynch with nuclear magnetic resonance; P. Pandit, L. Pissani, and H. Nejadnik for support with relaxivity measurements; and J. Shah for data analysis support. We also thank S. M. W. Y. Van de Ven and D. Ikeda for access to clinical ultrasound equipment, P. Chu for histology expertise, and C. Zheng for help with the DMMB assay.
Funding: Supported in part by National Cancer Institute grants U54CA119367 (S.S.G.) and P50CA114747 (S.S.G.). J.V.J. is grateful for fellowship support from the Stanford Molecular Imaging Scholars Program R25-TCA118681 and acknowledges the Burroughs Wellcome Fund (1011172).
Footnotes
Author contributions: J.V.J. and S.S.G. conceived the research and designed the experiments. J.V.J. and C.K. performed the experiments. J.V.J. and S.S.G. wrote the manuscript.
Competing interests: J.V.J. and S.S.G. hold a patent on nanoparticle-based tools for ultrasound imaging.
www.sciencetranslationalmedicine.org/cgi/content/full/5/177/177ra35/DC1
Materials and Methods
Fig. S1. Optimization and characterization of SiNP cell loading.
Fig. S2. Cell migration assay.
Fig. S3. Impact of SiNP fraction on ultrasound intensity.
Table S1. Serum toxicity tests.
Movie S1. Injection of hMSCs into the mouse LV wall.
Movie S2. Integration of hMSC bolus with the LV.
REFERENCES AND NOTES
- 1.Meyer GP, Wollert KC, Lotz J, Steffens J, Lippolt P, Fichtner S, Hecker H, Schaefer A, Arseniev L, Hertenstein B, Ganser A, Drexler H. Intracoronary bone marrow cell transfer after myocardial infarction: Eighteen months’ follow-up data from the randomized, controlled BOOST (BOne marrOw transfer to enhance ST-elevation infarct regeneration) trial. Circulation. 2006;113:1287–1294. doi: 10.1161/CIRCULATIONAHA.105.575118. [DOI] [PubMed] [Google Scholar]
- 2.Lee RH, Pulin AA, Seo MJ, Kota DJ, Ylostalo J, Larson BL, Semprun-Prieto L, Delafontaine P, Prockop DJ. Intravenous hMSCs improve myocardial infarction in mice because cells embolized in lung are activated to secrete the anti-inflammatory protein TSG-6. Cell Stem Cell. 2009;5:54–63. doi: 10.1016/j.stem.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolli R, Chugh AR, D’Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, Hosoda T, Sanada F, Elmore JB, Goichberg P, Cappetta D, Solankhi NK, Fahsah I, Rokosh DG, Slaughter MS, Kajstura J, Anversa P. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): Initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Williams AR, Hare JM. Mesenchymal stem cells: Biology, pathophysiology, translational findings, and therapeutic implications for cardiac disease. Circ Res. 2011;109:923–940. doi: 10.1161/CIRCRESAHA.111.243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranganath SH, Levy O, Inamdar MS, Karp JM. Harnessing the mesenchymal stem cell secretome for the treatment of cardiovascular disease. Cell Stem Cell. 2012;10:244–258. doi: 10.1016/j.stem.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parekkadan B, Milwid JM. Mesenchymal stem cells as therapeutics. Annu Rev Biomed Eng. 2010;12:87–117. doi: 10.1146/annurev-bioeng-070909-105309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gnecchi M, Zhang Z, Ni A, Dzau VJ. Paracrine mechanisms in adult stem cell signaling and therapy. Circ Res. 2008;103:1204–1219. doi: 10.1161/CIRCRESAHA.108.176826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries IJ, Lesterhuis WJ, Barentsz JO, Verdijk P, van Krieken JH, Boerman OC, Oyen WJ, Bonenkamp JJ, Boezeman JB, Adema GJ, Bulte JW, Scheenen TW, Punt CJ, Heerschap A, Figdor CG. Magnetic resonance tracking of dendritic cells in melanoma patients for monitoring of cellular therapy. Nat Biotechnol. 2005;23:1407–1413. doi: 10.1038/nbt1154. [DOI] [PubMed] [Google Scholar]
- 9.Bulte JWM. In vivo MRI cell tracking: Clinical studies. AJR Am J Roentgenol. 2009;193:314–325. doi: 10.2214/AJR.09.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Porcel M, Gheysens O, Chen IY, Wu JC, Gambhir SS. Image-guided cardiac cell delivery using high-resolution small-animal ultrasound. Mol Ther. 2005;12:1142–1147. doi: 10.1016/j.ymthe.2005.07.532. [DOI] [PubMed] [Google Scholar]
- 11.Frangioni JV, Hajjar RJ. In vivo tracking of stem cells for clinical trials in cardiovascular disease. Circulation. 2004;110:3378–3383. doi: 10.1161/01.CIR.0000149840.46523.FC. [DOI] [PubMed] [Google Scholar]
- 12.Pysz MA, Willmann JK. Targeted contrast-enhanced ultrasound: An emerging technology in abdominal and pelvic imaging. Gastroenterology. 2011;140:785–790. doi: 10.1053/j.gastro.2011.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casciaro S, Conversano F, Ragusa A, Malvindi MA, Franchini R, Greco A, Pellegrino T, Gigli G. Optimal enhancement configuration of silica nanoparticles for ultrasound imaging and automatic detection at conventional diagnostic frequencies. Invest Radiol. 2010;45:715–724. doi: 10.1097/RLI.0b013e3181e6f42f. [DOI] [PubMed] [Google Scholar]
- 14.Martinez HP, Kono Y, Blair SL, Sandoval S, Wang-Rodriguez J, Mattrey RF, Kummel AC, Trogler WC. Hard shell gas-filled contrast enhancement particles for colour Doppler ultrasound imaging of tumors. Med Chem Commun. 2010;1:266–270. doi: 10.1039/c0md00139b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsiao JK, Tsai CP, Chung TH, Hung Y, Yao M, Liu HM, Mou CY, Yang CS, Chen YC, Huang DM. Mesoporous silica nanoparticles as a delivery system of gadolinium for effective human stem cell tracking. Small. 2008;4:1445–1452. doi: 10.1002/smll.200701316. [DOI] [PubMed] [Google Scholar]
- 16.Memorial Sloan-Kettering Cancer Center. ClinicalTrials.gov. identifier NCT01266096. [Google Scholar]
- 17.Liu J, Levine AL, Mattoon JS, Yamaguchi M, Lee RJ, Pan X, Rosol TJ. Nanoparticles as image enhancing agents for ultrasonography. Phys Med Biol. 2006;51:2179–2189. doi: 10.1088/0031-9155/51/9/004. [DOI] [PubMed] [Google Scholar]
- 18.Rossi LM, Shi L, Quina FH, Rosenzweig Z. Stöber synthesis of monodispersed luminescent silica nanoparticles for bioanalytical assays. Langmuir. 2005;21:4277–4280. doi: 10.1021/la0504098. [DOI] [PubMed] [Google Scholar]
- 19.Amaral M, Costa M, Lopes M, Silva R, Santos J, Fernandes M. Si3N4-bioglass composites stimulate the proliferation of MG63 osteoblast-like cells and support the osteogenic differentiation of human bone marrow cells. Biomaterials. 2002;23:4897–4906. doi: 10.1016/s0142-9612(02)00249-1. [DOI] [PubMed] [Google Scholar]
- 20.Boddington SE, Sutton EJ, Henning TD, Nedopil AJ, Sennino B, Kim A, Daldrup-Link HE. Labeling human mesenchymal stem cells with fluorescent contrast agents: The biological impact. Mol Imaging Biol. 2011;13:3–9. doi: 10.1007/s11307-010-0322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makino S, Fukuda K, Miyoshi S, Konishi F, Kodama H, Pan J, Sano M, Takahashi T, Hori S, Abe H, Hata J, Umezawa A, Ogawa S. Cardiomyocytes can be generated from marrow stromal cells in vitro. J Clin Invest. 1999;103:697–705. doi: 10.1172/JCI5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derelanko MJ, Hollinger MA. Handbook of Toxicology. Informa HealthCare; 2001. [Google Scholar]
- 23.Wolford ST, Schroer RA, Gohs FX, Gallo PP, Brodeck M, Falk HB, Ruhren R. Reference range data base for serum chemistry and hematology values in laboratory animals. J Toxicol Environ Health. 1986;18:161–188. doi: 10.1080/15287398609530859. [DOI] [PubMed] [Google Scholar]
- 24.Chung TH, Wu SH, Yao M, Lu CW, Lin YS, Hung Y, Mou CY, Chen YC, Huang DM. The effect of surface charge on the uptake and biological function of mesoporous silica nanoparticles in 3T3-L1 cells and human mesenchymal stem cells. Biomaterials. 2007;28:2959–2966. doi: 10.1016/j.biomaterials.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Foster FS, Zhang MY, Zhou YQ, Liu G, Mehi J, Cherin E, Harasiewicz KA, Starkoski BG, Zan L, Knapik DA, Adamson SL. A new ultrasound instrument for in vivo microimaging of mice. Ultrasound Med Biol. 2002;28:1165–1172. doi: 10.1016/s0301-5629(02)00567-7. [DOI] [PubMed] [Google Scholar]
- 26.Ankrum J, Karp JM. Mesenchymal stem cell therapy: Two steps forward, one step back. Trends Mol Med. 2010;16:203–209. doi: 10.1016/j.molmed.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arifin DR, Long CM, Gilad AA, Alric C, Roux S, Tillement O, Link TW, Arepally A, Bulte JW. Trimodal gadolinium-gold microcapsules containing pancreatic islet cells restore normoglycemia in diabetic mice and can be tracked by using US, CT, and positive-contrast MR imaging. Radiology. 2011;260:790–798. doi: 10.1148/radiol.11101608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnett BP, Ruiz-Cabello J, Hota P, Ouwerkerk R, Shamblott MJ, Lauzon C, Walczak P, Gilson WD, Chacko VP, Kraitchman DL, Arepally A, Bulte JW. Use of perfluorocarbon nanoparticles for non-invasive multimodal cell tracking of human pancreatic islets. Contrast Media Mol Imaging. 2011;6:251–259. doi: 10.1002/cmmi.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nam SY, Ricles LM, Suggs LJ, Emelianov SY. In vivo ultrasound and photoacoustic monitoring of mesenchymal stem cells labeled with gold nanotracers. PLoS One. 2012;7:e37267. doi: 10.1371/journal.pone.0037267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jokerst JV, Thangaraj M, Kempen PJ, Sinclair R, Gambhir SS. Photoacoustic imaging of mesenchymal stem cells in living mice via silica-coated gold nanorods. ACS Nano. 2012;6:5920–5930. doi: 10.1021/nn302042y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deshpande N, Needles A, Willmann JK. Molecular ultrasound imaging: Current status and future directions. Clin Radiol. 2010;65:567–581. doi: 10.1016/j.crad.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grobner T. Gadolinium—A specific trigger for the development of nephrogenic fibrosing dermopathy and nephrogenic systemic fibrosis? Nephrol Dial Transplant. 2006;21:1104–1108. doi: 10.1093/ndt/gfk062. [DOI] [PubMed] [Google Scholar]
- 33.Penfield JG, Reilly RF. What nephrologists need to know about gadolinium. Nat Clin Pract Nephrol. 2007;3:654–668. doi: 10.1038/ncpneph0660. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Suzuki Y, Huang M, Cao F, Xie X, Connolly AJ, Yang PC, Wu JC. Comparison of reporter gene and iron particle labeling for tracking fate of human embryonic stem cells and differentiated endothelial cells in living subjects. Stem Cells. 2008;26:864–873. doi: 10.1634/stemcells.2007-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hare JM, Traverse JH, Henry TD, Dib N, Strumpf RK, Schulman SP, Gerstenblith G, DeMaria AN, Denktas AE, Gammon RS, Hermiller JB, Jr, Reisman MA, Schaer GL, Sherman W. A randomized, double-blind, placebo-controlled, dose-escalation study of intravenous adult human mesenchymal stem cells (prochymal) after acute myocardial infarction. J Am Coll Cardiol. 2009;54:2277–2286. doi: 10.1016/j.jacc.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunbar SA. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin Chim Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.