Summary
Dictyostelium discoideum is a useful cell model for studying protein–protein interactions and deciphering complex signaling pathways similar to those found in mammalian systems. Many of these interactions were analyzed using classical in vitro biochemical techniques. However, with the accessibility of fluorescently tagged proteins, extensive protein networks are now being mapped out in living cells using a variety of microscopic techniques. One such technique, fluorescent recovery after photobleaching (FRAP), has been used in Dictyostelium to investigate a number of cellular processes including actin and cytoskeleton dynamics during chemotaxis and cytokinesis (J. Muscle Res. Cell Motil. 23:639–649, 2002; Biophys. J. 81:2010–2019, 2001; Mol. Biol. Cell 16:4256–4266, 2005), to follow trafficking of proteins to organelles such as the membrane, nucleus, and endoplasmic reticulum (Development 130:797–804, 2003; J. Cell Biol. 154:137–146, 2001), and to understand the role of proteins in cell adhesion during motility and division (Mol. Biol. Cell 18:4074–4084, 2007; J. Cell Sci. 120:4302–4309, 2007). FRAP is a powerful tool that should provide a vast amount of information on the mobility of a number of proteins, not only in Dictyostelium, but in many organisms. This study will lay out the methods of conducting FRAP experiments in Dictyostelium and discuss the large amount of knowledge which can be gained by adopting this as a common technique.
Keywords: Dictyostelium, Photobleaching, Fluorescent proteins, FRAP, PTEN, PLC
1. Introduction
1.1. Overview of FRAP
Fluorescence recovery after photobleaching (FRAP) is not a novel technique. In fact, FRAP was first developed in the 1970s as a technique to study the mobility of proteins in living cells (8, 9). Experiments were initially aimed at investigating changes in lateral membrane transport as an indicator or consequence of changes in the physiological state of cells. Early success of FRAP experiments were limited to determining the rates of diffusion of molecules on or in the cell membrane. However, with the advent of fluorescently tagged proteins, which now allow the visualization of proteins in living cells, and the development of confocal microscopy, the use of FRAP experiments has expanded significantly. Today FRAP is used to investigate a plethora of biologic phenomena including motility, division, adhesion, transcription, signal transduction, and protein trafficking.
During FRAP experiments, fluorescent molecules are irreversibly photobleached in a geographically confined region of interest (ROI) using a tightly focused laser beam (Fig. 1). Photobleaching is the irreversible loss of the ability of a fluorophore to fluoresce due to photon-induced chemical damage and covalent modification. Some fluorophores bleach after only emitting a few photons, while others can undergo many millions of cycles before being covalently modified. Normally the fluorophore is photobleached at a wavelength that is not otherwise absorbed by cellular components, and therefore, should not damage other parts of the cell or specimen (8). The ROI can be in the form of a spot, multiple spots, or a specific area within the cell such as the cytoplasm, cell membrane, or whole organelles. Recovery of fluorescence can come only from molecules that move into the bleached area from outside and replace the bleached molecules within this area. The rate of fluorescent recovery over time in that specific area can be measured and these measurements can be used to calculate the mobility of a specific molecule/protein.
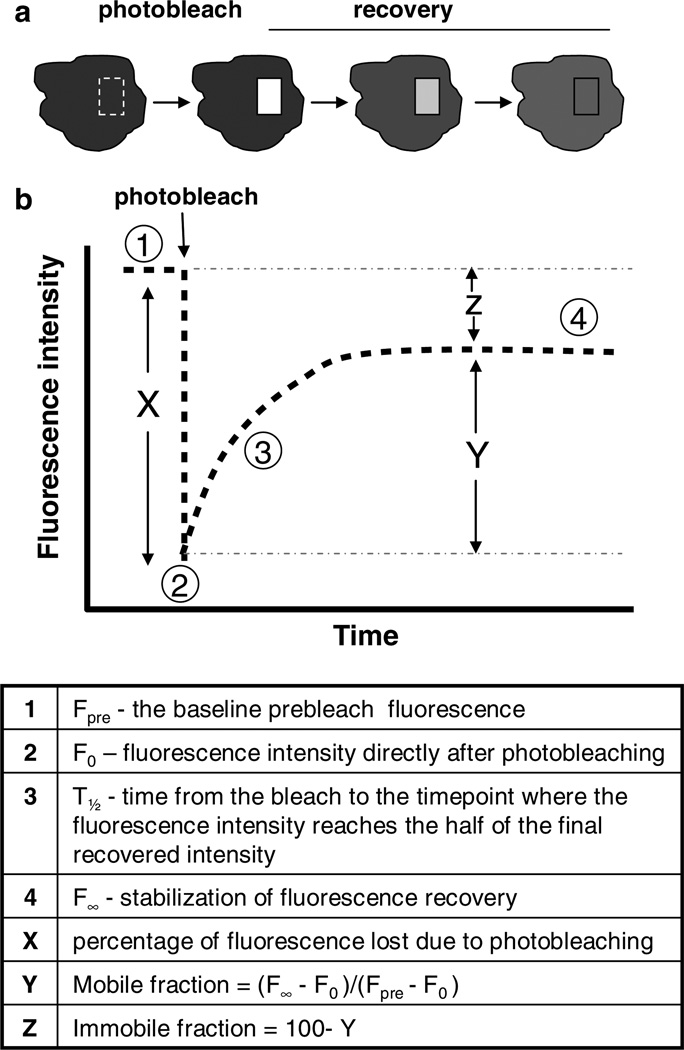
Fig. 1.
FRAP experiment. (a) Illustration of a cell undergoing spot photobleaching.The square box represents the region of interest where photobleaching occurs. Over time, the fluorescence in the region of interest recovers. (b) A characteristic recovery curve of fluorescent molecules within the region of interest. Graphing the prebleach intensity and the resulting recovery establishes the mobile fraction within that region. These numbers can all then be used to determine the half-time of recovery, as well as, the diffusion coefficient (see text for specific calculations).
The recovery process is dependent on the rates of diffusion and/or the transport through the cellular milieu. Barriers to diffusion can also be identified and analyzed and assessed using FRAP. The mobility of a molecule can be influenced by binding interactions to proteins, cell membranes, organelles or other changes that affect the local viscosity of the environment in which the molecule resides. Therefore, through careful data analysis, much information can be gained from FRAP including:
Mobility of a protein/molecule – the percentage of mobile vs. immobile populations
Recovery rates – how quickly the tagged protein/molecule moves within the cell
Type of transport – active versus diffusive, random diffusion versus uniform directed flow
Diffusion constants
1.2. Applications in Dictyostelium
Although many of the signaling networks in Dictyostelium are very similar to those in mammalian cells, Dictyostelium also provides unique differences that researchers can exploit. For instance, signaling and transport of molecules within Dictyostelium, as well as basic cellular processes such as phagocytosis, motility and division, often occur on a much faster time scale than in mammalian cells. These processes are easily induced during microscopic observation and allow for fast data acquisition and the ability to carry out more experiments in a shorter amount of time. These characteristics are very useful, but can sometimes be challenging during FRAP experiments. The value of using Dictyostelium will be discussed in the following subheaders, while the challenges will be discussed in Subheading 4.
1.2.1. Diffusion of Molecules and the Role of the Cytoskeleton
The importance of the simple kinetics of molecular diffusion within cells and the factors which might alter these kinetics are often overlooked in research. However, FRAP experiments have illuminated the significance of kinetics of molecules as they relate to changes in cell shape, developmental stage, cell cycle progression, and cellular environment. Early on, Potma et al. investigated many of these characteristics in Dictyostelium using the green fluorescent protein (GFP) (2). GFP when expressed alone had a 3.6-fold reduction in mobility within Dictyostelium as compared to its diffusion in other simple aqueous solutions. The filamentous structures of the cytoskeleton, collisions with macromolecular solutes, and confined motional freedom due to microcompartments within the cell were all likely contributors to this reduction in mobility. In fact, it was shown that the actin cytoskeleton alone accounted for 53% of the restrained molecular diffusion of GFP (2). Thus, changes in the cytoskeleton have profound effects on the diffusion of molecules within the cell and should be taken into account when conducting FRAP experiments. Additionally, cytoplasmic changes that in turn affect the meshwork of actin should also be taken into consideration. For instance, diffusion of GFP was faster in polarized cells than nonpolarized cells. Specific differences in mobility have been noted in the fronts versus the backs of polarized cells (2). Similarly, differences at the cleavage furrow compared with the poles of a dividing cell have also been reported (10). Osmotic properties of the medium have also elicited differences in molecular diffusion, as cells placed in a hypertonic medium showed a decrease in GFP diffusion (2).
Although a significant amount of knowledge in Dictyostelium has been gained using GFP alone, the use of FRAP to determine the diffusion of specific proteins in Dictyostelium has been somewhat underutilized, especially considering the high number of fluorescently tagged proteins available. Additionally, it is possible to examine the involvement of binding interactions of a protein (see Note 1)through comparisons of a GFP-tagged protein to GFP alone. When the FRAP recovery of a GFP-tagged protein is slower than that of GFP alone, binding is implicated and the degree of slowdown of recovery is a measure of the strength of binding. However, an important caveat is that FRAP recovery rates from diffusion are weakly dependent on protein mass (11).
1.2.2. Membrane FRAP
FRAP is a powerful tool for investigating transmembrane proteins or proteins associated with the cell membrane. For instance, FRAP of molecules on the cell membrane can provide information about the molecules’ size, environment, and participation in intermolecular interactions, including ligand-driven associations (12). FRAP of transmembrane proteins (such as serpentine receptors) can occur only by the lateral diffusion or directed motion of unbleached proteins in the membrane. Conversely, proteins which interact only with the inner membrane leaflet (G-proteins, PTEN, etc.) will typically have a cytoplasmic population as well. These proteins can recover from FRAP by both lateral diffusion in the membrane and also by exchange with cytoplasmic stores. Therefore, the recovery time after FRAP of transmembrane proteins (lateral diffusion only) is proportional to the area illuminated by the laser beam. Whereas the recovery of membrane-associated proteins will also include the chemical relaxation time, which is constant regardless of membrane area and thus beam size (13).
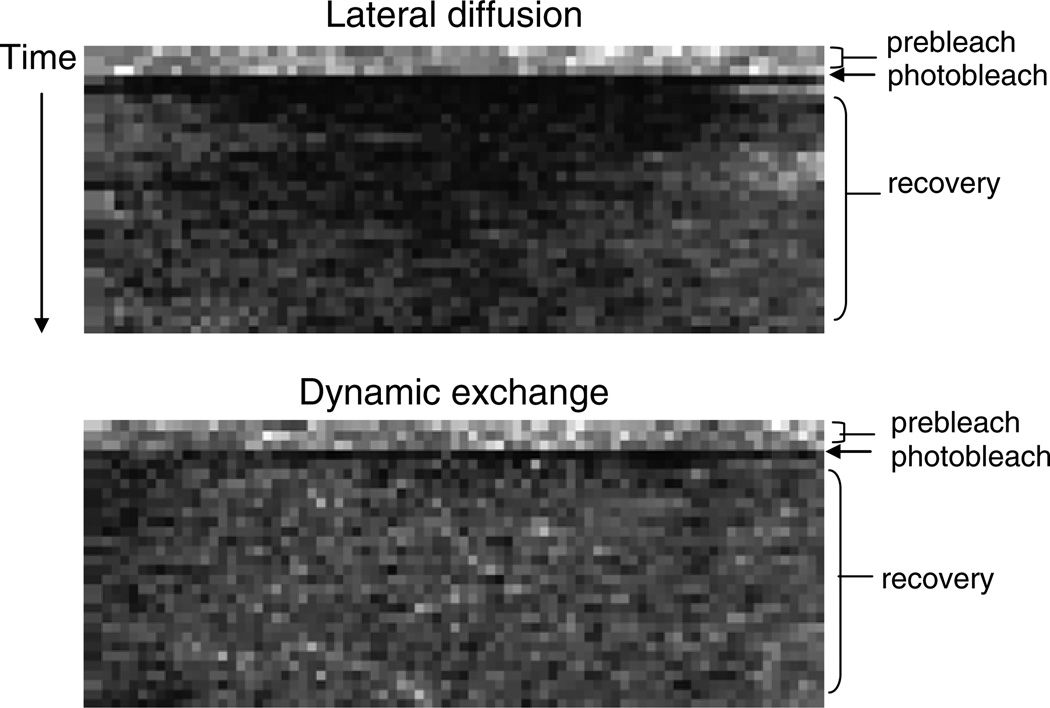
Thus, one can determine if a protein is recovering from lateral exchange versus exchange plus diffusion by varying the size of the bleached region of interest on the membrane (14). Recovery rates will be faster for a smaller region of interest rather than a larger one, when analyzing FRAP of transmembrane proteins. On the contrary, recovery rate should not be affected by alterations in bleach spot size for proteins which are undergoing active exchange with the membrane. Additionally, recovery from lateral diffusion only will typically be slower than dynamic exchange. For example, the half-time of recovery of the cAMP receptor cAR1 is about 17 s (data not shown), while the recovery of cortexillin is around 3 s (10). Lateral diffusion versus dynamic exchange can be further analyzed through kymography. A kymograph (also called a time space plot) measures velocities of moving structures in an image time series. Thus, a kymograph of the membrane after FRAP will appear differently from a protein like cAR1 which undergoes only lateral diffusion when compated to PTEN, which is involved in dynamic exchange (Fig. 2). As both these are major players involved in chemotaxis, it would be interesting to investigate whether the recovery rates would differ for these proteins, as well as others, in the presence of cAMP.
Fig. 2.
Characteristic FRAP kymographs from proteins undergoing either lateral diffusion or dynamic exchange to the membrane. For each frame of a time series, gray values along a line type from the region of interest on the membrane were collected. From these lines of gray values, a new image (the kymograph or time-space plot) was assembled with black boxes representing low fluorescence and white areas being maximal fluorescence. The line read from the first frame of the time series is put down as the first line in the kymograph, the line from the second frame is the second line of the kymograph, and so forth. In this way, the y axis of the kymograph becomes a time axis and the unit is the time interval of the sequence. The x axis is the distance along the line ROI and the unit is the pixel size of your sequence. Notice that the recovery (light boxes) from lateral diffusion of cAR1 occurs first from the sides of the kymograph, then with time, fills in the middle. However, dynamic exchange of PTEN recovers uniformly across the region of interest with recovery occurring immediately from the center and from the sides.
1.2.3. Selective FRAP
With advanced software technology and precise mechanical control of the laser spot, photobleached areas are no longer limited to regions of interest in the shapes of circles or squares. Today, many confocal microscopes have an ROI selection tool which allows for bleaching of regions within the cell of varying sizes and shapes. Therefore, entire subcellular compartments can be bleached and their recovery analyzed. For instance, organelles such as the nucleus, endoplasmic reticulum, Golgi apparatus, and mitochondria can now be subjected to FRAP analysis (see Note 2). Data from this type of selective photobleaching can be combined with kinetic analysis to determine the rate of cycling between two compartments. Exchange rates between the Golgi and endoplasmic reticulum have already been measured in mammalian cells (15). In general, compartmental exchange can occur via diffusion, exchange, vesicular transport or any combination of them.
The newly developed ability to now FRAP complex structures has opened up a new avenue of research for understanding the mechanisms which control protein trafficking through cellular compartments. For example, in Dictyostelium, the kinetics of nuclear export of the protein Dd-STATc have been investigated using FRAP of the cell nucleus. Factors that regulate nuclear export were also explored using DIF, a differentiation-inducing factor, and further demonstrated how external factors could in turn regulate transport of proteins within the cell (4).
1.2.4. Combining Molecular Techniques with FRAP
FRAP alone as a technique is exciting, but combining it with molecular biology tools makes it a very powerful technique. The recovery of proteins in the presence of various inhibitors of cellular processes can provide information on the regulatory mechanisms of diffusion, recruitment from cytoplasmic pools, or transport of that protein. For example, the inhibition of actin cytoskeletal dynamics in Dictyostelium was validated when the diffusion rate of GFP increased significantly after cells were treated with the actin inhibitor, Latrunculin A (2). The inhibitor Nocodazole could be used to investigate the role of microtubules, while other drugs such as LY294002 or Wortmannin could help characterize the participation of signaling molecules like PI3-kinase in regulating the mobility of other signaling molecules and effector proteins.
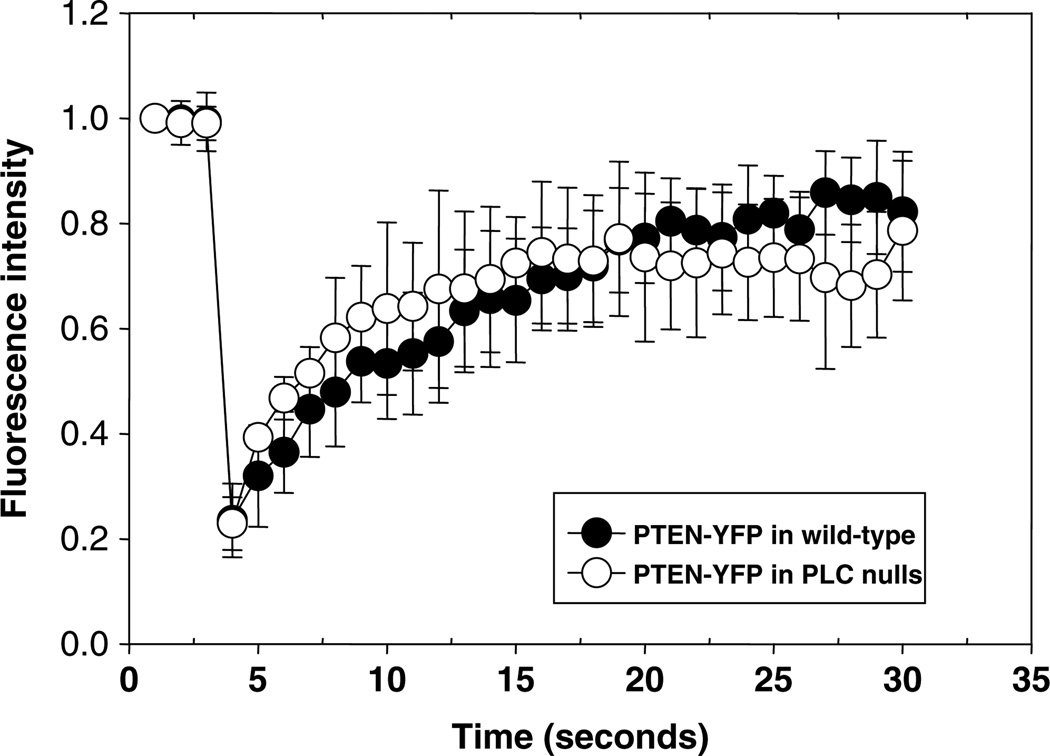
Another utility of FRAP is the comparison of mutant cell lines to wild type. In one case, investigators who varied the levels of talinA in cells affected the dynamics of membrane-associated myosin VII (6). In another example, the kinetics of actin crosslinkers were shown to be different in myosin II null cells compared to wild type (10). Conversely, the absence of differences in recovery rates in mutant cells can also be telling. For instance, we have found that the recovery of PTEN is virtually identical in wildtype and phospholipase C (PLC) null cells (Fig. 3), suggesting that the loss of PLC activity in cells has no detectable effect on the basal rate of PTEN membrane association.
Fig. 3.
FRAP recovery of PTEN on the membrane of wild-type cells and PLC null cells. FRAP analyzes of plasma membrane PTEN-YFP expressed in wild-type (AX3) and PLC null cells. No significant difference in recovery was seen between the cell types. For each cell line, the halftime of PTEN-YFP was 2.3 s and the mobile fraction was 77%. Data were collected over two separate days with a total of 18 cells.
The effect of changes in protein structure, sequence, or activity can also be investigated using FRAP through truncation mutants or point mutations. By performing FRAP experiments on truncation mutants, Fukuzawa et al. were able to identify the sequence required for nuclear efflux of the protein Dd-STATc (4). The removal of key threonine residues on myosin II completely impaired its recovery after photobleaching, suggesting the importance of phosphorylation in the dynamic turnover of this motor protein (5).
2. Materials
HL5 growth medium: 0.5% (w/v) proteose peptone, 0.5% (w/v) thiotone E peptone, 55.5 mM glucose, 0.5% (w/v) yeast extract, 1.3 mM Na2HPO4 and 2.57 mM KH2PO4. Bring to a volume of 1 l. Adjust pH with HCl to pH 6.4–6.6. Autoclave to sterilize.
Developmental buffer (DB): 5 mM Na2HPO4, 5 mM KH2PO4, 1 mM CaCl2, and 2 mM MgCl2. Prepare the phosphate solution as 25 mM (5×), adjust the pH to 6.5, and autoclave. Make 10× CaCl2 (10 mM) and MgCl2 (20 mM) solutions each separately and autoclave. To make 1 l of DB, mix 600 ml of distilled, autoclaved water with 200 ml of 5× phosphate solution and 100 ml of 10× CaCl2 and 10× MgCl2 solution, respectively.
Glass bottom culture dishes: 35-mm glass bottom dishes No. 1, uncoated and γ-irradiated (MatTek Corporation).
Cells: AX3 wild-type and HD1.19 PLC null cells (Dicty-Base Stock Center http://www.dictybase.org/StockCenter/OrderInfo.html).
Constructs: PTEN-YFP and cAR1-GFP plasmids with G418 resistance (Devreotes lab).
H-50 transformation buffer: 20 mM HEPES, 50 mM KCl, 10 mM NaCl, 1 mM MgSO4, 5 mM NaHCO3 and 1.3 mM NaH2PO4 in 1 l of distilled water. Adjust pH to 7.0 with HCl or NaOH as appropriate. Autoclave and store cold or frozen.
G418: sulfate used at 20 µg/ml in the HL5 growth medium of the transformants (Invitrogen).
cAMP: 50 nM for developing cells.
3. Methods
For Subheading 3.1–3.7 general methods for performing a FRAP experiment in Dictyostelium are described. Subheading 3.8 includes detailed steps for investigating PTEN recovery on the membrane using FRAP.
3.1. Preparation of Cells
Dicytostelium can be prepared in a variety of ways for FRAP experiments depending on the interest of the investigator. Because axenic media is highly autofluorescent, cells are typically washed and placed in a salt buffer such as development buffer (DB) described earlier. Cells should be allowed to adhere to a glass coverslip at a low density in order to prevent cells from touching one another. This is not a requirement for cytosolic or internal membrane FRAP, but can be critical for plasma membrane FRAP if cells were to contact or overlap each other during the data analysis. Typically it is advisable to have at least one other cell in the image that is not undergoing FRAP to serve as a control for general photobleaching.
3.2. Instruments Used
These techniques require a confocal microscope equipped with a YFP filter and the capability to perform time-lapse imaging in conjugation with photobleaching. In addition, a high numerical aperture objective is necessary for higher resolution and maximum signal detection. This allows the laser transmission to be reduced in order to minimize non-specific photobleaching of the entire cell. The following protocols are specific to FRAP experiments and are outlined under the assumption that the operator is familiar with the basic operation of the confocal microscope.
3.3. Determining Cell Imaging Parameters for Confocal FRAP
The light intensity and the duration of the bleaching period are adjusted accordingly for the specific fluorophore, developmental stage, and molecule under examination. Investigators should experimentally determine the appropriate duration of irradiation required to reach the extent of photobleaching, appropriate for different specimens (see Notes 3–5). Typically photobleaching should result in a 60–80% loss of fluorescence intensity in the ROI. To achieve this, one can vary either the laser power strength or the number of iterations that the laser fires. For best results, set the laser at maximum power and then adjust the number of iterations accordingly. This approach minimizes the time required for the actual photobleaching.
The molecule of interest will dictate the objective, filter sets, laser excitation, and zoom required for adequate FRAP. EGFP is commonly used because of its brightness and resistance to photobleaching. Additionally most microscopes are equipped with the proper filter set for GFP. However, other fluorophores can be used successfully with Dictyostelium including RFP, YFP, and CFP (data not shown). Other new fluorescent protein variants will certainly become useful for FRAP as microscopes acquire the proper laser lines and filter sets. With repetitive scans some overall photobleaching can occur (see Note 6). To minimize this effect, it is important to set the laser power and detector gains to the minimum settings required for obtaining images with high signal-to-noise ratios. Additionally the scan speed, pinhole settings, excitation intensity, detector gain, and line averaging should be adjusted for each experimental condition (see Note 7).
3.4. Selecting the Region of Interest
The confocal microscope software is used to define the bleaching ROI; an often-used geometry is a rectangle, typically with an area of about 100 µm2. However, the choice of size will differ depending on the molecule of interest, its distribution in the cell, the recovery speed, the size of the cell, and the method of data analysis. It is important when choosing the size of the ROI to keep in mind that larger areas will typically recover slower than smaller areas. However, if the area is too small, then a significant amount of recovery may occur during the photobleaching phase, thereby skewing your results. Nevertheless, this same phenomenon of masking the recovery can occur with large bleach spots due to the longer time required to bleach the entire area. Therefore, a medium must be established for each molecule examined. However, it may not be necessary to measure recovery of the whole cell and in this case, only the ROI can be measured. This will speed up the data acquisition but will also sacrifice information about overall photobleaching and/or focal plane drifts (14).
For measurements of lateral diffusion on the membrane, it is especially important to use the same bleach size for each experiment to allow for direct comparisons between data sets and to facilitate data analysis. Most software programs will allow the user to either copy and paste the ROI from one cell to another or permit one to type in the exact specifications of the bleach region. Often, the ROI specifications are measured in pixels; therefore, it is imperative to always use the same zoom parameters before photobleaching to maintain consistent bleach data. For selective photobleaching of an entire compartment or organelle, a freehand drawing tool may be required to outline the ROI.
3.5. Determining Proper FRAP Experimental Conditions
Once the bleaching parameters have been established, it is then important to determine the proper temporal settings to capture the recovery. Full recovery will be in the form of a plateau regardless of the percentage of recovery. The speed of recovery will vary depending on the molecule. Examples of recovery times for Dictyostelium are shown in Table 1. Typically for proteins which recover quickly, images were collected at the speed of the scan time (usually around 1 s). The acquisition speed can be slowed for proteins such as membrane receptors that are less diffusive. Regardless, it is important to minimize the overall nonspecific photobleaching that occurs when collecting multiple images over time. For a 70-s recovery, a reasonable sample rate is 75 images at a rate of 1 image/s (14). To establish a proper baseline for data analysis, it is also important to take a few (2, 3) prebleach images.
Table 1.
Characteristics of FRAP on published Dictyostelium proteins
| Protein | Half-time (s) | Diffusion coefficient (µm2/s) |
Immobile fraction (%) |
Reference |
|---|---|---|---|---|
| GFP (cytoplasm) | – | 87 ± 2 | 0.0 ± 0.3 | (2) |
| GFP (cytoplasm) + Latrunculin | – | 42 ± 2 | 3.2 ± 0.3 | |
| GFP (nucleus) | – | 22 ± 2 | 4.0 ± 0.3 | |
| Myosin VII (cytoplasm) | 1.0 ± 0.4 | – | ≈50a | (6) |
| Mysoin VII (membrane) | 2.8 ± 1.7 | – | ≈80a | |
| Dynacortin | 0.45 | – | 26 | (10) |
| Fimbrin | 0.26 | – | 33 | |
| Cortexillin I | 3.3 | – | 32 | |
| Myosin II (cortical ring during interphase) | 7.28 ± 1.95 | – | – | (5) |
| Myosin II (contractile ring) | 7.01 ± 2.62 | – | – | |
| MHCK B (contractile ring) | 1.72 ± 0.3 | – | – | (3) |
| MHCK C (contractile ring) | 2.32 ± 0.25 | – | – | |
| Actin (mitosis-specific dynamic actin structures) | 2.15 ± 0.89 | – | – | (7) |
| Coronin | 4.8 ± 1.3 | 2.2 ± 0.2 | – | (1) |
| cAR1 | 18 ± 0.41b | – | ≈60b | Unpublishedb |
| PTEN (membrane) | 2.3 ± 1.6 | – | 23 | Reported here |
“–”Values not reported
Average interpretations based on graphs reported
Unpublished data from the Janetopoulos lab
3.6. Performing the Actual Experiment
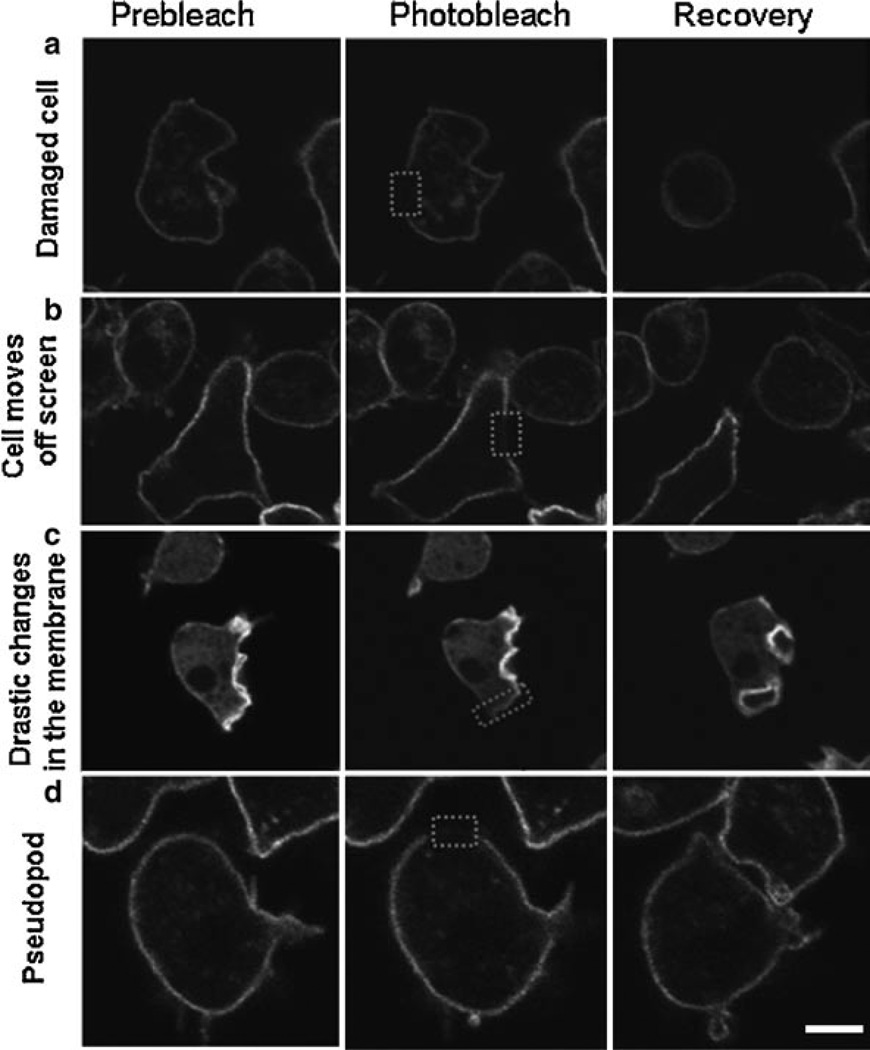
Performing the actual experiment is the easy part once the optimal conditions for imaging, bleaching, and recovery have been established. To obtain accurate data, each experiment should include prebleach images for normalization. Typically three images are acquired prior to photobleaching. Cells should be chosen that have representative expression levels of your molecule of interest. One should obtain an image that has the experimental cell to be photobleached and another cell which is not manipulated. The second cell can serve as a good control for general photobleaching effects. A minimum of ten cells per day should be collected, as it is likely not all will be used because of cell movement and/or the introduction of recovery artifacts. Once the movie has been acquired, it should be examined for artifacts such as changes in focus, cell movement, or significant changes in fluorescent intensities which could confound the data analysis. Examples of these artifacts are shown in Fig. 4 and should be excluded from further data analysis. Images should be saved in the format specific to the software as well as in a general format (such as TIFF) that can be used with other software programs. Additionally, it may be useful to save the image with and without the ROI for aiding the analysis.
Fig. 4.
Cellular artifacts seen with FRAP. Images of cells prior to photobleaching, immediately after, and at the end of the recovery are shown. These cells represent data that would be discarded from analysis. (a) The cell completely rounds up indicating excessive damage or possible death. (b) The cell moved off the screen by the end of the time-lapse acquisition. (c) The FRAPped region on the membrane was hard to distinguish. (d) A pseudopod formed on the membrane where FRAP occurred.
3.7. Analyzing the Data
Three separate measurements of fluorescent intensity need to be obtained from the data set: pixel values of the ROI, the entire cell, and the background. Depending on the software used, these are typically saved as individual masks which can then be exported to a spreadsheet for further calculations. The larger the background chosen, the more accurate it usually is. However, the background region should be examined for the entire movie to ensure that an artifact or cell doesn’t travel through it. Typically the background should not change more than 2–3% during the entire time-lapse series.
Once these raw values are obtained, the following calculations should be done:
Subtract the background fluorescence from the whole cell fluorescence for each frame.
Subtract the background fluorescence from the ROI for each frame.
Divide the adjusted ROI value (#2) by the adjusted whole cell value (#1) in order to correct for general photobleaching within the cell.
Normalize the data to the prebleach intensity by dividing each frame value (#3) by the prebleach intensity of frame 1 and multiplying by 100. This will give you the percentage of initial fluorescence recovered (see example of data from one cell in Table 2).
Table 2.
Example data set from FRAP on PTEN-YFP of one cell
| Time (s) | Background | Whole cell | ROI | Step #1 | Step #2 | Step #3 | Step #4 |
|---|---|---|---|---|---|---|---|
| Whole cell – background |
ROI – background |
#2/#1 | (#3/2.35) × 100 |
||||
| 0 | 183.14 | 1,004.56 | 2,115.49 | 821.42 | 1,932.35 | 2.35 | 100 |
| 1 | 181.69 | 971.05 | 2,087.98 | 789.36 | 1,906.29 | 2.41 | 102.66 |
| 2 | 177.92 | 939.32 | 2,031.46 | 761.4 | 1,853.54 | 2.43 | 103.48 |
| 3 | 169.79 | 728.15 | 448.55 | 558.36 | 278.76 | 0.50 | 21.22 |
| 4 | 173.55 | 714.7 | 658.45 | 541.15 | 484.9 | 0.90 | 38.09 |
| 5 | 175.43 | 705.82 | 755.98 | 530.39 | 580.55 | 1.09 | 46.53 |
| 6 | 171.54 | 700.87 | 846.26 | 529.33 | 674.72 | 1.27 | 54.18 |
| 7 | 174.33 | 671.91 | 947.19 | 497.58 | 772.86 | 1.55 | 66.03 |
| 8 | 175.44 | 655.2 | 960.02 | 479.76 | 784.58 | 1.64 | 69.52 |
| 9 | 168.56 | 668.03 | 989.62 | 499.47 | 821.06 | 1.64 | 69.88 |
| 10 | 173.51 | 665.53 | 973.33 | 492.02 | 799.82 | 1.63 | 69.10 |
| 11 | 171.38 | 664.92 | 858.81 | 493.54 | 687.43 | 1.39 | 59.21 |
| 12 | 179.16 | 660.32 | 986.38 | 481.16 | 807.22 | 1.68 | 71.32 |
| 13 | 178.68 | 656.7 | 993.51 | 478.02 | 814.83 | 1.70 | 72.46 |
| 14 | 181.44 | 654.89 | 964.72 | 473.45 | 783.28 | 1.65 | 70.33 |
| 15 | 185.63 | 654.35 | 1,185.08 | 468.72 | 999.45 | 2.13 | 90.64 |
| 16 | 176.58 | 658.79 | 1,189.66 | 482.21 | 1,013.08 | 2.10 | 89.31 |
| 17 | 186.04 | 646.84 | 1,071.14 | 460.80 | 885.1 | 1.92 | 81.65 |
| 18 | 182.12 | 645.09 | 1,145.35 | 462.97 | 963.23 | 2.08 | 88.44 |
| 19 | 178.58 | 658.39 | 1,156.40 | 479.81 | 977.82 | 2.04 | 86.63 |
| 20 | 179.66 | 661.27 | 1,143.36 | 481.61 | 963.80 | 2.00 | 85.97 |
| 21 | 179.68 | 647.25 | 1,049.53 | 467.57 | 869.85 | 1.86 | 79.08 |
| 22 | 173.48 | 644.72 | 1,107.93 | 471.24 | 934.45 | 1.98 | 84.29 |
| 23 | 177.55 | 654.29 | 1,145.78 | 476.74 | 968.23 | 2.03 | 86.33 |
| 24 | 182.07 | 642.61 | 1,141.64 | 460.54 | 959.57 | 2.08 | 88.57 |
| 25 | 180.52 | 645.71 | 1,176.73 | 465.19 | 996.21 | 2.14 | 91.03 |
| 26 | 175.02 | 647.05 | 1,201.81 | 472.03 | 1,026.79 | 2.18 | 92.47 |
| 27 | 174.68 | 641.86 | 1,112.38 | 467.18 | 937.7 | 2.01 | 85.32 |
| 28 | 176.89 | 653.04 | 1,159.17 | 476.15 | 982.28 | 2.06 | 87.69 |
| 29 | 173.93 | 651.99 | 1,182.13 | 478.06 | 1,008.2 | 2.11 | 89.65 |
Once calculated, the final numbers can be graphed on any graphing program to determine the percent bleached, the recovery, and the plateau. From this graph, both the mobile and the immobile fractions of the molecule can be determined (see Fig. 1). A kymograph can also be made to qualitatively visualize the recovery and to ascertain whether just lateral diffusion is occurring on the membrane or whether there is dynamic exchange (see Fig. 2).
The mobile fraction (Mf) is the percentage of molecules which are free to recover from photobleaching during the time course (Fig. 1). The fraction of molecules that cannot exchange between bleached and nonbleached regions is called the immobile fraction. The mobile fraction can also be calculated from your data set by using the following equation: Mf = (F∞ − F0)/Fpre−F0), where Fpre is the prebleach intensity, F0 is the intensity directly after photobleaching, and F∞ s the final postbleach intensity which should be within the plateau. The immobile fraction is then calculated by subtracting the Mf from 100.
The halftime of recovery is the time from the bleach to the time point where the fluorescence intensity reaches the half (t1/2) of the final recovered intensity (F∞). The halftime serves as a relevant marker for comparative FRAP analyses, e.g., to measure the mobility of a labeled protein under different physiological conditions. The shorter the halftime, the higher the mobility will be of the observed protein. This allows you to determine the effective diffusion coefficient of the molecule.
Many methods have been described to calculate the diffusion coefficient depending on the type of diffusion occurring and on the nature of the ROI that was photobleached. The two-dimensional diffusion equation by Axelrod et al. is most commonly used for simple diffusion of molecules within the cytoplasm or membrane (8). For examples of other calculations see ref. 14. However, caution should be taken when comparing diffusion coefficients between different cell lines or compartments as the diffusion of the same molecule in different compartments may vary significantly due to the architecture of that compartment (16).
3.8. Detailed Methods for Membrane FRAP of PTEN-YFP
Transform wild-type AX3 and HD1.19 PLC null cells with PTEN-YFP by electroporation with 5.0 µg of plasmid using established methods (17, 18). G418-resistant clones are selected in 2–3 weeks. Culture cell lines axenically in HL5 medium at 22°C in the presence of G418 to maintain selection of transformants.
Prior to FRAP, develop cells by washing them twice with DB buffer followed by 5.5 h of starvation with continuous shaking (19) (see Note 9). Pulse cells with 50 nM cAMP every 6 min for the last 4.5 h of starvation.
Seed cells in DB on glass bottom dishes and allow them to firmly attach for 15 min prior to FRAP.
For these particular experiments, an Olympus FV-1000 confocal microscope and its corresponding software were used. Although this microscope is equipped with a separate laser for imaging and bleaching, the data were collected using a single HeNe laser at 515 nm for both, as this is currently the more accessible approach.
Perform the experiments with a 60×/1.45 Plan-Apochromat oil objective.
For adequate bleaching of PTEN-YFP use a 515-nm laser for ten iterations at 100% power (see Note 3).
Set the pinhole at 1 Airy unit and do not use line averaging. These settings allow you to photobleach approximately 80% of the initial fluorescence without creating any visible signs of damage to the cell.
For FRAP of the membrane, use a freehand drawing tool to create the ROI. This allows you to create the appropriate ROI when there are significant curves along the cell perimeter. However, to maintain consistency, you should maintain the maximum width along the membrane constant at 4.5 µm and the average depth below and above the membrane between 1 and 1.5 µm depending on how much the cell moved before bleaching.
Images should be acquired at the speed of the scan (about 1 s) due to the quick recovery of PTEN. The amount of time taken to bleach the ROI is about 3 s. Full recovery will occur about 9 s after bleaching, so it is possible that some recovery was actually occurring during the 3-s bleach and remains unaccounted for with this method of acquisition.
Analyze the recovery over 30 s, which is sufficient to reach a plateau (Fig. 3). During this time the average of overall photobleaching in the cells undergoing photobleaching should be calculated (approximately 40–50% for PTEN-YFP cells). This number should be accounted for in the data analysis.
Collect three prebleach images of cells for normalization of the data.
Collect images of 15–20 cells of varying intensities on three different days.
Exclude cells that move too much, have significant changes in the shape of their membrane, or have substantial changes in fluorescent intensity from further data analysis (Fig. 4).
To analyze PTEN recovery to the plasma membrane, draw an individual line on the membrane (instead of taking a mask of the entire ROI). The entire cell including the membrane should be used for normalization. Choose a background near the cell that is roughly half the size of the cell. Examples of each of these can be seen in Fig. 5) All measurements were obtained were analyzed using Slidebook software, but many other forms of software are available.
Graph PTEN recovery using any standard graphing software. We use SigmaPlot software and perform the kymographs using the kymograph plug from ImageJ, which is available to download for free from the NIH.
The mobile fraction of PTEN in both wild-type cells and in PLC nulls cell is 77%. This number was generated by using 99.4 as Fpre, 23.5 as F0 and 82.2 as F∞. Thus, the immobile fraction is 23%.
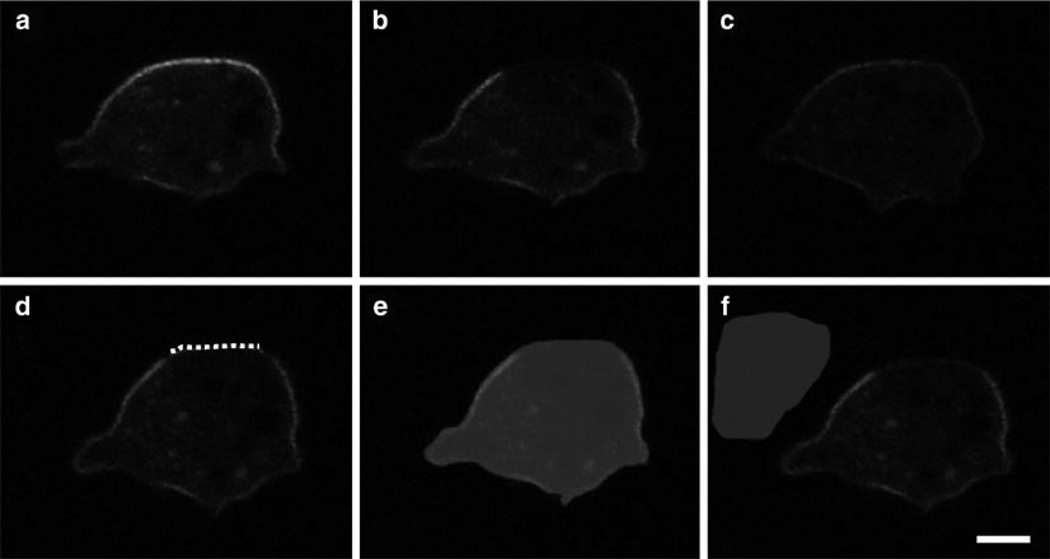
Fig. 5.
Examples of masks from the data analysis of PTEN-YFP in AX3 cells. Images of PTEN-YFP expressed in AX3 wild-type cells are shown prior to photobleaching (a), immediately after (b) and at the end of recovery (c). Example masks for analysis are shown for the membrane (dotted line) (d), whole cell (shaded area) (e), and background (shaded area) (f). Bar equals 3 mm.
3.9. The Future of FRAP
In the coming years we can look forward to widespread use of confocal FRAP as well as the implementation of new techniques which enhance the imaging and/or data acquisition of these experiments. Already gaining momentum is the use of multicolor FRAP which allows for the simultaneous analysis of two or more fluorescent molecules that have minimal to no spectral overlap (such as eGFP and mCherry). The use of two-colored FRAP could be used to study the dynamics of two proteins in different but overlapping cellular compartments and of two proteins of the same complex. Moreover, mutant proteins could be directly compared with wild-type versions within the same cell (20).
Another exciting technique combines FRAP with total internal reflection fluorescence microscopy. This technique, called total internal reflection interference fringe fluorescence photobleaching recovery (TIRIF-FPR), has been advantageous for sparsely expressed membrane species or when high expression in the Golgi or ER masks membrane expression. Additionally it can be used to measure cell receptors labeled with fluorescent antibodies or ligands (21). It should also be noted that FRAP can and should be used to complement data generated using single molecule imaging. Single molecule imaging, usually done using total internal reflection microscopy, often suffers from artifacts resulting from the blinking and bleaching of fluorophores. Half times and diffusion rates for molecules that shuttle from one compartment to another should be determined using both technologies.
The recovery of molecules in one plane using confocal microscopy presents unique challenges for structures that are moving and also is confounded by the fact that the profile of the bleach spot in the z direction cannot be determined looking at a single plane. One way to visualize the full recovery is to do a rapid z scan and watch the recovery in three dimensions. Brighter fluorescent proteins, more sensitive cameras, and more precise z stepping will make this standard practice in the near future. We are also experimenting with another solution. We have developed mirrored pyramidal wells that allow the imaging of a cell after photobleaching from multiple perspectives (22). In addition, we can direct the bleaching laser off the side reflections and also bleach the cell in the x and y directions and watch the recovery from multiple perspectives (Fig. 6). This gives unprecedented flexibility for bleaching and will allow the experimenter to visualize the bleached area with much higher spatial resolution.
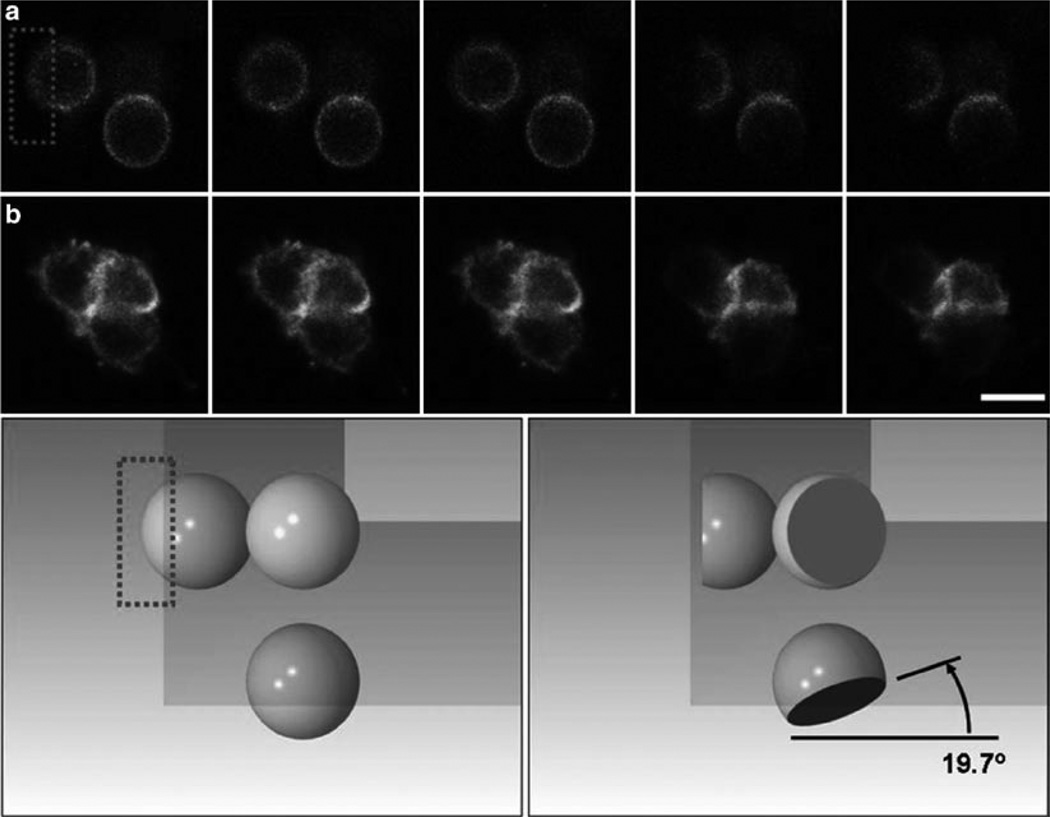
Fig. 6.
Fluorescence recovery after photobleaching (FRAP) experiments in mirrored pyramidal wells (MPWs). Shown is a series of two individual cells expressing cAR1-GFP along with their reflections prior to and after selective photobleaching in an MPW. Top panel: Five successive frames of a Latrunculin-treated, immobilized cell (top row, a) and an untreated cell (bottom row, b), each of which was positioned in the corner of an inverted MPW. In each case, laser scanning of the upper left reflected image leads to selective photobleaching of the dorsal surface of the cell, as can be seen in reflected images in the same frame (bleaching occurs after frame 3 in each series). The angle of the photobleached section is very near the expected angle. Bottom panel: A SolidWorks simulation of the photobleach experiment illustrating the cell before bleaching (left) and after bleaching (right), and the shape of the bleached section of the sphere as seen from above and from both reflections. Image is courtesy of Kevin Seale. Bar is 15 µm.
The benefits of these advanced techniques will be an improved knowledge of protein-binding interactions and trafficking in Dictyostelium. This knowledge combined with that gathered in other cell systems will further our understanding of complex signaling networks and their relation to cell behavior.
Acknowledgments
The authors would like to acknowledge the Vanderbilt University Cell Imaging Shared Research Core for the use of their microscopes and software, and in particular, Sean Schaeffer, for his training and knowledge. We would also like to thank Peter van Haastert for the PLC null cell lines, Morgan Sammons for his kymograph analyses, Karl Aufderheide and other members of the Janetopoulos lab for critical analysis of the manuscript, and the DictyBase Stock Center for cells. CAE was supported by the Vanderbilt Biomedical Research and Education Training Office and NIGMS/NIH grants R01-GM080370 (to CJ) and K12-GM068543.
Footnotes
It should be noted that many fluorophores including eCFP, eGFP, and eYFP can undergo light-induced and pH-dependent reversible photobleaching (25). This could potentially generate artifacts during FRAP. If you bleach the whole cell and no recovery occurs, then this can typically be ruled out as an issue.
For FRAP experiments on organelles with complex threedimensional shapes (such as the endoplasmic reticulum (ER)), additional z-planes should probably be imaged. Several papers have discussed ways to incorporate these calculations (26–28).
Inadequate photobleaching may occur when either the number of laser iterations or the strength of the laser is too low. It is recommended that the laser is set to full power and then alter the number of iterations to avoid general photobleaching and to minimize the amount of time taken to perform the actual photobleaching.
If no recovery is observed, then it is possible that the recovery is slow and the time lapse should be extended. When doing long time lapses, it is important to space out the number of images taken in order to avoid excess exposure. Alternatively, a time delay can be set to increase the interval between images. No recovery may also result from a cell being irreversibly damaged or killed.
If the recovery is very fast, then part of it may be masked by the acquisition parameters. To correct for this, try using a scope with two separate lasers – one for imaging and one for bleaching. If a separate laser is not available, then try increasing the speed of data acquisition by only collecting data from the ROI versus the entire cell. However, it is important to note that this method sacrifices information about overall photobleaching and/or focal plane drifts which may impact your data. Alternatively, the size of the ROI can be increased.
During prebleach and postbleach image acquisition, the lowest possible illumination intensity should be used. This minimizes bleaching of the sample during monitoring. For detection purposes “Gain” and “Offset” should be set to ensure the dynamic range is exploited to the fullest.
It is important to be aware of the fact that both temperature and pH can affect diffusion, so these parameters should be kept constant (24).
Photobleaching can be damaging to cells; therefore, it is important to make sure that the cell is not too damaged or dead at the end of your time lapse. To do this make sure the cell is still moving throughout the time lapse and hasn’t rounded up (see example Fig. 4a).
Highly mobile cells are hard to FRAP and analyze; this is particularly true of cells grown on bacteria and of highly polarized cells that have been developed. To compensate for this, cells are typically only developed for 5.5 h so they are responsive to cAMP, but not quite polarized. However, this may be cell type specific and may not be possible if you are interested in cells characterized by polarized morphology. Actin inhibitors, like Latrunculin, will immobilize cells and make FRAP easier. However, as noted earlier, dissolution of actin can have significant effects on the diffusion of molecules within the cell. One alternative is to compress cells between glass and an agar layer in order to flatten them and to reduce their mobility (1). Another is to mechanically compress them in a device such as a rotocompressor (23).
References
- 1.Bretschneider T, Jonkman J, Kohler J, Medalia O, Barisic K, Weber I, et al. Dynamic organization of the actin system in the motile cells of Dictyostelium. J. Muscle Res. Cell Motil. 2002;23:639–649. doi: 10.1023/a:1024455023518. [DOI] [PubMed] [Google Scholar]
- 2.Potma EO, de Boeij WP, Bosgraaf L, Roelofs J, van Haastert PJ, Wiersma DA. Reduced protein diffusion rate by cytoskeleton in vegetative and polarized Dictyostelium cells. Biophys. J. 2001;81:2010–2019. doi: 10.1016/s0006-3495(01)75851-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yumura S, Yoshida M, Betapudi V, Licate LS, Iwadate Y, Nagasaki A, et al. Multiple myosin II heavy chain kinases: roles in filament assembly control and proper cytokinesis in Dictyostelium. Mol. Biol. Cell. 2005;16:4256–4266. doi: 10.1091/mbc.E05-03-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuzawa M, Abe T, Williams JG. The Dictyostelium prestalk cell inducer DIF regulates nuclear accumulation of a STAT protein by controlling its rate of export from the nucleus. Development. 2003;130:797–804. doi: 10.1242/dev.00303. [DOI] [PubMed] [Google Scholar]
- 5.Yumura S. Myosin II dynamics and cortical flow during contractile ring formation in Dictyosteliumcells. J. Cell Biol. 2001;154:137–146. doi: 10.1083/jcb.200011013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galdeen SA, Stephens S, Thomas DD, Titus MA. Talin influences the dynamics of the myosin VII-membrane interaction. Mol. Biol. Cell. 2007;18:4074–4084. doi: 10.1091/mbc.E06-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itoh G, Yumura S. A novel mitosis-specific dynamic actin structure in Dictyostelium cells. J. Cell Sci. 2007;120:4302–4309. doi: 10.1242/jcs.015875. [DOI] [PubMed] [Google Scholar]
- 8.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koppel DE, Axelrod D, Schlessinger J, Elson EL, Webb WW. Dynamics of fluorescence marker concentration as a probe of mobility. Biophys. J. 1976;16:1315–1329. doi: 10.1016/S0006-3495(76)85776-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reichl EM, Ren Y, Morphew MK, Delannoy M, Effler JC, Girard KD, et al. Interactions between myosin and actin crosslinkers control cytokinesis contractility dynamics and mechanics. Curr. Biol. 2008;18:471–480. doi: 10.1016/j.cub.2008.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Hagen GM, Roess DA, de Leon GC, Barisas BG. High probe intensity photobleaching measurement of lateral diffusion in cell membranes. J. Fluoresc. 2005;15:873–882. doi: 10.1007/s10895-005-0012-5. [DOI] [PubMed] [Google Scholar]
- 13.Niv H, Gutman O, Kloog Y, Henis YI. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J. Cell Biol. 2002;157:865–872. doi: 10.1083/jcb.200202009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodwin JS, Kenworthy AK. Photobleaching approaches to investigate diffusional mobility and trafficking of Ras in living cells. Methods. 2005;37:154–164. doi: 10.1016/j.ymeth.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Zaal KJ, Smith CL, Polishchuk RS, Altan N, Cole NB, Ellenberg J, et al. Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell. 1999;99:589–601. doi: 10.1016/s0092-8674(00)81548-2. [DOI] [PubMed] [Google Scholar]
- 16.Gaudet P, Pilcher KE, Fey P, Chisholm RL. Transformation of Dictyostelium discoideum with plasmid DNA. Nat. Protoc. 2007;2:1317–1324. doi: 10.1038/nprot.2007.179. [DOI] [PubMed] [Google Scholar]
- 17.Vazquez F, Matsuoka S, Sellers WR, Yanagida T, Ueda M, Devreotes PN. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc. Natl. Acad. Sci. USA. 2006;103:3633–3638. doi: 10.1073/pnas.0510570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fey P, Kowal AS, Gaudet P, Pilcher KE, Chisholm RL. Protocols for growth and development of Dictyostelium discoideum. Nat. Protoc. 2007;2:1307–1316. doi: 10.1038/nprot.2007.178. [DOI] [PubMed] [Google Scholar]
- 19.Reits EA, Neefjes JJ. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 2001;3:E145–E147. doi: 10.1038/35078615. [DOI] [PubMed] [Google Scholar]
- 20.Aufderheide KJ. An overview of techniques for immobilizing and viewing living cells. Micron. 2008;39:71–76. doi: 10.1016/j.micron.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Kenworthy AK, Nichols BJ, Remmert CL, Hendrix GM, Kumar M, Zimmerberg J, et al. Dynamics of putative raft-associated proteins at the cell surface. J. Cell Biol. 2004;165:735–746. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinnecker D, Voigt P, Hellwig N, Schaefer M. Reversible photobleaching of enhanced green fluorescent proteins. Biochemistry. 2005;44:7085–7094. doi: 10.1021/bi047881x. [DOI] [PubMed] [Google Scholar]
- 23.Sbalzarini IF, Mezzacasa A, Helenius A, Koumoutsakos P. Effects of organelle shape on fluorescence recovery after photobleaching. Biophys. J. 2005;89:1482–1492. doi: 10.1529/biophysj.104.057885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olveczky BP, Verkman AS. Monte Carlo analysis of obstructed diffusion in three dimensions: application to molecular diffusion in organelles. Biophys. J. 1998;74:2722–2730. doi: 10.1016/S0006-3495(98)77978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Partikian A, Olveczky B, Swaminathan R, Li Y, Verkman AS. Rapid diffusion of green fluorescent protein in the mitochondrial matrix. J. Cell Biol. 1998;140:821–829. doi: 10.1083/jcb.140.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picard D, Suslova E, Briand PA. 2-color photobleaching experiments reveal distinct intracellular dynamics of two components of the Hsp90 complex. Exp. Cell Res. 2006;312:3949–3958. doi: 10.1016/j.yexcr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 27.Hagen GM, Roess DA, Barisas BG. Fluorescence photobleaching recovery using total internal reflection interference fringes. Anal. Biochem. 2006;356:30–35. doi: 10.1016/j.ab.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 28.Seale KT, Reiserer R, Markov DA, Ges IA, Wright C, Janetopoulos CJ, Wikswo JP. Mirrored pyramidal wells for simultaneous multiple vantage point microscopy. J. Microsc. 2008;232:1–6. doi: 10.1111/j.1365-2818.2008.02110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]