Abstract
The mammalian immune system has the ability to discriminate between pathogenic and non-pathogenic microbes to control inflammation. Here we investigated ubiquitinylation profiles of host proteins after infection of macrophages with a virulent strain of the intracellular bacterium Legionella pneumophila and a non-pathogenic mutant. Only infection with pathogenic Legionella resulted in ubiquitinylation of positive regulators of the metabolic checkpoint kinase mTOR leading to diminished mTOR activity. Detection of pathogen signatures resulted in translational biasing to proinflammatory cytokines through mTOR-mediated regulation of cap-dependent translation. Thus, there is a pathogen detection program in macrophages that stimulates protein ubiquitinylation and degradation of mTOR regulators, which suppresses mTOR function and directs a proinflammatory cytokine program.
Pattern recognition receptors (PRRs) are used by mammalian cells to detect conserved molecular patterns presented by microbes. When PRRs bind microbial ligands they induce transcriptional responses that promote inflammation through the production of cytokines by the responding cells.1 The magnitude of the host inflammatory response can be greater in response to pathogenic microbes when compared to non-pathogenic organisms. This is in part because many pathogenic organisms manipulate host processes by either entering the host cytosol to deliver proteins or by using specialized secretion systems to deliver bacterial effectors across a membrane barrier into the cytosol. When membrane barriers are breached by pathogens, microbial signatures are often detected by cytosolic PRRs that activate additional innate immune pathways to control infection. Although PRRs play an important role in detecting pathogens, they are not sufficient to account for the differences in host responses to pathogenic and non-pathogenic microbes, which indicates that other signaling pathways must be involved.2
The signaling mechanisms mediating pathogen responses are complex and involve many different proteins that rapidly transduce molecular information in the cell.1 Typically, rapid signaling responses involve post-translational modifications (PTMs) to proteins in a signaling cascade.3 PTMs often serve as molecular ‘on/off’ switches that modulate the activity of a signaling network. For instance, signaling pathways of the innate immune system are controlled by PTMs that include phosphorylation and ubiquitinylation of proteins in the regulatory circuits.1, 3, 4 For this reason, PTM mapping of cellular networks5 has been implemented successfully in several studies to elucidate novel aspects of macrophages responses to lipopolysaccharide (LPS), lysine acetylation-regulated cellular pathways and SUMO-dependent heat-shock responses.3, 6, 7 Thus, it stands to reason that similar approaches would provide new insight into how cells discriminate between pathogenic and non-pathogenic bacteria.
The bacterium Legionella pneumophila has been used to investigate innate immune signaling pathways directed against intracellular pathogens.8 Legionella is a common inhabitant of fresh water and soil ecosystems, where the organism proliferates inside of protozoan hosts that feed on bacteria. Legionella has the ability to replicate inside of macrophages even though it has not co-evolved with mammalian hosts.9 A bacterial type IV secretion system called Dot/Icm is required for Legionella replication inside of protozoan hosts and mammalian macrophages.10, 11 The Dot/Icm system delivers effector proteins into the host cell that enables the vacuole containing Legionella to avoid fusion with lysosomes and to develop into a specialized vacuole that supports bacterial replication.12
There is a robust cytokine response detected shortly after the infection of macrophages with Legionella, and this response is attenuated when macrophages are infected with a non-pathogenic strain of Legionella having the Dot/Icm system inactivated.13 Cytosolic PRRs such as Nod1, Nod2, Rig-I and Naip5 play a role in discriminating between pathogenic Legionella and mutant Legionella having the Dot/Icm system inactivated13–15; however, studies using knockout mice deficient in these canonical innate immune pathways suggest that stimulation of these cytosolic PRRs cannot account for all of the differences observed when macrophages are infected with virulent or Dot/Icm-deficient Legionella. Thus, to gain new insight into pathogen-specific signaling cascades that are differentially regulated during infection we have analyzed the ubiquitin PTM network during infection of macrophages induced by virulent and Dot/Icm-deficient Legionella. These data revealed that infection of macrophages by pathogenic Legionella induced ubiquitinylation-dependent degradation of proteins in the mTOR pathway. Downregulation of mTOR activity during infection of macrophages by pathogenic Legionella suppressed cap-dependent translation, which enhanced a proinflammatory cytokine response that provided host defense against infection.
RESULTS
Analysis of pathogen-induced macrophage responses
To investigate pathogen-specific responses we used liquid chromatography tandem mass spectrometry (LC-MS/MS) to compare patterns of host protein ubiquitinylation in response to virulent Legionella and an isogenic ΔdotA mutant that is non-pathogenic because it is deficient in Dot/Icm function. A mouse macrophage-like cell line (RAW267) was used that stably produced either a tandem affinity-tagged version of ubiquitin (Ub) or the UbG76V protein, a mutant version of Ub that is defective for conjugation (Supplementary Fig. 1). After infection, proteins containing the tagged-Ub were isolated under stringent denaturing conditions to prevent post-lysis processing of the Ub moieties, and peptides containing Ub conjugates were identified by LC-MS/MS analysis. These data were analyzed to identify proteins that were differentially ubiquitinylated in cells infected with pathogenic Legionella compared to proteins from cells infected with the non-pathogenic strain. The ubiquitinylated host proteins identified were superimposed onto known molecular networks in silico (Supplementary Fig. 2 and Supplementary Tables 1–3). Pathway over-representation analysis revealed that innate immune cascades were enriched within the pathogen-specific response. Moreover, cascades previously shown to respond differently in macrophages following infection by pathogenic Legionella, such as the NF-kB, MAPK and IFNα/β pathway,13, 14, 16 were represented within the pathogen-specific response network (Supplementary Fig. 2). Thus, the analysis was successful at identifying pathways differentially regulated in response to pathogenic Legionella.
Infections by virulent Legionella suppress mTOR function
The mTOR pathway was highly represented in the analysis of proteins ubiquitinylated after infection of macrophages with virulent Legionella. This pathway plays an important role in regulating host metabolism and protein translation in response to changing environmental conditions,17 and there is growing evidence suggesting that mTOR is involved in immune regulation.18 The proteins phosphatidylinositol 3-kinase (PI3K) and Akt are important activators of mTOR. Our data indicated that infection of macrophages with virulent Legionella favored ubiquitinylation of PI3K, Akt and mTOR, which suggests that this pathway was differentially regulated in response to pathogen infection (Supplementary Fig. 2). Thus, we investigated whether the regulation of this pathway varied in response to virulent Legionella compared to avirulent mutants by measuring phosphorylation of Akt, mTOR and mTOR substrates as indicators of a functional protein kinase cascade in the cell.
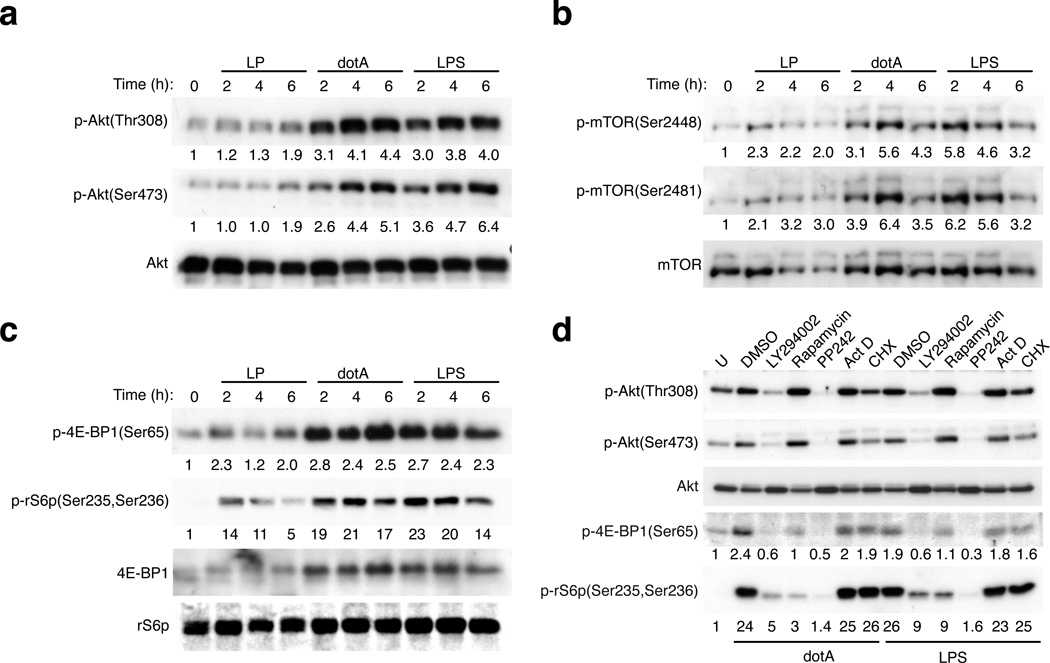
Robust phosphorylation of Akt (Fig. 1a), mTOR (Fig. 1b) and the mTOR substrates 4E-BP1 and the ribosomal protein S6 (Fig. 1c) were observed after infection of macrophages with the avirulent ΔdotA mutant of Legionella indicating an active mTOR pathway. The mTOR pathway was activated to a similar extent by LPS, which is a known mTOR pathway agonist that served as a positive control (Fig. 1). Chemical inhibition of PI3K activity (Supplementary Fig. 2) blocked mTOR activation by the dotA mutant but inhibition of transcription with actinomycin D or translation inhibition by cycloheximide did not (Fig. 1d). Thus, activation of mTOR during infection by avirulent Legionella was independent of host transcription and translation but required PI3K signaling.
Figure 1. PI3K-AKT-mTOR pathway is suppressed in pathogenic Legionella infections.
(a) Immunoblot analysis of AKT-mTOR pathway activation in BMMs infected with pathogenic Legionella (LP) or the avirulent dotA mutant or treated with LPS (100 ng/ml) as indicated. Phosphorylation of (a and d) Akt, (b) mTOR, (c and d) 4E-BP1 and the ribosomal S6 protein (rS6p) (d) BMMs were pre-treated with inhibitors for 30min prior to infection or stimulation with LPS (100 ng/ml) for 4hrs. The following inhibitors were used: PI3K – LY294002 [10µM], mTOR - Rapamycin [100 nM], mTOR - PP242 [2.5 µM], transcription - actinomycin D (Act D) [500ng/ml], translation – cycloheximide (CHX) [50 µg/ml]. Densitometry of signal intensity normalized to a respective loading control are listed for each immunoblot. One of six biological replicates is shown.
To measure mTOR activation in response to virulent Legionella (LP) with a functional Dot/Icm system we used an isogenic strain that had the flagellin gene deleted (ΔflaA), which prevented activation of a rapid cell death response in mouse macrophages via pyroptosis. This virulent strain also had a defective thymidylate synthetase gene (thyA), which limited intracellular replication and allowed virulent and avirulent bacteria to be maintained at similar amounts after infection of macrophages. Phosphorylation of substrates in the mTOR pathway was significantly lower in macrophages infected with virulent Legionella (Figs. 1a–c) and suppression of the mTOR pathway was dose dependent (Supplementary Fig. 3a). MAPK activation in macrophages was equivalent in response to virulent and avirulent Legionella (Supplementary Fig. 3b), which indicated that these bacteria stimulated the Toll-like receptor (TLR) pathway similarly. Thus, increased ubiquitinylation of proteins in the mTOR pathway represents a host response that negatively regulates this signaling cascade following infection by Legionella that display signatures of being a pathogen.
Akt-mTOR control cytokine bias independent of autophagy
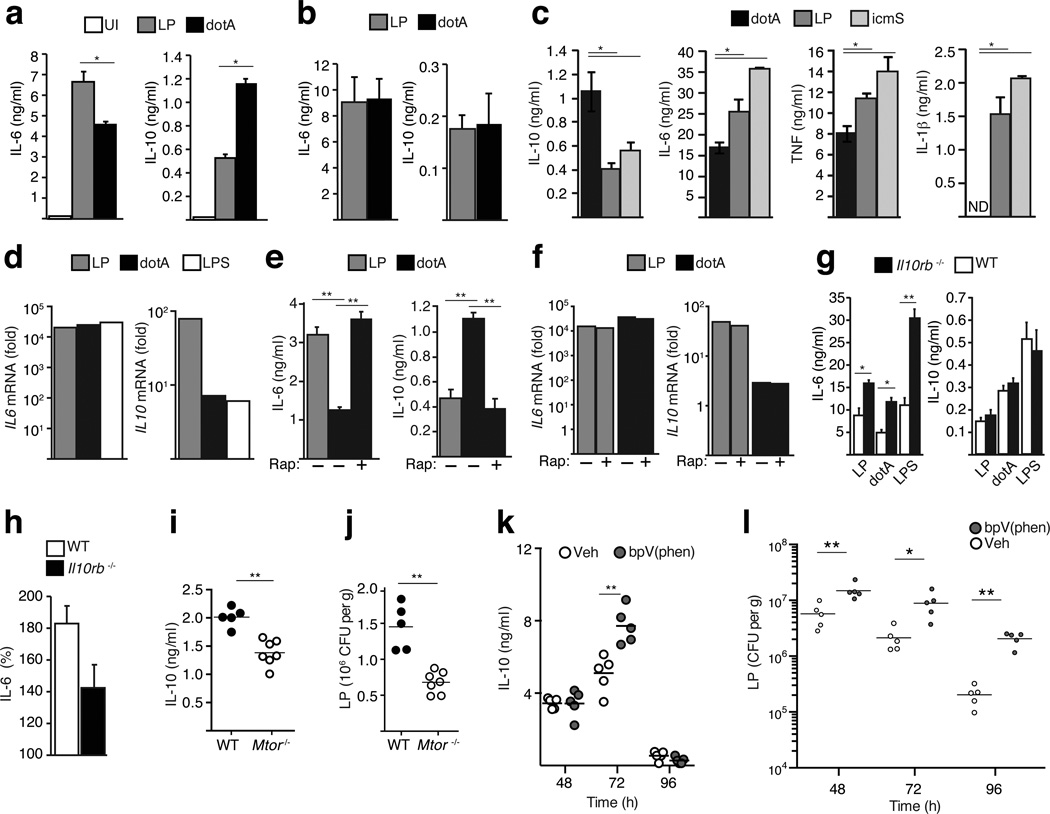
Macrophages produce proinflammatory cytokines such as IL-6, TNF, IL-1β, and also cytokines such as IL-10, IFNα/β and TGFβ that suppress inflammation.1, 19, 20 A strong proinflammatory response as measured by IL-6 secretion was observed when bone marrow-derived macrophages (BMMs) were infected with virulent Legionella; however, lower concentrations of proinflammatory cytokines and higher amounts of the immunosuppressive cytokine IL-10 were observed in BMMs infected with the avirulent dotA mutant (Fig. 2a). Heat-killed bacteria elicited responses similar to the dotA mutant, which indicated that the robust proinflammatory response to virulent Legionella required bacterial viability (Fig. 2b). A Legionella ΔicmS mutant induced a response that was similar to virulent Legionella (Fig. 2c). Although the icmS mutant has a functional type IV system, the IcmS protein is required for efficient translocation of multiple effectors,21 which prevents the icmS mutant from trafficking correctly (Supplementary Fig. 3c) and replicating in primary macrophages.22 Thus, the robust proinflammatory response triggered by virulent Legionella required a functional Dot/Icm system but was independent of efficient effector translocation and establishment of the specialized vacuole that supports bacterial replication.
Figure 2. Suppression of mTOR mediates cytokine biasing in response to pathogenic Legionella.
(a–c) ELISA analysis of cytokine responses in uninfected (UI) and infected BMMs as indicated with live (a and c) or heat-killed bacteria (b). (d) qPCR expression analysis in BMMs infected as indicated for 4hrs. (e and f) Cytokine responses in BMMs pre-treated with Rapamycin [100 nM] or vehicle alone for 60min then infected as indicated. ELISA analysis (e) of secreted cytokines and qPCR analysis (f) of mRNA induction. (g and h) Cytokine responses (g) in C57BL/6 (WT) and Il10rb−/− BMMs infected as indicated. (h) Change in the ratio (LP response/dotA response) of IL-6 secretion by WT and il10rb−/− BMM. For all ELISA analysis average values of technical replicates ± s.d are presented in all panels. One of six biological replicates is shown (*) p<0.05, (**) p<0.005 (paired T-test) (i–l) IL-10 concentration (i) and colony-forming units (CFU) (j) were measured in lungs isolated from mice with macrophages deficient in mTOR (Mtor−/−) and WT littermates control mice 48 hours after infection with virulent Legionella. (k and l) C57BL/6 mice injected with bpV(phen) or vehicle control were infected with virulent Legionella and IL-10 levels (k) and CFU (l) data were obtained from lungs isolated at the times indicated. (i–l) Each point represents data obtained from a single mouse and lines indicate the means for each group of mice. (*) p<0.05, (**) p<0.005 (Mann-Whitney U-test) One of two biological replicates is shown.
Although there were significant differences in cytokine production in response to virulent Legionella and dotA mutants, these differences did not correlate with gene expression. The level of Il6 mRNA was similar under all infection conditions, and Il10 mRNA levels were higher after infection by virulent Legionella even though IL-10 production was lower (Fig. 2d). Thus, the cytokine biasing observed in response to virulent Legionella involved post-transcriptional regulation of gene expression.
Genetic and pharmacological agents that perturb signaling in the mTOR pathway will increase production of proinflammatory cytokines in innate immune cells following activation by microbial products.23–25 Thus, it was possible that differential regulation of mTOR signaling in response to pathogenic and non-pathogenic Legionella was involved in the cytokine biasing observed during infection of macrophages. Consistent with this hypothesis, a significant increase of IL-6 secretion concomitant with a decrease in IL-10 production was observed when mTOR signaling through the TORC1 complex was blocked using rapamycin in macrophages infected with the dotA mutant (Fig. 2e). Similar to what was observed during infection of macrophages by virulent Legionella, rapamycin treatment enhanced dotA-induced inflammation without affecting gene expression (Fig. 2f). Additionally, mTOR signaling was suppressed in macrophages infected with the icmS mutant (Supplementary Fig. 3d). Moreover, cytokine biasing was still observed in macrophages deficient in the IL-10 receptor (Figs. 2g and 2h), indicating that this is an intrinsic response that does not require feedback inhibition mediated by IL-10. Because Legionella potently inhibits autophagy using the effector protein RavZ, it was predicted that cytokine biasing would not require autophagy, even though autophagy is typically induced when mTOR function is suppressed. Indeed, cytokine biasing was unaffected in macrophages deficient for Atg5 (Atg5−/−) – a protein critical for autophagy - and infection of macrophages with a ΔravZ mutant of Legionella did not augment cytokine biasing, which indicates that cytokine biasing by mTOR suppression does not require autophagy (Supplementary Fig. 3e and 3f). Thus, cytokine biasing involved a host response that suppressed mTOR activity after macrophage infection by virulent Legionella.
To determine whether mTOR suppression would enhance host protection against infection we used mice where the Mtor gene was deleted specifically in cells of the myeloid lineage, which included macrophages (Supplementary Figs. 3g and 3h). IL-6 and IL-10 secretion by Mtor-deficient BMMs infected with the avirulent dotA strain were similar to those produced by control BMMs infected with pathogenic Legionella (Supplementary Fig. 3h), which indicated that BMMs from the Mtor-deficient mice had a defect in cytokine biasing triggered by pathogen detection. Mtor-deficient mice were more resistant to pulmonary infection by virulent Legionella as indicated by lower numbers of bacteria and an enhanced pro-inflammatory cytokine response in the lung compared to Mtor-sufficient littermate control mice (Figs. 2i and 2j). Conversely, mice were more sensitive to Legionella infection when mTOR activation was augmented by maintaining Akt activation, which was achieved by treating mice with bisperoxo(1,10-phenanthroline)oxovanadate [bpV(phen)] to interfere with the phosphatase PTEN that negatively regulates Akt (Figs. 2k and 2l). Thus, the ability to respond to pathogenic Legionella and suppress mTOR function enables the host to efficiently combat infection.
Pathogenic infections inhibit cap-dependent translation
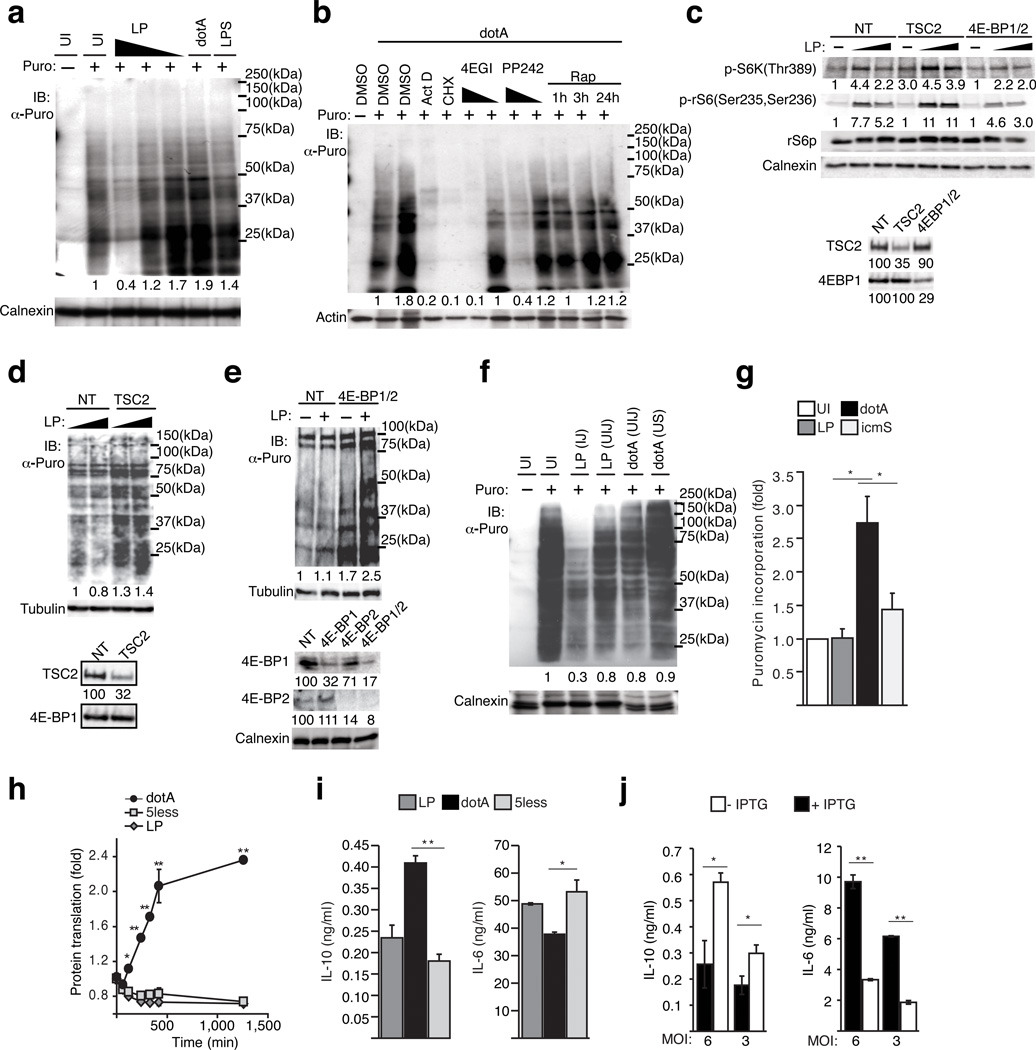
The mTOR pathway regulates ribosome biogenesis and the initiation of cap-dependent translation in mammalian cells.17 When macrophages were stimulated with LPS or infected with the non-pathogenic dotA there was a measurable increase in the rate of protein synthesis as determined by puromycin incorporation26 into nascent polypeptides (Fig. 3a). An increase in host translation stimulated by LPS or non-pathogenic Legionella was not detected when mTOR activity was blocked using the specific inhibitors rapamycin or PP24227 (Fig. 3b and Supplementary Fig. 4). Similarly, increased translation induced by LPS or non-pathogenic Legionella was blocked using 4EGI-128 (Fig. 3b), which is an inhibitor that binds eIF4E to prevent formation of the cap-dependent translation initiation complex eIF4F (Supplementary Fig. 2f). In contrast to the increase in translation detected after infection by a dotA mutant, there was a dose-dependent decrease in host protein translation when macrophages were infected with virulent Legionella (Fig. 3a and Supplementary Fig. 4). To test whether suppression of the mTOR pathway was sufficient to interfere with host translation after infection by virulent Legionella we used siRNA to silence either TSC2 or the proteins 4EBP-1 and 4EBP-2, which derepressed mTOR activation (Fig. 3c) and eIF4F assembly respectively. TSC2 functions as a negative regulator of mTOR, and the proteins 4EBP-1 and 4EBP-2 are inhibitors of cap-dependent translation that are inactivated when phosphorylated by mTOR (Supplementary Fig. 2f). Translational suppression after infection by virulent Legionella was not observed in macrophages when TSC2 or 4EBP-1 and 4EBP-2 were silenced in macrophages (Figs. 3d and 3e). The amounts of phospho-eIF2α in BMMs and epithelial cells infected with pathogenic Legionella did not increase compared to the levels after infection with non-pathogenic Legionella, which indicates that suppression of mTOR is not mediated by a host starvation response (Supplementary Figs. 4g and 4h). Thus, negative-regulation of mTOR activity is sufficient to suppress host translation following infection by virulent Legionella.
Figure 3. Suppression of mTOR by pathogenic Legionella inhibits cap-dependent translation.
(a, b, d–g) Host translation measured by puromycin incorporation. (a) Translation rates in BMMs uninfected (UI), infected with Legionella [MOI = 20, 10, 5], dotA [MOI= 20] or stimulated with LPS. (b) BMMs pre-treated with 4EGI-1 [25 µM, 2.5 µM], actinomycin D [500ng/ml], cycloheximide [50µg/ml], PP242 [25 µM, 2.5 µM] for 30min or Rapamycin [100 nM] as indicated and infected with dotA. (c) Depletion of TSC2 restored mTOR activation in pathogenic infections. Immunoblot analysis of S6 kinase and rS6p phosphorylation in BMMs silenced with non-targeting (NT) siRNA or siRNA against TSC2 or 4EBP1 and 4EBP2. Silencing efficiency for each gene is quantified. (d and e) Puromycin incorporation by BMMs knocked down with siRNA against TSC2 (d), 4EBP-1 and 4EBP-2 (e) or non-targeting (NT) control following Legionella infection [MOI=10 and 30]. Silencing efficiency is quantified. (f) Puromycin incorporation rates in macrophages injected (IJ) and uninjected (UIJ) by the Legionella T4SS (see Supplementary Fig. 5). dotA infections produced only UIJ cells and unsorted (US) dotA infected cells are shown as control. (g) Puromycin incorporation in BMMs quantified by immunoblots densitometry and normalized to uninfected control. Averages from biological triplicates ± s.d. (*) p < 0.05 (paired T-test). (h) Kinetic analysis of fluorescent translation reporter macrophages (see Supplementary Fig. 4) infections. Averages from technical triplicates + s.d (*) p < 0.05 (ANOVA). (i and j) Cytokine responses in BMMs infected with (i) Legionella, 5less, dotA strains or (j) dotA expressing LLO under an IPTG-inducible promoter. Averages from technical triplicates ± s.d. (*) p<0.05 (**) p<0.005 (paired T-test)
A pathogen signature triggers translational repression
After macrophages were infected by virulent Legionella we used a fluorescence-based cell-sorting protocol to separate macrophages that were injected by the Legionella Dot/Icm system from macrophages that were not injected (Supplementary Fig 5).29 Puromycin incorporation assays showed equivalent levels of protein translation in the population of uninjected macrophages as in macrophages that were infected with a dotA mutant (Fig. 3f). By contrast, the macrophage population that had been injected by the Dot/Icm system had reduced rates of protein translation compared to the uninjected macrophages from the same culture (0.3 vs. 0.8) (Fig. 3f). Thus, an autonomous response that interfered with host translation was triggered only in cells that received a signal from Legionella having a functional Dot/Icm system.
We next addressed whether the cell-autonomous response that limits host translation was mediated directly by Legionella effector proteins or whether translation inhibition was a general host response to bacteria that display a pathogenic signature. Although Legionella icmS mutants do not deliver effectors into host cells efficiently, and this results in vacuoles that fail to mature into ER-like compartments (Supplementary Fig 3c), the icmS mutant activates several effector-independent innate immune pathways that respond to the pathogenic signature displayed by a functional Dot/Icm system (Fig. 2c).13, 15, 16 Similarly, macrophages infected with the icmS mutant suppressed translation to an extent that was equivalent to macrophages infected with virulent Legionella (Fig. 3g), which suggests that translation inhibition is a cell-autonomous response that does not require the direct activities of Legionella effector proteins. There are five Legionella effectors (Lgt1, Lgt2, Lgt3, SidI and SidL) having biochemical activities that interfere with host protein synthesis when these proteins are expressed ectopically in eukaryotic cells.30 To test whether these effectors were required for the general suppression of host translation observed during infection, we used a Legionella Δ5less mutant that has the genes encoding these five effectors deleted from the chromosome.30 The Legionella 5less mutant suppressed cap-dependent translation to an extent that was indistinguishable from the parental strain (Fig. 3h). The cytokine profile displayed by macrophages infected with the 5less mutant was similar to that of macrophages infected with virulent Legionella (Fig. 3i). Thus, cytokine biasing did not require these effectors.
To investigate whether cytokine-biasing might be an effector-independent process that is triggered in response to bacteria with pathogenic traits, a plasmid was introduced into the dotA mutant of Legionella that produces the pore-forming toxin Listeriolysin O under the control of an IPTG-inducible promoter. It has been shown that non-pathogenic bacteria such as E. coli that harbor this plasmid will damage the vacuoles in which they reside when grown under inducing conditions, and this will trigger host cytosolic immune responses.31 Indeed, the cytokine profile for macrophages infected with dotA mutant bacteria grown under non-inducing conditions (−IPTG) favored IL-10 production over IL-6 production, whereas, macrophages infected with dotA mutant bacteria grown under conditions that promote LLO production (+IPTG) showed a bias toward more IL-6 production and lower IL-10 production (Fig. 3j). Ubiquitinylation of membrane regions surrounding dotA mutant bacteria were not apparent in the infected cells after induction of LLO, which suggests this response is not dependent on recruitment of adapter proteins such as NDP52 that signal and direct autophagy in response to cytosolic pathogens.32 This is consistent with studies showing Galectin-3 is not accessible to most virulent Legionella after infection of macrophages, which means these bacteria rarely escape the vacuole and enter the cytosol.33 Thus, vacuole damage caused by LLO production and signaling by the Dot/Icm system are likely to facilitate the release of microbial-associated molecular patterns into the host cytosol, and detection of microbial determinant in the cytosol then suppresses mTOR activity to modulate cytokine production.
Cytokine biasing is mediated by mTOR regulation of eIF4E
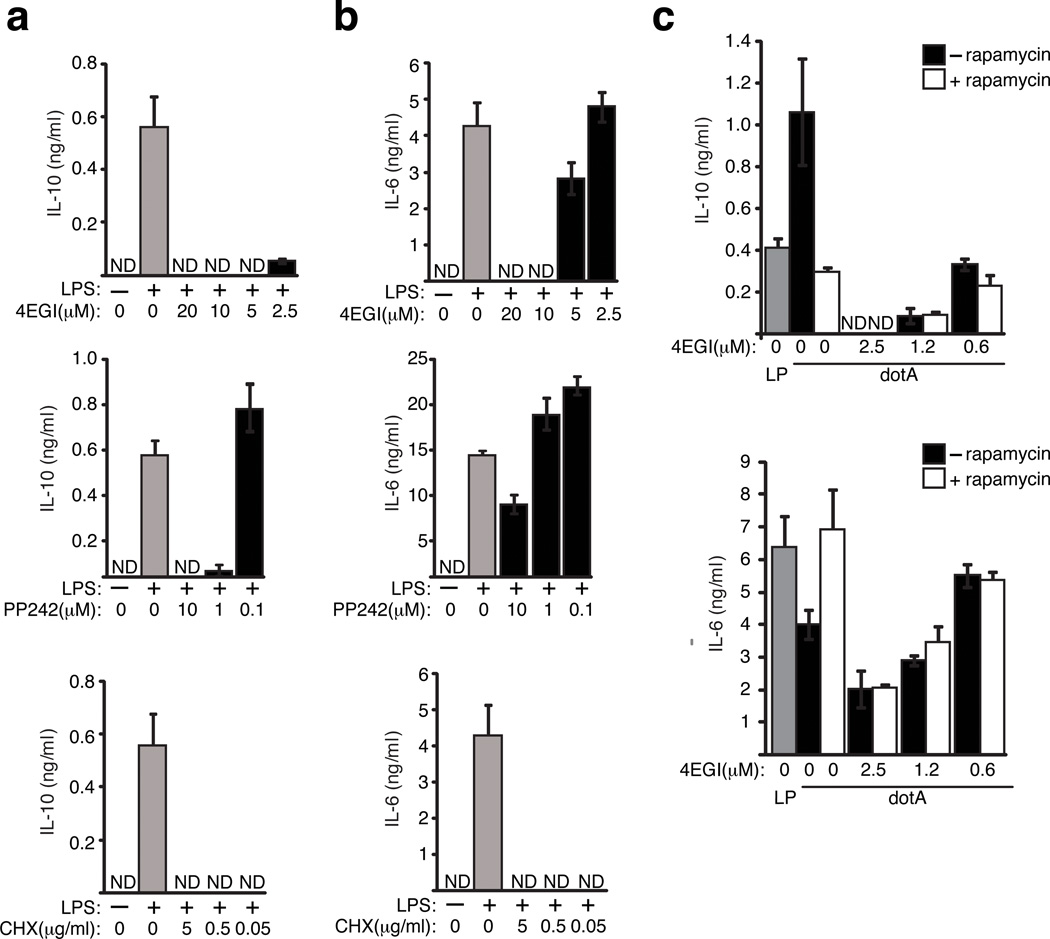
One mechanism by which mTOR regulates translation is through phosphorylation of the negative regulator 4E-BP1, which releases the translation initiation factor eIF4E to assemble into the eIF4F complex to initiate cap-dependent translation.17 The peptidomimetic inhibitor 4EGI-1 blocks translation by interfering specifically with the interaction between eIF4E and eIF4G (Supplementary Fig. 2f). Cytokine production was inhibited in a dose-dependent manner upon treating LPS-stimulated macrophages with 4EGI-1 or PP242 (Figs. 4a and 4b), which indicates that mRNAs encoding these cytokines were translated by a cap-dependent mechanism requiring mTOR function. From these studies it became evident that IL-10 production was more sensitive to eIF4E inhibition than IL-6 production. IL-10 production was not detectable when macrophages were treated with 4EGI-1 [2.5 µM] or with PP242 [1µM] (Fig. 4a), whereas, the production of IL-6 was not compromised at the same concentrations of these inhibitors (Fig. 4b). By contrast, cytokine production was completely abolished when cyclohexamide was used to interfere with translation at the elongation stage (Figs. 4a and 4b). Thus, cytokine biasing in LPS-treated macrophages was achieved by modulating cap-dependent translation initiated by eIF4E.
Figure 4. Cytokine biasing results from inhibition of eIF4F assembly.
ELISA analysis of IL-10 (a) and IL-6 (b) secreted by BMMs stimulated with LPS [100 ng/ml] for 18hrs in the presence of 4EGI-1, PP242 or cycloheximide. (c) Cytokine responses in BMMs pre-treated with increasing amounts of 4EGI-1 in the presence or absence of Rapamycin [100 nM] for 60 min and infected [MOI = 6] for 18hrs. Average values of three technical replicates + s.d are presented in all panels. The data presented in all panels is one of three biological replicates.
If cytokine biasing observed during infection by pathogenic Legionella is mediated primarily by downregulating the ability of mTOR to promote eIF4E-dependent translation, then rapamycin and 4EGI-1 should have an epistatic relationship in biasing cytokine responses following macrophage infection by a dotA mutant. Indeed, 4EGI-1 treatment of macrophages infected with the dotA mutant resulted in dose-dependent cytokine biasing but this effect was not augmented by the addition of rapamycin (Fig. 4c). The absence of synergy in this assay further supports that cytokine biasing is mediated by mTOR regulation of eIF4E function.
eIF4E inhibition favors abundant transcripts translation
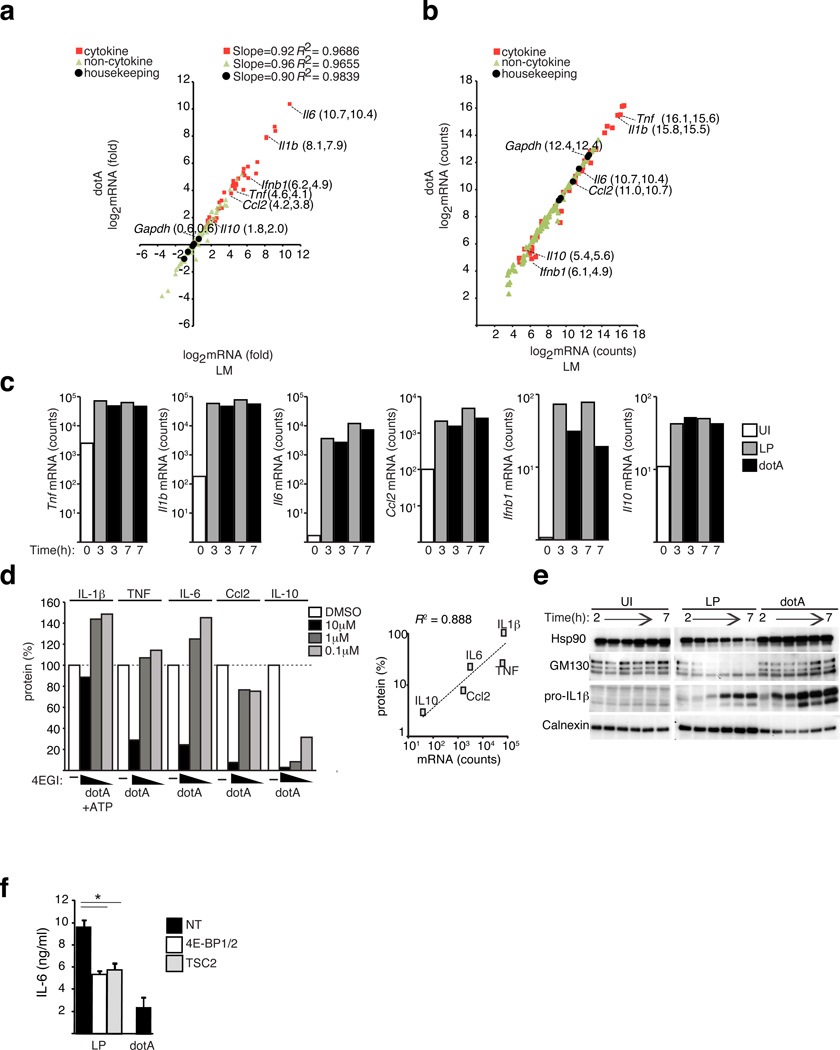
There was no evidence suggesting that structural cis-acting elements in the untranslated regions of the cytokine mRNAs could account for selective translation that displayed an expression bias during infection by virulent Legionella (Supplementary Fig. 6). Thus, we investigated whether mRNA abundance might account for mTOR-regulated biasing toward the translation of proinflammatory genes during infection of macrophages by pathogenic Legionella. Transcript levels were determined for 179-different genes involved in immune regulation following macrophage infection with virulent Legionella and compared to the levels induced following macrophage infection by the dotA mutant (Supplementary Table 4). This comparison revealed no differences in the mRNA levels that would explain cytokine biasing (Fig. 5a). Expression levels for the mRNA transcripts encoding different cytokines spanned four orders of magnitude (Fig. 5b). Many proinflammatory transcripts (Il1b, Tnf, Il6) were highly abundant, and Il10 transcripts were less abundant (Fig. 5b). Following infection the cytokine mRNA levels peaked at 3 h and were maintained at 7 h (Fig. 5c). The amount of Il6 mRNA detected after infection by Legionella was 85-fold higher than Il10 mRNA (3502 vs. 41 counts) (Fig. 5c). The sensitivity of cytokine production to inhibition by 4EGI-1 correlated significantly with mRNA quantity for multiple proteins tested (Pearson coefficient 0.888, p<0.05) (Fig. 5d). A decrease in multiple cellular proteins resulting from the mTOR-regulated block in translation was observed during infection by virulent Legionella but not during infection by dotA mutant bacteria (Fig. 5e). By contrast, infection by virulent Legionella did not interfere with the synthesis of pro-IL-1β (Fig. 5e). Similar results were obtained for other highly abundant transcripts encoding proinflammatory cytokines (Fig. 2c and Supplementary Figs. 7a–c); however, translational biasing was not cytokine specific, as messages encoding calnexin were also highly abundant in macrophages and calnexin synthesis was not adversely affected during infection of macrophages by virulent Legionella (Supplementary Figs. 7d–f).
Figure 5. Transcript abundance determines sensitivity to eIF4F inhibition.
(a–c) nanoString analysis of induction (a) and mRNA abundance (b) of immune and housekeeping genes induced in BMMs infected with Legionella. or dotA for 3hrs [MOI=10]. Linear regression of logarithmic transformed data and R-squared values are shown for the different gene categories. One of two biological replicates is shown. (c) Kinetic analysis of cytokine mRNA abundance in BMMs infected as indicated [MOI=10]. One of two biological replicates is shown. (d) 4EGI-1 sensitivity of cytokine or chemokine synthesis induced by dotA infections of BMMs for 18hrs [MOI = 6]. For IL-1β release, ATP was added for 30min before sample collection. 4EGI-1 was added 30min prior to infection. The secreted proteins were measured by ELISA and are shown as a percentage of DMSO treated controls. The correlation between cytokine synthesis in cells treated with 4EGI-1 (10 µM) as percentage of DMSO controls vs. mRNA abundance is shown, Pearson R2=0.888 (e) Kinetics of protein expression in synchronized BMMs infections by immunoblot analysis. (f) ELISA of IL-6 secretion in BMMs knocked down with siRNA targeting TSC2 or 4E-BP1 and 4E-BP2 or non-targeting (NT) control and infected as indicated for 18hrs [MOI = 6], averages of technical triplicates ± s.d. (*) p<0.05 (paired T-test). Unless indicated, presented data is representative of at least three biological replicates.
Consistent with previous studies indicating that the Dot/Icm system induces the expression of type I interferon genes, we found that transcripts encoding IFN-β (Ifnb1) were higher following infection of macrophages with virulent Legionella (Fig. 5c). The induced levels of Ifnb and Il10 transcripts were less abundant than many of the proinflammatory gene transcripts (Fig. 5b). Similar to what was observed for the cytokine IL-10, bioassays revealed higher amounts of bioactive type I interferon were produced by macrophages infected with the dotA mutant compared to macrophages infected with virulent Legionella (Supplementary Fig. 7g). When the proteins 4E-BP1/2 or TSC2 were silenced in macrophages, which was shown to eliminate translational suppression by mTOR (Fig. 3d and 3e), cytokine biasing was no longer observed as indicated by the reduction in IL-6 production following infection with virulent Legionella to concentrations that were similar to macrophages infected with the dotA mutant (Fig. 5f). Thus, downregulation of mTOR function following infection by virulent Legionella results in suppression of cap-dependent translation, which mediates cytokine biasing by favoring the translation of highly abundant messages encoding proinflammatory cytokines such as IL-6 and IL-1β and interfering with the synthesis of the less abundant transcript encoding IL-10 and IFN-β.
Legionella stimulates ubiquitinylation of activated Akt
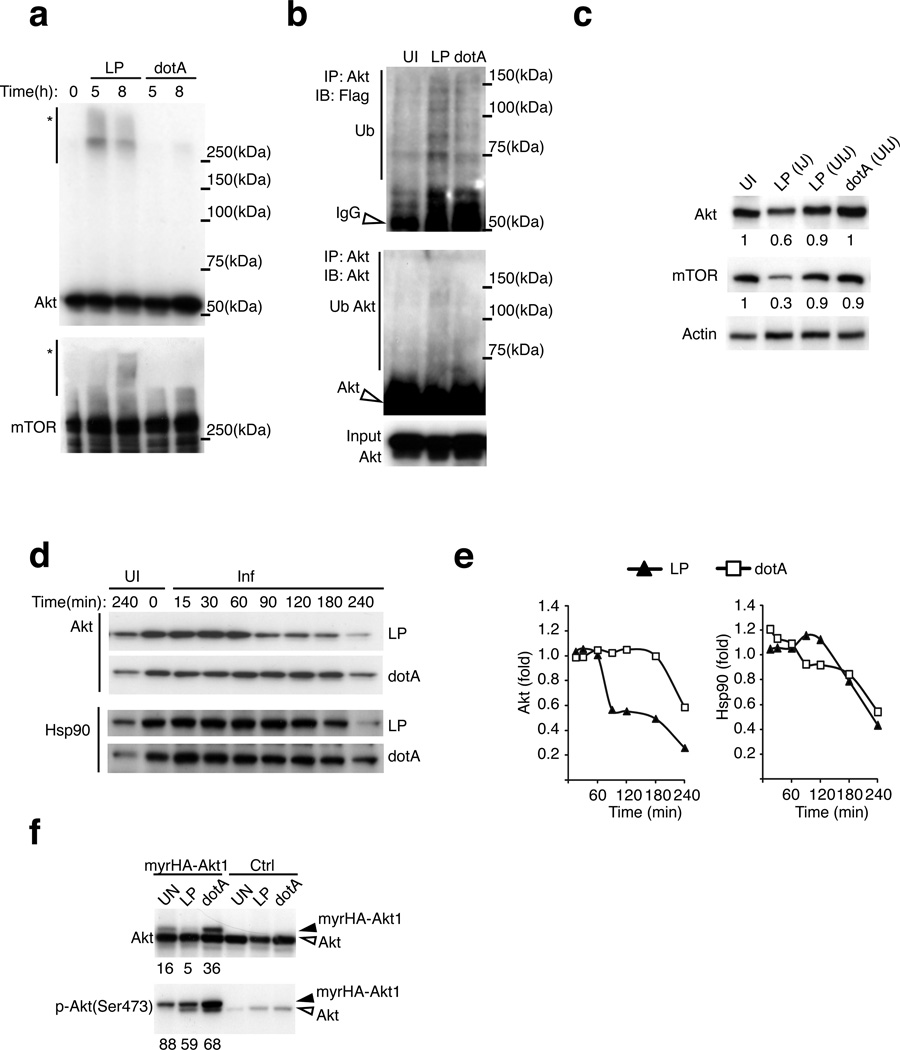
Akt ubiquitinylation was detected preferentially in the PTM analysis of macrophages infected with virulent Legionella. In agreement with the PTM analysis, mobility shift and immunoblot assays revealed that infection of macrophages by virulent Legionella, but not by the dotA mutant, resulted in ubiquitinylation of Akt and mTOR (Figs. 6a and 6b). Akt and mTOR ubiquitinylation coincided with reductions in the amounts of these proteins in macrophages following infection by virulent Legionella (Fig. 6c). When de novo protein synthesis was blocked using cyclohexamide the degradation of Akt was accelerated in cells infected with virulent Legionella, whereas, no enhanced degradation was observed for Hsp90 (Figs. 6d and 6e), which indicates that pathogen-induced Akt ubiquitinylation stimulates protein degradation.
Figure 6. Activated Akt is degraded by a pathogen-induced ubiquitinylation pathway.
(a) Appearance of high molecular weight forms (*) of Akt and mTOR. Immunoblot analysis of total cell lysates of BMMs infected as indicated [MOI = 20] and lysed in 8M Urea buffer to preserve PTMs. (b) Ubiquitination of Akt immunoprecipitated from lysates of RAW264.7 macrophages stably expressing 3XFlag-ubiquitin infected as indicated for 4hrs [MOI=50]. (c) Immunoblot analysis of Akt and mTOR expression in BMMs injected (IJ) and uninjected (UIJ) by the T4SS. Densitometry of band intensity normalized to a respective loading control are indicated. (d and e) Turnover of Akt and HSP90 proteins in BMMs infected as indicated in the presence of cycloheximide [50 µg/ml]. Densitometry analysis (e) of the protein amounts detected by immunoblot in (d). (f) Akt expression in BMMs transduced with HA-tagged myrAkt1 and infected as indicated for 4hrs [MOI = 20]. The signal ratio of myrHA-Akt1 to endogenous Akt was determined by densitometry and is shown below each panel. The data presented in all panels is one of three biological replicates.
Activation of Akt requires generation of phosphatidylinositol (3,4,5)-trisphosphate (PIP3) at the plasma membrane, which is recognized by a pleckstrin homology domain in Akt. Appending a myristoylation motif to Akt (myrAkt) results in PIP3-independent membrane recruitment and activation.34 When produced in macrophages the majority of the myrAkt protein was phosphorylated, which indicates myrAkt is constitutively active (Fig. 6f). After infection of macrophages with virulent Legionella the degradation of myrAkt occurred more rapidly when compared to endogenous Akt (Fig. 6f), which indicates that the active pool of membrane-associated Akt is the primary target for pathogen-induced ubiquitinylation and degradation.
Pathogen-induced Akt ubiquitinylation suppresses mTOR
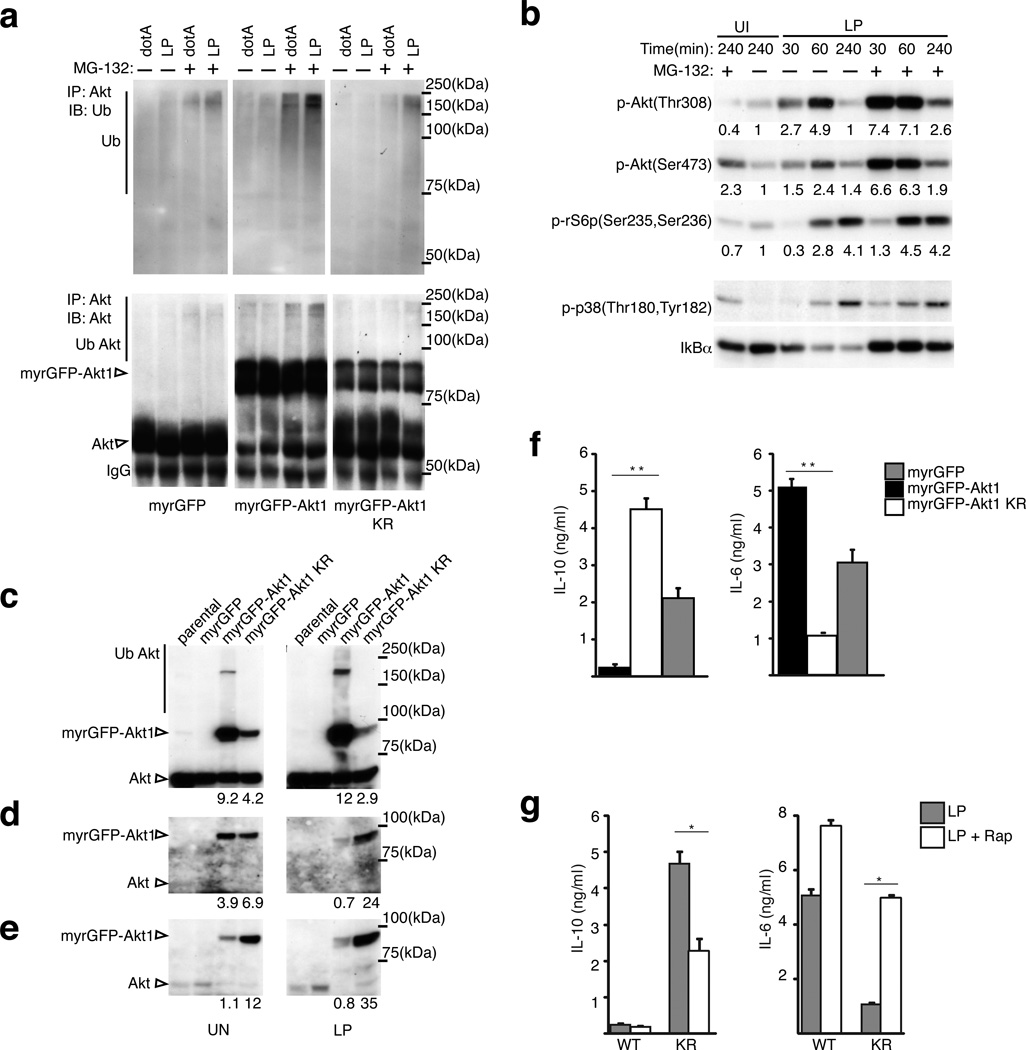
All lysine residues in Akt that were identified in the PTM analysis as being potential sites for pathogen-induced ubiquitinylation (K8, K111, K112, K154) were changed to arginine residues to generate the Akt1KR allele, and J774 macrophage-like cell lines were established that produce either myrGFP, myrGFP-Akt1 or myrGFP-Akt1KR (Supplementary Fig. 7h). Robust ubiquitinylation of myrGFP-Akt1 occurred when cells were infected with virulent Legionella, whereas, myrGFP-Akt1KR and myrGFP were not ubiquitinylated to similar extents after infection (Fig. 7a). Blocking degradation of ubiquitinylated proteins using the proteasome inhibitor MG132 enhanced detection of ubiquitinylated Akt proteins and validated that myrGFP-Akt1KR evades pathogen-induced ubiquitinylation (Fig. 7a). Furthermore, the addition of MG132 during infection by virulent Legionella increased the amount of activated Akt detected in the cells and augmented phosphorylation of the ribosomal S6 protein (Fig. 7b). The addition of MG-132 also increased the amount of phosphorylated myrGFP-Akt1, however, MG-132 did not affect the amount of phosphorylated myrGFP-Akt1KR (Supplementary Fig. 7i). Consistent with the Akt1KR protein evading pathogen-induced ubiquitinylation, the levels of phosphorylated myrGFP-Akt1KR remained high following infection of host cells by virulent Legionella (Figs. 7c–e). The expression of myrGFP-Akt1KR resulted in increased production of IL-10 and decreased production of IL-6 following infection of macrophages by virulent Legionella, which was in contrast to results from control cells that produced either myrGFP-Akt1 or myrGFP (Fig. 7f). The defect in cytokine biasing demonstrated by the macrophages producing myrGFP-Akt1KR was overcome when mTOR function was directly inhibited using rapamycin (Fig. 7g). Thus, pathogen-induced ubiquitinylation of Akt results in downregulation of mTOR activity, which promotes cytokine biasing by favoring the synthesis of highly abundant transcripts through suppression of cap-dependent translation (model in Supplementary Fig. 7j).
Figure 7. Akt ubiquitination is important for cytokine biasing.
(a) Akt ubiquitination in response to Legionella. Immunoblot analysis of Akt immunoprecipitated with a pan-Akt antibody from J774 macrophage cell lines stably expressing myrGFP, myrGFP-Akt1 or myrGFP-Akt1 KR allele (Supplementary Fig. 7h) infected as indicated for 4hrs [MOI = 20] in the presence or absence of the proteasome inhibitor MG-132. Immunoblot analysis of (b) Akt, rS6p and p38 phosphorylation as well as IkBα levels in J774 macrophages infected as indicated [MOI = 20] in the presence or absence of proteasome inhibitor MG-132. Akt amount (c) and phosphorylation (d and e) in uninfected and Legionella infected J774 cell stably expressing the indicated alleles [4hrs at MOI = 20] immunoblotted with (d) anti-p-Akt(Thr308) and (e) anti-p-Akt(Ser473). (f and g) ELISA of IL-6 and IL-10 secreted by J774 cells stably expressing the indicated Akt alleles infected for 18hrs [MOI = 10] in the absence (f) or presence (g) of Rapamycin [100 nM]. (*) p<0.05 (**) p<0.005 (paired T-test). The data presented in all panels is one of three biological replicates.
DISCUSSION
Macrophages can modulate cytokine responses according to the pathogenic potential of the organism detected. Here, we found that detection of pathogenic Legionella resulted in ubiquitinylation of regulators that maintain mTOR activity in the cell. Downregulation of mTOR activity suppressed translation and promoted a proinflammatory cytokine response where IL-6 levels were high when macrophages were infected with virulent Legionella and IL-10 levels were higher when macrophages were infected with the dotA mutant. The drop in IL-10 production observed during infection by virulent Legionella was in contrast to an increase in gene transcription, which indicated that cytokine biasing in response to pathogenic Legionella was regulated at the post-transcriptional level. The following experiments provided evidence that this was due to suppression of mTOR function: i) inhibition of mTOR function enabled dotA mutant bacteria to stimulate responses similar to virulent Legionella; ii) inhibition of mTOR-regulated cap-dependent translation enabled dotA mutant bacteria to stimulate responses similar to virulent Legionella; iii) constitutive activation of mTOR function resulted in macrophages responding similarly to virulent and dotA mutant Legionella.
It has been suggested that Legionella suppressed host translation by delivering bacterial effectors that interfere with the elongation phase of translation.30 However, because a Legionella mutant deficient in all of the effectors predicted to target the host translational machinery was still capable of blocking host translation, it remained unclear if effector-mediated translational suppression was responsible for cytokine biasing during infection. Our data reveal that efficient translational suppression can be triggered by an effector-independent pathogen detection pathway that downregulates mTOR activity and that this pathway is sufficient to mediate cytokine biasing. This was supported by data showing that a non-pathogenic dotA mutant induced a cytokine profile similar to the one generated by virulent Legionella when mTOR function was inhibited pharmacologically or when the vacuole containing the dotA mutant was disrupted. Although it remains possible that translation-suppressing Legionella effectors may augment cytokine biasing by enhancing the abundance of proinflammatory cytokine transcripts, our data indicate that modulating mTOR function to suppress cap-dependent translation promotes host defense because it interferes preferentially with the synthesis of immunosuppressive cytokines encoded on the less abundant transcripts to direct a proinflammatory cytokine program that protects against Legionella infection.
Pathogen-detection selectively ubiquitinylated the active pool of Akt located at the membrane. One of the residues found to be ubiquitinylated during infection was K8. K8 ubiquitinylation has been reported to increase membrane association and signaling of Akt in response to growth factors.35, 36 It was found that myrAkt completely bypassed the requirement for ubiquitin-dependent Akt activation36, which is consistent with the capacity of the myrAktKR allele we generated to initiate and sustain mTOR activation in response to Legionella. These data suggest that ubiquitinylation is an important mechanism for regulating Akt activity. Host receptor stimulation will activate Akt through ubiquitinylation of the K8 residue, and pathogen-induced ubiquitinylation of other residues will lead to deactivation of Akt by promoting protein degradation.
Pathogenic signatures presented by Legionella suppressed mTOR activity, which decreased the availability of eIF4E for assembly of the translation initiation complex eIF4F. This established a translational bias against low abundance transcripts in favor of the highly abundant transcripts encoding many of the proinflammatory cytokines. Production of proinflammatory cytokines encoded by the most highly abundant transcripts in the cell was maintained when eIF4F function was limited. Translation of viral mRNA can be augmented when translation factors are released from host mRNA through inhibition of eIF4F37, which by analogy might be a mechanism to enhance translation of the proinflammatory messages that are initiated under conditions of low mTOR activity.
Suppression of mTOR during pathogen discrimination should also benefit the host through the induction of autophagy - a process important for clearance of intracellular bacterial pathogens and damaged organelles.38 Consistent with our data, it has been shown that infection of epithelial cells with pathogenic Salmonella or Shigella inhibited mTOR function and enhanced autophagy, which was suggested to be triggered by an amino-acid starvation response.39 Amino acid sensing and Akt regulate mTOR by distinct pathways that converge on the small GTPase Rheb.17 Our data indicate that Akt-dependent regulation of TSC2, but not amino acid starvation, is critical for mTOR suppression in response to Legionella, which suggests this response is independent of nutrient sensing because amino acid starvation suppresses mTOR function independent of TSC2 activity.40 Thus, there may be several pathways that integrate pathogen sensing with mTOR suppression.
Inhibition of the Akt-mTOR pathway in vivo and in vitro augments inflammation23–25, 41, however, the mechanism by which this might occur is unclear. Our data indicate that mTOR functions as a translational rheostat that responds to microbial infection, which provides new insight into the inverse relationship between protein translation and inflammation observed in several experimental systems.42–44 We propose that the high transcriptional output observed for most proinflammatory cytokines could have evolved as a mechanism to preserve the ability of innate immune cells to respond robustly to pathogens under conditions where mTOR-dependent translational initiation is suppressed. Indeed, mice with myeloid cells deficient in mTOR were more resistant to Legionella infection, which indicates that mTOR suppression favors host defense. This model could explain how a robust inflammatory response is triggered in a mouse model of Leishmania major infection under conditions in which mTOR proteolysis is observed,44 and would fit with data obtained in Drosophila that show bacterial infection correlates with reduced host protein translation and a more robust systemic anti-microbial response.45
Several negative feedback loops for cytokine mRNA transcription46 and turnover47 require de novo protein synthesis and perhaps have evolved to augment abundance of these mRNAs when translation is inhibited. However, our data indicate that an enhanced transcriptional response resulting from infection might not always lead to an increase in protein production because translational suppression could be dominant over an increase in transcription if mRNA abundance does not reach the threshold needed for efficient translational initiation. Thus, the finding that pathogen discrimination triggers a ubiquitinylation pathway that suppresses mTOR function provides a new paradigm for how the mammalian immune system is able to regulate inflammation in response to different microbes.
ONLINE METHODS
Reagents
Lipopolysaccharides (LPS) from E. coli 0111:B4 were purchased from Sigma. The pharmacological agents used in the study were obtained from the following suppliers Cell Signaling (LY294002, rapamycin), Sigma (PP242, actinomycin D, cycloheximide, puromycin), Cayman Chemical (bpV(phen)) and Enzo Life Sciences (MG132, epoxomicin, 4EGI-1). The following antibodies were purchased from Cell Signaling – α-pan-Akt (clone 11E7), α-pAkt(T308) (clone C31E5E), α-pAkt(S473) (clone D9E), α-mTOR (clone 7C10), α-pmTOR(S2481) (polyclonal), α-pmTOR(S2448) (clone D9C2), α-4EBP1 (clone 53H11), α-4EBP2 (polyclonal), α-p4EBP1(S65) (polyclonal), α-rS6p (clone 54D2), α-prS6p(S235/236) (clone 91B2), α-pS6K(T389) (clone 108D2), α-TSC2 (clone D93F12), α-HSP90 (clone C45G5), α-p38 (polyclonal), α-pp38(T180/Y182) (clone 12F8), α-ikBα (polyclonal), α-pERK1/2(T202/Y204) (clone D13.14.4E), α-Calnexin (clone C5C9)); Enzo Life Sciences - α-Ubiquitin (clone P4D1), α-Ubiquitinated proteins (clone FK2); Sigma - α-FLAG, α-Actin, α-tubulin; Santa Cruz Biotechnology - α-IL1β; BD Biosciences - α-p115, α-GM130. The anti-puromycin antibody was a kind gift from Peter Walter (UCSF).
Bacterial strains
L. pneumophila serogroup 1, strain Lp0111, and the isogenic dotA mutant strain48 were used for the proteomics studies. Legionella pneumophila (L.p.) flaA thyA strain is a Lp02 derived strain with a clean deletion of the flaA gene and is used throughout this study unless otherwise noted. The Lp02 strain11 is a pathogenic thymidine auxothroph strain derived from Lp01. The isogenic dotA11, ΔicmS13, Δ5less30 and ΔravZflaA29 mutants were derived from Lp02. The plasmid pJB1806 was used to produce LLO (NP_463733.1) and BlaM-RalF (AAL23711.1) in Legionella. The BlaM-RalF expressing strains used for the sorting of T4SS-injected macrophages were described previously29. Legionella strains were grown on charcoal yeast extract plates (1% yeast extract, 1% N-(2-acetamido)-2-amino-ethanesulphonic acid (pH 6.9), 3.3 mM L-cysteine, 0.33 mM Fe(NO3)3, 1.5% bacto-agar, 0.2% activated charcoal). Plates for Lp02 derived strains were supplemented with 100µg/ml thymidine. For all experiments, Legionella were harvested from charcoal yeast extract plates after growth for 2 days at 37 °C.
Mice
To generate mice where the gene encoding mTOR was deleted in cells of the myeloid lineage we crossed B6.129S4-Mtortm1.2Koz/J mice, (Jackson Labs, 011009) having a floxed mTOR gene49 with B6.129P2 Lyz2tm1(cre)Ifo/J mice (Jackson Labs, 004781) producing Cre recombinase under the control of the myeloid-specific Lyzs promoter. Loss of mTOR in myeloid lineage cells of Cre-expressing mice was confirmed by immunoblot analysis. The littermates of mTOR−/− mice that lacked Cre recombinase were used as WT controls for in vivo experiments. C57BL/6 purchased from Jackson Laboratories were used in the in vivo PTEN studies. The il10rb−/− mice on C57BL/6 background were purchased from Jackson Laboratories. The atg5−/− mice were produced as described29. All procedures relating to the care and treatment of the animals were performed in accordance with NIH guidelines.
In vivo infections
Age (14 to 16 weeks old) and sex matched littermates were used for the in vivo infections. Mice were anesthetized by subcutaneous injection of a ketamine (100 mg/kg) - xylazine (10 mg/kg) PBS solution and infected with L. pneumophila strain JR32 ΔflaA by intranasal inoculation of 1×106 bacteria in 40 µl of PBS. For the PTEN inhibition studies, freshly prepared 62mM stock solution of bpV(phen) in DMSO was diluted down to 2mM with PBS and administrated 30min prior to infection (400nmol in 200µl PBS solution), at 3hrs and at 24hrs post infection (500nmol in 300µl solution) by intraperitoneal injection. A PBS solution containing an equivalent amount of DMSO was administrated to the “vehicle control” group. At the times indicated mice were euthanized by CO2 asphyxiation and the lungs were removed. Tissue homogenates were prepared from the lungs using a BulletBlender (Next>>>Advance) at a weight to volume ratio of 100µl PBS per 100mg lung tissue. The number of Legionella colony-forming units (CFUs) in each sample was determined by spreading dilutions of each homogenate onto CYE agar plates. ELISA assays were used to determine the cytokine concentrations in each sample.
Bone marrow-derived macrophages
Bone marrow was collected from the femurs and tibiae of mice and cultured as previously described50. Bone marrow progenitors were differentiated for 7days with RPMI media containing 15% fetal bovine serum (FBS) and 20% (v/v) RPMI media containing macrophage colony-stimulating factor produced by L929 cells.
Expression constructs
pYIC (YFP-IRES-CFP) translational reporter construct was obtained from Addgene (plasmid#18673). Retroviral expression construct encoding hemagglutinin (HA)-tagged human Akt1 fused to the Src myristoylation sequence was obtained from Addgene (pLNCX::myr-HA-Akt1, plasmid#9005). The pLNCX::myr-GFP-HA-Akt1 was generated by in-frame insertion of eGFP ORF at the BamHI site between the myr sequence and the HA epitope. The KR allele was subsequently generated by sequential site-directed lysine to arginine mutagenesis of residues 8, 111, 112 and 154 using the Pfu Turbo DNA Polymerase (Stratagene) followed by DpnI digestion (NEB). Tandem-affinity tagged (TAP)-Ubiquitin construct was generated by cloning a 3X Flag epitope, 6X Histidine epitope and a 75-aa biotinylation motif derived from a Propionibacterium shermanii transcarboxylase in the pcDNA 4/TO vector. The Ub G76V mutant was generated by site-directed mutagenesis. The 5’UTR sequence of il6 and il10 mRNA was PCR amplified from cDNA prepared from BMMs infected with L.p. and cloned upstream of the Renilla luciferase gene in pcDNA 4/TO expression vector. The internal ribosome entry site (IRES) from picornavirus was PCR amplified using pYIC as template and cloned upstream of the Renilla luciferase gene in pcDNA 4/TO. Listeriolysin O ORF (lmo0202) was PCR amplified from Listeria monocytogenes strain SLCC5850 genomic DNA and cloned in the prokaryotic expression vector pJB1806 under the control of the IPTG-inducible tac promoter using the KpnI and PstI restiction enzyme sites. All expression constructs were verified by sequencing.
Cell culture and generation of stable cell lines
To generate RAW-Ub and RAW-UbG76V cell lines RAW264.7 macrophage-like cell line purchased from ATCC was transfected with either (TAP)-Ub or the (TAP)-UbG76V expression vector using Lipofectamine2000 (Invitrogen) and selected in the presence of Zeocin (500µg/ml) to derive single colonies, which were subsequently screened for (TAP)-Ub expression to establish cell lines with uniform protein expression. To generate the RAW-YIC translation reporter cell lines RAW264.7 cells were transfected the pYIC expression vector with lipofectamine 2000, selected with G418 (400µg/ml) and cell showing comparable reported expression were isolated using flow cytometry. Cell lines expressing myrAkt were generated by culturing J774 macrophage-like cells purchased from ATCC with virus-containing supernatants produced by the 293E packaging cell line transfected with the packaging vector pCL-Eco and the various retroviral constructs pLNCX::myr-GFP-HA-Akt1, pLNCX::myr-GFP-HA-Akt1 K/R or pLNCX::myr-GFP. GFP expressing J774 macrophages were selected for stable expression with G418 (400µg/ml) and sorted by flow cytometry. Protein levels were analyzed by FACS and immunoblot analysis.
Analysis of BMMs responses to infection
For analysis of cytokine secretion BMMs were added to 48-well plates (1×105cells/well) and infected at the MOI as indicated for each experiment for 18hrs. Supernatants were collected and analyzed by ELISA according to the manufacturers’ instructions BD Biosciences (IL-1α, IL-1β, IL-6 and IL-10), eBioscience (Ccl2) and R&D Systems (TNFα). Type I IFN secretion was determined using the interferon reporter cell line HL116 expressing the luciferase gene under the control of the IFN-inducible 6–16 promoter. Cultured supernatants from infected BMMs were treated with gentamicin to kill remaining bacteria and added to the HL116 reporter cells for 8hrs to induce luciferase expression. In parallel titrations of recombinant IFNα A/D (InvivoGen) was used to generate standards. Following incubation with cultured supernatants HL116 were lysed and luminescence was quantitated by the addition of a luciferase substrate (Promega). For immunoblot analysis of signaling pathways BMMs were added to 12-well plates (5×105cells/well) and stimulated as indicated in each experiment. Unless indicated, pharmacological inhibitors were added for 60min prior to stimulation.
RNA interference
On-target siRNA oligos for tsc2 (NM_011647.2), 4ebp1(NM_007918.3), 4ebp2 (NM_010124.2) and GFP (a non-targeting control) were purchased from Thermo Scientific. Pools of four individual RNAi oligos per gene (250nM) were transfected into differentiating BMMs by nucleofection using the 4D-Nucleofector System (Lonza AG) according to the manufacturer’s instruction using the “P3 primary cells transfection kit” and the program “EW_113”. Bone marrow progenitors (5×106 cells) differentiated for 5days with RPMI supplemented with M-CSF (20% v/v) produced by L929 cells were nucleofected with 250nM siRNA in a 100µl reaction and subsequently expanded for additional 3 days in M-CSF-containing RPMI. For each experiment, protein expression of the targeted gene was assessed by immunoblot analysis to determine knockdown efficiency.
Cell lysate preparation, immunoblotting and immunoprecipitation
For isolation of ubiquitinylated proteins cell were washed with PBS and lysed under stringent conditions using Urea buffer (8M Urea, 100mM NaH2PO4, 20mM HEPES, 2mM N-Ethylmaleimide, pH 7.6). For immunoblot analysis cells were washed with cold PBS and lysed with RIPA buffer (20mM Tris-HCl pH7.5, 200mM NaCl, 1mM EDTA, 1% NP-40, 1% sodium deoxycholate, 2mM N-Ethylmaleimide, 2.5mM sodium pyrophosphate, 1mM β-glycerophosphate, 1mM Na3VO4, 1µg/ml leupeptin). Total cell lysates (20µg) were resolved by SDS-PAGE, transferred onto PVDF membranes blocked with 1% BSA and incubated with primary antibody for either 3hrs at room temperature or overnight at 4C for phospho-antibodies. Primary antibody dilutions were used as per manufacturers’ instructions. Secondary horseradish peroxidase-conjugated antibodies (Invitrogen) were used at 1:5000 in 2hrs incubations at room temperature. Signals were visualized with ECL on ImageQuant LAS4000 (General Electric). For IP analysis of Akt, 400µg of precleared total cell lysates were incubated overnight with 1µg of rabbit polyclonal α-panAkt antibody (Cell Signaling), followed by the addition of α-rabbit IgG and recombinant proteinG conjugated sepharose for 60min. Sepharose was washed 5 times with 20X excess RIPA buffer (v/v) and the immunoprecipitated proteins were resolved by SDS-PAGE and analyzed via immunoblot analysis.
Assays for protein translation analysis
For the puromycin incorporation assay cells were added to 12-well plates (5×105cells/well) and infected as indicated. Inhibitors were added for 60min prior to infection where indicated. At 2hrs post infection, puromycin was added to the medium at final concentration of 100µg/ml for 60min, and then cell were either lysed and processed for immunoblot analysis or sorted by flow cytometry to isolate the T4SS injected vs. uninjected cell populations. Puromycin incorporation in nascent polypeptides was detected with anti-puromycin antibody. For kinetic analysis of protein translation RAW264.7 cells stably expressing pYIC were added to 96-well black plates with clear bottoms (1×105 cells/well) in RPMI media lacking phenol red and infected as indicated. In-plate CFP (excitation 440±10 / emission 485±10) and YFP (excitation 515±5 / emission 540±15) fluorescence from live cells was acquired with the Tecan Infinite M1000 plate reader. 16 overlapping areas (4×4 square pattern) were acquired and averaged for each well. Minimum of three technical replicates per condition were acquired in each experiment. Initial reading (T0) was acquired immediately after infection. Where applicable inhibitors were add at the time of infection.
Gene expression analysis
For real-time PCR analysis of gene expression, BMMs added to 12-well plates (5×105cells/well) were infected as indicated. Total RNA was isolated using the Rneasy kit (Qiagen) and first-strand cDNA synthesis was performed using the SuperScript™ II Reverse Transcriptase (Invitrogen) and oligo(dT) 12–18 primer (Invitrogen). mRNA levels for each gene were determined by real-time qPCR using the Universal ProbeLibrary (Roche) and LightCycler 480 Probes Master (Roche) in the LightCycler 480 instrument (Roche Diagnostics). For nanoString analysis of mRNA levels, BMMs were added to 12-well plates (3.5×105cells/well), infected at MOI of 10 for 3hrs or 7hrs and lysed in 350µl of RNeasy Lysis “RTL” Buffer (Qiagen). The mRNA molecular counts for 179 immune genes were determined simultaneously from volume equivalent to 10,000 cells (10µl) using the nCounter Mouse Inflammation Kit in the nCounter Analysis system (nanoString Technologies).
Isolation and processing of ubiquitinated proteins for proteomics analysis
Total of 1×108 Raw-Ub or Raw-UbG76V were seeded in 10cm dishes (2×107cells/plate) and infected with the Lp01 or dotA strains for 1hrs MOI of 40, then washed 3X with warm media to remove extracellular bacteria and cultured in the presence of 1nM epoxomicin (Enzo Life-sciences) for additional 3hrs. The urea buffer was freshly prepared for each experiment and precleared with Amberlite IRN-150L mixed bead resin (GE Healthcare) for 60min at room temperature to remove impurities. Cells were lysed in 1ml of 8M Urea buffer, scraped, lysates were passed through QIAshredder columns (Qiagen) to shred the genomic DNA. Lysate supernatants from a high-speed spin (~4mls) were combined with 100µl of Streptavidin-agarose beads (Thermo scientific) pre-equilibrated in urea buffer and rotated at room temperature for 2hrs. Beads were washed 6X with 8M urea buffer, resuspended in 50µl of 8M urea buffer containing 5mM DTT and incubated at 60C for 30min, followed by addition of 2-chloroacetamide to 50 mM final concentration and incubation at room temperature for 60 min. On-beads trypsin digestion were performed by diluting samples to 2M urea, adding 100ng of sequencing-grade modified trypsin (Promega) and incubating for 24hrs at 37C. LC-MS/MS and subsequent analysis was carried out at the Yale W.M. Keck Foundation Proteomics Center.
Assembly and analysis of the pathogen response network
The nodes of the pathogen response network are derived from: (1) proteins identified by mass spectrometry that are specific for Legionella infections and (2) their known experimentally validated interacting partners. Proteins identified by LC-MS/MS in pull-downs of ubiquitinylated proteins derived from lysates of Lp01-infected Raw-Ub cells were filtered against data sets from uninfected Raw-Ub cells, dotA-infected Raw-Ub cells, uninfected Raw-UbG76V cells and Lp01-infected Raw-UbG76V cells to generate the list of 1573 proteins modified specifically in response to pathogenic infections listed in Table S1. InnateDB database (www.innatedb.ca) analysis revealed 608 proteins had at least one experimentally validated physical/functional interacting protein and these binary relationships are shown in Table S2 and were compiled with Cytoscape software to generate the pathogen-response network. Pathway over-representation analysis of the network was completed with the “Pathway analysis” function of InnateDB using the hypergeometric algorithm and the Benjamini Hochberg correction method.
Isolation of T4SS injected macrophages
L.p. and control isogenic dotA strain expressing BlaM-RalF were used to infect 1×107 BMMs or Raw264.7 cells at MOI of 40 for 2hrs. The media was removed and cells were loaded with the fluorescent substrate CCF4/AM, using the LiveBLAzer-FRET B/G kit (Invitrogen) with 15mM probenecid, in the dark at room temperature for 60min. Cells were then washed and sorted based on the 460nm vs. 535nm signal ratio when excited at 415nm using an iCyt Reflection cell sorter. Lysates of injected and uninjected cells were generated from equivalent number of cells and analyzed by immunoblot analysis.
Analysis of UTR-reporter translation
To determine the translation efficiency of the il6, il10 and IRES 5’UTR-luciferace reporter, HK293 cells seeded in 6 well plates at 1.5×106 cell/well were transfected with 1µg reporter DNA using Fugene6 (Roche) and allowed to uptake the DNA for 60min, each population of transfected cells was split into 9 wells of 48 well plate containing either DMSO (3 wells), rapamycin (3 wells) or cycloheximide (3 wells) and cultured for 6hrs. Cells were collected, washed with PBS (2X), lysed, protein concentration was measured using Bradford assay to ensure equal input and luciferase expression was quantified by the addition of a luciferace substrate (Promega) in a Tecan Infinite M1000 plate reader.
Data statistical analysis
The statistical tests performed for each data set are listed in the figure legends and were derived using the Prism 6 statistical analysis software (GraphPad Software Inc.)
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Peter Walters (UCSF) for the α-puromycin antibody, Russell Vance (UC Berkeley) for the Δ5less Legionella strain, Peter Kaiser (UC Irvine) for the biotinylation sequence expression vector and Walther Mothes (Yale University) for the HL116 interferon reporter cell line. We would like to thank Ana-Maria Dragoi (Yale University) for insightful suggestions and manuscript critique, Cameron Godecke (Yale University Core Facility) and Franz Sewald (Yale University) for technical assistance with the flow cytometry and Xiaoyun Liu for assistance with the analysis of the proteomics data. This work is supported by grants from the NIH (AI097847 and AI048770) to CR.
Footnotes
AUTHORS CONTRIBUTIONS
S.S.I. designed and performed the experiments, interpreted the data and wrote the manuscript. C.R.R. helped interpret the data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weintz G, et al. The phosphoproteome of toll-like receptor-activated macrophages. Mol Syst Biol. 2010;6:371. doi: 10.1038/msb.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol. 2011;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 6.Golebiowski F, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 7.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 8.Vance RE. Immunology taught by bacteria. J Clin Immunol. 2010;30:507–511. doi: 10.1007/s10875-010-9389-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadosky AB, Wiater LA, Shuman HA. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun. 1993;61:5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 12.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 13.Shin S, et al. Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 2008;4:e1000220. doi: 10.1371/journal.ppat.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monroe KM, McWhirter SM, Vance RE. Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS Pathog. 2009;5:e1000665. doi: 10.1371/journal.ppat.1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamboni DS, et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 16.Losick VP, Isberg RR. NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J Exp Med. 2006;203:2177–2189. doi: 10.1084/jem.20060766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2011;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benveniste EN, Qin H. Type I interferons as anti-inflammatory mediators. Sci STKE. 2007;2007:pe70. doi: 10.1126/stke.4162007pe70. [DOI] [PubMed] [Google Scholar]
- 20.Banchereau J, Pascual V, O'Garra A. From IL-2 to IL-37: the expanding spectrum of anti-inflammatory cytokines. Nat Immunol. 2012;13:925–931. doi: 10.1038/ni.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cambronne ED, Roy CR. The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 2007;3:e188. doi: 10.1371/journal.ppat.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coers J, et al. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol Microbiol. 2000;38:719–736. doi: 10.1046/j.1365-2958.2000.02176.x. [DOI] [PubMed] [Google Scholar]
- 23.Fukao T, et al. PI3K-mediated negative feedback regulation of IL-12 production in DCs. Nat Immunol. 2002;3:875–881. doi: 10.1038/ni825. [DOI] [PubMed] [Google Scholar]
- 24.Aksoy E, et al. The p110delta isoform of the kinase PI(3)K controls the subcellular compartmentalization of TLR4 signaling and protects from endotoxic shock. Nat Immunol. 2012;13:1045–1054. doi: 10.1038/ni.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weichhart T, et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29:565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 27.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moerke NJ, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 29.Choy A, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fontana MF, et al. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 2011;7:e1001289. doi: 10.1371/journal.ppat.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgins DE, Shastri N, Portnoy DA. Delivery of protein to the cytosol of macrophages using Escherichia coli K-12. Mol Microbiol. 1999;31:1631–1641. doi: 10.1046/j.1365-2958.1999.01272.x. [DOI] [PubMed] [Google Scholar]
- 32.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 33.Creasey EA, Isberg RR. The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc Natl Acad Sci U S A. 109:3481–3486. doi: 10.1073/pnas.1121286109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andjelkovic M, et al. Role of translocation in the activation and function of protein kinase B. J Biol Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 35.Yang WL, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan CH, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 149:1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svitkin YV, et al. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol Cell Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Randow F. How cells deploy ubiquitin and autophagy to defend their cytosol from bacterial invasion. Autophagy. 7:304–309. doi: 10.4161/auto.7.3.14539. [DOI] [PubMed] [Google Scholar]
- 39.Tattoli I, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 11:563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 40.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 41.Schabbauer G, et al. Myeloid PTEN promotes inflammation but impairs bactericidal activities during murine pneumococcal pneumonia. J Immunol. 2010;185:468–476. doi: 10.4049/jimmunol.0902221. [DOI] [PubMed] [Google Scholar]
- 42.Cao W, et al. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol. 2008;9:1157–1164. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colina R, et al. Translational control of the innate immune response through IRF-7. Nature. 2008;452:323–328. doi: 10.1038/nature06730. [DOI] [PubMed] [Google Scholar]
- 44.Jaramillo M, et al. Leishmania repression of host translation through mTOR cleavage is required for parasite survival and infection. Cell Host Microbe. 2011;9:331–341. doi: 10.1016/j.chom.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Chakrabarti S, Liehl P, Buchon N, Lemaitre B. Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe. 12:60–70. doi: 10.1016/j.chom.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Ruland J. Return to homeostasis: downregulation of NF-kappaB responses. Nat Immunol. 2011;12:709–714. doi: 10.1038/ni.2055. [DOI] [PubMed] [Google Scholar]
- 47.Anderson P, Phillips K, Stoecklin G, Kedersha N. Post-transcriptional regulation of proinflammatory proteins. J Leukoc Biol. 2004;76:42–47. doi: 10.1189/jlb.1103536. [DOI] [PubMed] [Google Scholar]
- 48.Roy CR, Berger KH, Isberg RR. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol Microbiol. 1998;28:663–674. doi: 10.1046/j.1365-2958.1998.00841.x. [DOI] [PubMed] [Google Scholar]
- 49.Gangloff YG, et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol. 2004;24:9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ivanov SS, Roy CR. Modulation of ubiquitin dynamics and suppression of DALIS formation by the Legionella pneumophila Dot/Icm system. Cell Microbiol. 2009;11:261–278. doi: 10.1111/j.1462-5822.2008.01251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.