Abstract
Sharks and skates represent the earliest vertebrates with an adaptive immune system based on lymphocyte antigen receptors generated by V(D)J recombination. Shark B cells express two classical immunoglobulins (Ig), IgM and IgW, encoded by an early, alternative gene organization consisting of numerous autonomous miniloci, where the individual gene cluster carries a few rearranging gene segments and one constant region, μ or ω. We have characterized eight distinct Ig miniloci encoding the nurse shark omega heavy (H) chain. Each cluster consists of VH, D and JH segments and six to eight constant (C) domain exons. Two interspersed secretory exons, in addition to the 3’-most C exon with tailpiece, provide the gene cluster with the ability to generate at least six secreted isoforms that differ as to polypeptide length and C domain combination. All clusters appear to be functional, as judged by the capability for rearrangement and absence of defects in the deduced amino acid sequence. We previously showed that IgW VDJ can perform isotype switching to μ C regions; in this study we found that switching also occurs between ω clusters. Thus C region diversification for any IgW VDJ can take place at the DNA level, by switching to other ω or μ C regions, as well as by RNA processing to generate different C isoforms. The wide array of pathogens recognized by antibodies require different disposal pathways, and our findings demonstrate complex and unique pathways for C effector function diversity that evolved independently in cartilaginous fishes.
Keywords: evolution, RNA processing, isotype switching
INTRODUCTION
B lymphocytes in jawed vertebrates express IgM as a cell surface receptor and as secreted protein. Although the quaternary structure can vary among species, the basic μ (mu) heavy (H) chain is highly conserved in structure with its rearranged variable (V) region and four constant (C) region domains [1]. The ubiquity and constancy of IgM suggest a strongly preserved function, in contrast to a second Ig class called IgD or IgW, also present in most vertebrate classes although intriguingly absent in some species [for a review see 2]. The IgW H chain (ω, omega) in cartilaginous fishes and lungfish [3] is an ortholog of the IgD H chain (δ, delta) in bony fishes and tetrapods [4]. Delta is often characterized by its position 3’ of the IgM H chain C exons and dependence on μ transcription, which was also key to its classification in bony fishes [5] and the amphibian Xenopus [4]. In this study the name IgW will be retained for the systems like sharks where the IgW H chain genes rearrange and are expressed autonomously. Thus IgD/IgW appears to be as old as IgM although its function remains unclear. Mouse and human IgD bind to basophils that upon crosslinking induces proinflammatory activity; catfish IgD also binds a subset of granulocytes, suggesting conservation across species of a common immune function that has yet to be fully elucidated [2, 6].
Very little is known about IgW in cartilaginous fishes, where it has been variously called IgX, IgNARC, IgW in various species of skates and sharks [7–12]. Some genomic sequence obtained from the clearnose skate [13] confirmed that the IgW H chain was encoded by multiple genes of the “cluster” type that consisted of genetic elements similar to the IgM H chain clusters [14]. Each μ gene contains VH, D and JH gene segments all within a 1–2 kb distance that can somatically recombine, and the rearranged VDJ is transcribed with a set of C region exons [15]. The μ clusters in nurse shark function autonomously and are as isolated from each other, >120 kb, as from the ω clusters [16, 17].
The multi-cluster organization is considered an early alternative form evolved from a primordial Ig gene. The number of μ clusters varies greatly among species, from 100–200 in horned shark [18] to 15 in nurse shark [16]. Antibody combining site diversity is due to junctional diversity and generated by rearrangement. But with apparently only three kinds of serum Ig classes (IgM, IgW, IgNAR [8]) there would appear to be a limit on shark C region effector function, compared to the eight Ig isotypes in mouse. Comparison of the five subfamilies of μ sequences in nurse shark showed that, whereas CH3 and CH4 were highly conserved among clusters, CH2 and VH were under strong positive selection for amino acid diversity [16]. This observation suggested that C region function could differ among the Ig clusters, a notion reinforced when it was found that VDJ from one cluster could switch to the C region of another cluster through recombination within the J-C intron [17]. The frequency of switching coincided with the expression of activation-induced cytidine deaminase (AID) and the switch junctions mostly appeared to be the results of double-strand DNA breakage repaired by non-homologous end-joining.
Not only could μ clusters switch among themselves but VDJ from ω clusters could also acquire μ C regions. These observations demonstrated that the elasmobranch Ig cluster system was not as static as originally appeared. Since the ω C region is so entirely different from that of μ [11, 12], the shark inter-class switching strongly suggested C effector function selection. We embarked on the characterization of nurse shark ω genes with the long-term view of clarifying their status in an IgM-expressing B lymphocyte. Eight distinct ω clusters have been identified; with the genomic organization elucidated for the first time, we discovered that most ω clusters are each capable of producing six secreted H chain isoforms.
Given the existence of shark immune cells (neutrophils, eosinophils, monocytes, macrophages, dendritic cells [19, 20]) that in other vertebrates mediate antibody-dependent cellular cytotoxicity, opsonization, and cell degranulation, it is anticipated that shark antibodies likewise recruit effector cells against pathogens. The wide array of pathogens recognized by antibodies require different disposal pathways, and this need for a flexible array of processing was met with by uncoupling the antigen recognition site from a fixed C-terminus/effector region. The importance of this principle is demonstrated by the phenomena of AID-induced switching at the DNA level in tetrapods and sharks [21, 17]. The current study reports on extensive C region diversity generated by RNA processing, a phenomenon uniquely evolved in multi-cluster gene organization of sharks.
MATERIALS and METHODS
Animals
Nurse sharks (Ginglymostoma cirratum) were captured off the coast of the Florida Keys. Shark-AQ was one week old after caesarian section. Other animals, aged 3–7 years at the time of sacrifice [23], are considered adults with respect to the state of their immune system [20].
Libraries
The shark-Y BAC library [22] was screened in a joint effort with Yuko Ohta (University of Maryland) to look for immune-related genes. Seventy-two clones purchased from Arizona Genomics Institute (http://www.genome.arizona.edu) are identified by their grid positions (plate addresses) in supplemental Table 1. cDNA libraries constructed from neonatal spleen [23] and shark-Y PBL [24], gifts of Martin Flajnik (University of Maryland), and shark-JS spleen [25] were described previously.
Subcloning and mapping
The BAC clone Ig fragments were subcloned into pUC19 and sequenced. Their relative positions on the BAC were ascertained by long-extend PCR. BAC DNA insert ends were identified through PCR analyses including primers at either end of the Hind III cloning site (T7, 5’-TAATACGACTCACTATAGGG-3’ or BES_HR, 5’-CACTCATTAGGCACCCCA-3’) [22]. Pulse field electrophoresis was performed by Arizona Genomic Institute.
PCR, RT-PCR
The bacteria carrying BAC clones were streaked out and two colonies were selected from each sample to be used in a colony-PCR assay to detect VH gene segments for subfamilies W1–5 (JWF1 5’-GATTGCTCCWAATCTCKG-3’ and JWR2 5’-AGTCACTTCCAGRAAGGT-3’) and W6 (IgW-LG6A 5’-TGATGGGGATTGCTAT-3’ and JWR2). If multiple colonies from one BAC stab were negative both by PCR and Southern blotting, it was deemed a false positive.
First strand cDNA was primed using oligo dT as previously described [25]. Long template PCR (Roche) was performed according to manufacturer’s instructions and optimized for the amount of input DNA. PCR primers amplified the secreted (JWF1 and UTR2 5’-GAATTTTGTTCATGTTGCAC-3’ ) or transmembrane (JWF1 and TMR 5’-CCTTGACAACAGTTACAAAG-3’) forms of ω for subfamilies W1–5. In parallel, forward primer W-LG6c (5’-TCTGTCTCTGCTTCCACG-3’) with reverse primer G6CH6BR (5’-GTGCACTACTACCAGGAG-3’) or with TMR amplified the respective species for the W6 subfamily. In experiments designed to isolate secreted forms from subfamily W1GL (1.8 kb, 2.5 kb, 3.1 kb), specific primers in CDR2 (IgWLG1-F2, 5’-TTCAGCCTCAGCAGCTACG-3’) and 5’ of CH6 (G1UTR1, 5’-CAGGAGAGAGGTTTCCGG-3’) were used.
Probes
Probes to IgM derived from Group 2 cDNA (vh, cμ1, cμ2, cμ3-cμ4) have been described elsewhere [16]. Probes to IgW were derived from cDNA or selected BAC clones as follows. wV (333 bp, JWF1 5’-GATTGCTCCWAATCTCKG-3’ and IgWFR3R2 5’-GACACCGCCATCTATTAC-3’) and transmembrane (wTM) with 3’UT (472 bp, WTMF 5’-GGTCCCTCCAGATGTGAA-3’ and WTMUTR 5’-GGAATCAGTCCTCCACCC-3’) were amplified from pup spleen cDNA primed with oligo dT. wCH1/2 (561 bp, IgWCH1F1 5’-AAGATGAGATCAGCCTCC-3’ and W543R 5’-GATCCTGGTACTGAAGCT-3’), wCH3/4 (508 bp, WC3F 5’-CCACCCTGGTGTGTACA-3’ and WC4R 5’-GCAGGAAGGAGGTGAAGA-3’), wCH5 (260 bp, WC5F 5’-CTGGAGGAACAGAACAGC-3’ and WC5R 5’-GGGCTTTCCTGATGAACC-3’) and wCH6 (231 bp, W1580F 5’-AACTGCTCATACTGCGAC-3’ and W1792R 5’-GTTGTGACTGACTCGTGT-3’) from amplified from a cloned W6 sequence (C region identical to GenBank accession number U51450). All of these probes cross-hybridized among the IgW genes except for W1 or W1GL, where a specific probe (wG1CH2) was used to detect CH2 (303 bp, WCH2G1F 5’-TTCAAGAGCCGAACATCA-3’ and WCH2-1R 5’-CCTCTTGGTATTTCATGC-3’).
When the filters with BAC DNA were sequentially hybridized, the previous probe was removed using a solution of 0.1% SDS brought to boiling; this was allowed to cool to room temperature and the filters were rinsed in 2X SSC. The blots were examined by autoradiography for residual signal before embarking on the new hybridization.
Accession Numbers
Genomic DNA (W2B: KC920789, W6: KC920790, W1: KC920791; eight VH gene segments KF192877 - KF192884) and cDNA clones (KC920792 - KC920803, KF184389, KF184390) and RT-PCR products (KC920804 - KC920812) were deposited at GenBank (http://www.ncbi.nlm.nih.gov/genbank/).
RESULTS
Sixty-two nurse shark BAC clones with ω genes were characterized. Eight distinct genomic ω clusters were determined (W1, W1GL, W2A, W2B, W3, W3GL, W5, W6), seven of them in various linkage relationships to each other (Fig. 1–3). Four clusters (W1GL, W1, W2B, W6) were entirely or partially sequenced, some revealing two secretory exons in addition to the 3’-most C exon with tailpiece (tp) (Fig. 4A). The complex ω gene organization involving eight C domains and three secretory exons presented potential for a wide variety of isoforms generated by RNA processing, which was confirmed by isolating clones from cDNA libraries or by RT-PCR (Fig. 4B, C). All clusters appear to be functional, as judged by the capability for rearrangement and absence of defect in the deduced amino acid sequence and in splicing. Isotype switching between two ω clusters was observed (Fig. 5).
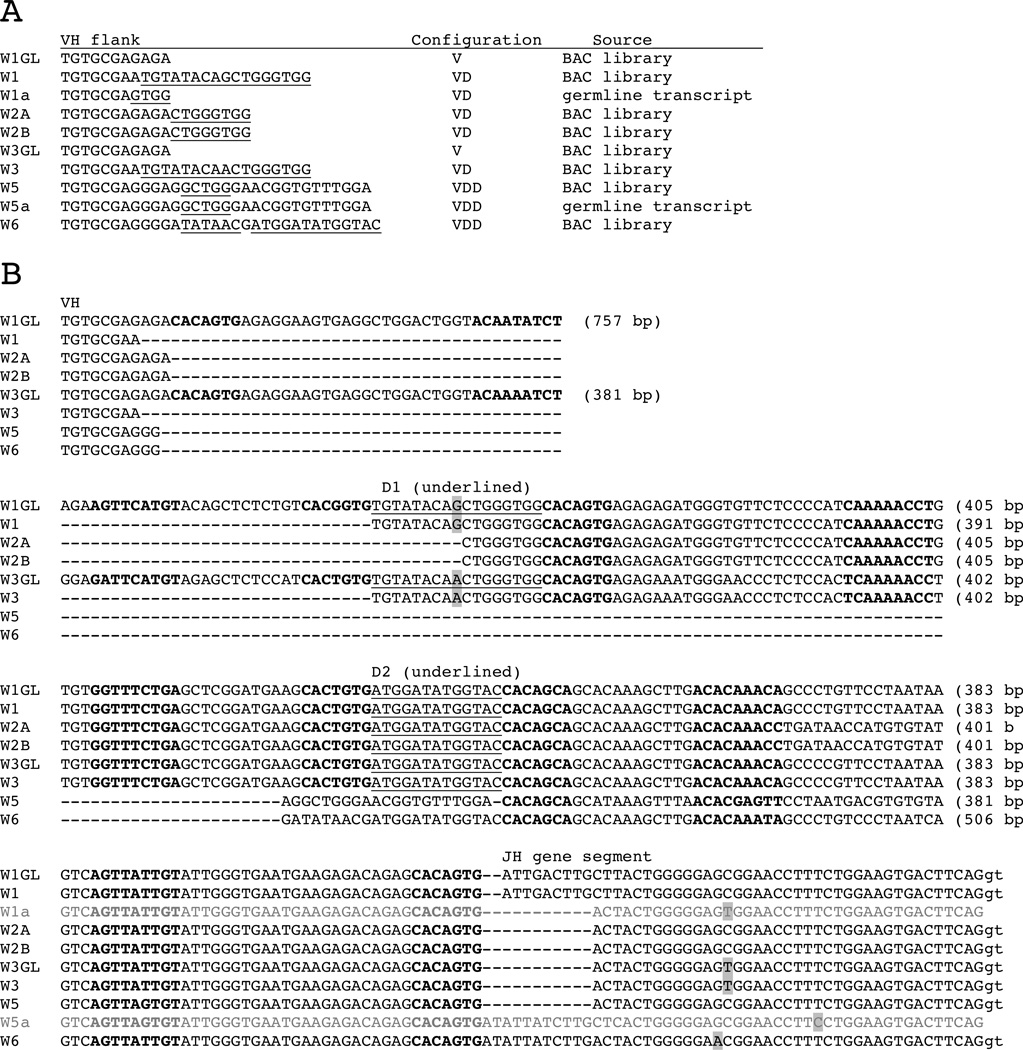
Figure 1.
Omega germline gene segments. A. VH gene flanks 5’ of RSS. The D segments identified the germline rearranged VD and VDD are underlined. No D2 can be discerned in the W5 or W5a flank. W1a and W5a sequences were from a cDNA library. B. Alignment of the VH, D1, D2 and JH sequences of IgW clusters with flanking RSS. The D genes that exist as independent gene segments are underlined. The 7-mer and 9-mer of the RSS are bolded. Distances between gene segments, including the RSS are in parentheses. The difference between W1GL/W1 and W3GL/W3 is highlighted in gray, as are those distinguishing the JH gene segments. The splice donor is in lower case at the end of JH. (Accession numbers KF192887-884), W1a and W5a are non-rearranged transcripts with JH and spliced to CH (KC920792-3). Gaps (dashes) are inserted to enable comparison of the genes.
Figure 3.
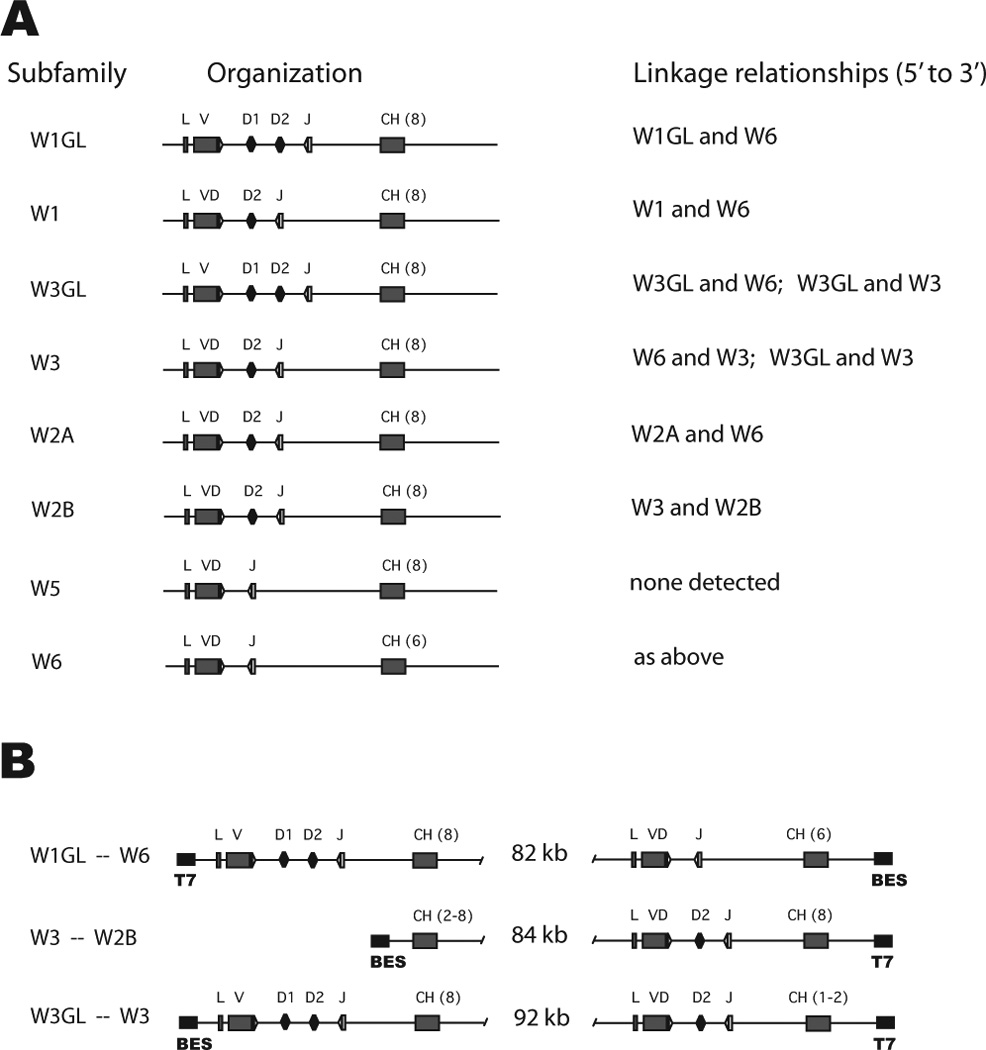
Linkage of Ig clusters. A. Summary of the organization of the eight ω subfamilies is shown, where L is leader, V is VH, VD is germline-joined VH-D1 or VH-D1-D2, J is JH, CH is C domain exon with the number of units in parentheses. At right are the associated clusters found among the BAC clones (see supplemental Table S1). B. Intergenic distances in three BAC clones. The BAC50 (Gc_Ba 214H18) contains a 134.1 kb insert; clusters W1GL and W6 occupied respectively 27.6 kb and 24.1 kb at either cloning site so that the intergenic distance is calculated to be 82 kb. BAC45 (Gc_Ba 183D24) contains a 128.4 kb insert with W3/W2B; BAC55 (Gc_Ba 238C10) contains a 130.7 kb insert with W3GL/W3. BAC55 can delete to a 28 kb insert containing only W3GL. Some coding elements are indicated (VH gene segments and number of C exons) as is the orientation with respect to the cloning site (black boxes with primers BES, T7).
Figure 4.
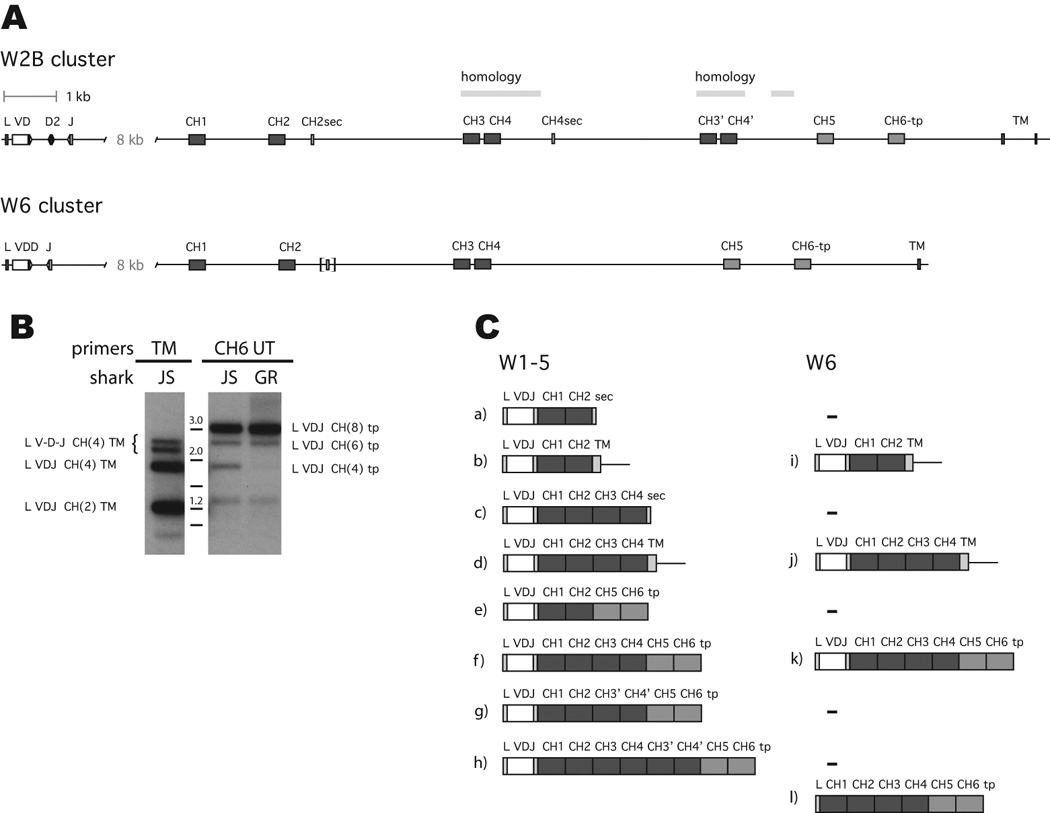
The organization of ω clusters and their differentially spliced transcripts
A, top. Organization of cluster W2B, with 1 kb scale. White box is VH, filled boxes C exons; CH5 and CH6 are lighter shade. CH2sec and CH4sec indicate the secretory exons. The extent of sequence homology is indicated between CH3 and CH4 and the duplicated CH3’ and CH4’. Bottom. Organization of cluster W6. See legend to W2B. Brackets indicate CH2sec pseudogene. The TM exon 2 was not isolated for W6. GenBank accession numbers KC920789 (W2B) and KC920790 (W6).
B. Two kinds of Tm and four kinds of Sec for ω. RT-PCR performed on oligo dT-primed spleen cDNA from sharks-JS and GR, using primers in the leader (JWF1) and in the transmembrane region (TMR) or 3’ of the CH6 exon (UTR2), with PCR products shown in panels TM and CH6 UT, respectively. The filters were hybridized with probe wV. Panel TM, various bands labeled at left, after cloning. Panel CH6 UT, three bands labeled at right characterized after cloning. Molecular size markers 1–3 kb shown between panels. Unlabeled bands are non-Ig.
C. Differentially spliced ω transcripts isolated to date from the subfamilies W1GL, W2A, W2B, W3GL and W5 (left) and W6 (right). All forms have been observed for W1GL and W6 in this study. Accession numbers for some sequences isolated in this study are as follows: (a) KF184389 (b) ref. 28 (c) KC920796, KC920794 (d) ref. 28 (e) KC920804 - KC920809 (f) KC920795, KC920810, KF184390 (g) KC920812 (h) KC920811, KC920799, KC920800, KC920801 (I,j) ref. 28 (k) KC920797, KC920798 (l) ref. 8.
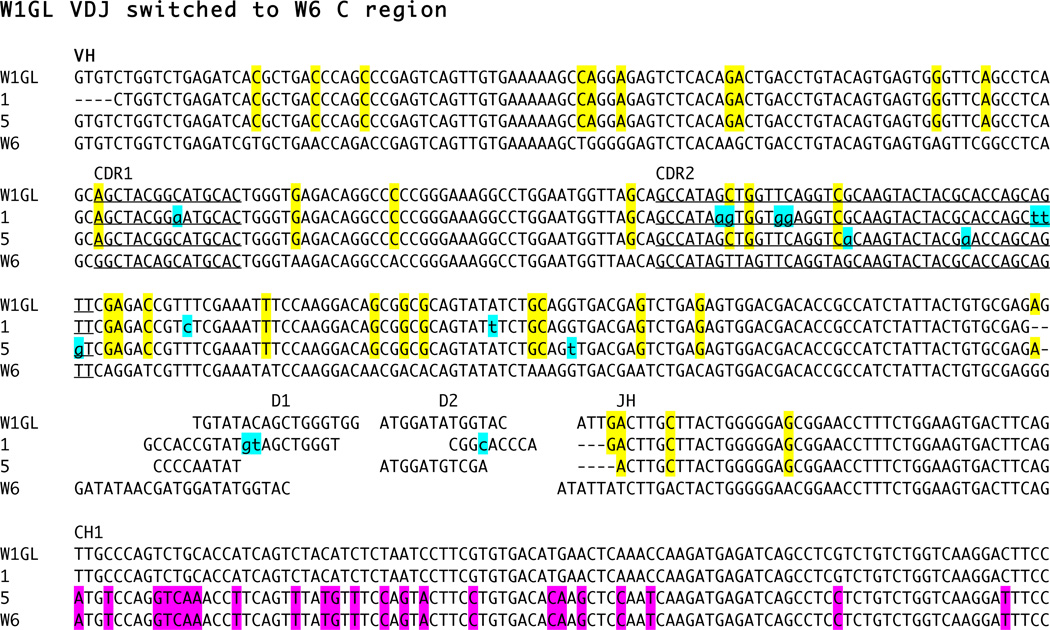
Figure 5.
C region switching takes place between ω clusters. cDNA clone 5, W1GL VDJ with W6 C region, is compared to non-switched W1GL clone 1 and to germline VH gene segments and partial CH1 sequence from W1GL and W6 clusters. Nucleotide identities shared between the cDNA sequences and W1GL are highlighted in yellow for VH and JH, and identities between clone 5 and W6 C region are shaded in pink. Mutations are marked by lower case and highlighted in blue. The rearranging germline gene segments consist of VH gene segment, two D genes and JH gene segment, as labeled; the V-D1, D1-D2 and D2-JH intersegmental sequences not shown for W1GL and that of VDD-JH not shown for W6. The CDR1 and CDR2 in the V gene segment are underlined and labeled. Dashes indicate gapping. Accession numbers KC920802 (clone 5) and KC920803 (clone 1).
Classification of IgW clusters
Seventy-two clones were selected at random from 204 BAC clones that hybridized to ω probe. Sixty-two (86%) proved to carry ω clusters, as determined by Southern blotting and PCR (supplementary data, Table S1). Eight kinds of ω genes were distinguished. The clusters W1/W1GL, W2A/B, W3/W3GL, W5, and W6 were named according to VH identity with partial cDNA sequences previously characterized as subfamilies [27]. Each genomic gene cluster consists of one VH, two D and one JH (W1GL, W3GL) that in six cases are germline-rearranged as VD (W1, W2A, W2B, W3) or VDD (W5, W6). Canonical recombination signal sequences (RSS) flank the gene segments (Fig. 1B).
Judging by the germline joints, one can deduce that the six VD and VDD genes arose five times independently, and one pair (W2A, W2B) was probably derived from one germline-rearrangement event. Although W1 and W3 appear to have identical joints, they differ in one nucleotide (Fig. 1 B, shaded). Since these nucleotides are the same as in their respective germline counterparts, W1 must have derived from W1GL and W3 from W3GL. D1 or D2 can be identified in the V flanks of the VD and VDD (Fig. 1A underlined). In the course of screening spleen cDNA libraries two germline transcripts (W1a, W5a) carrying non-rearranged gene segments from other ω clusters were found (accession numbers KC920792 – 93). It is not clear these are variant alleles in the population.
Linkage of the clusters
Southern blots with BAC DNA were hybridized to six probes in succession to detect VH, CH1/CH2, CH3/CH4 and the duplicated CH3’/CH4’, CH5, CH6 and the transmembrane exon 1 (TM1) that is 2–3 kb 3’ of CH6. Twenty out of 62 BAC clones carry two VH, as determined by separate PCR assays for W1-W5 and W6 (Table S1); in most cases the second gene was W6. On further analysis using CH probes, we found more BAC clones carrying two clusters but lacking the VH gene (and 5’ C exons) of the second cluster. In total, 29/62 BAC clones contained two clusters.
The absence of VH or CH6 or TM on any BAC clone containing two ω clusters was used to determine their relative positions. For example, of eight BAC clones with W2A VH, six also carried the W6 cluster, sometimes missing the 5’ portions, W6 VH or CH1/CH2 (Fig. 2). By the same token, two of the eight lack W2A downstream components CH1/2, CH3/4 and CH6 (Fig. 2, lanes 23, 34). These results indicate that the W6 cluster is upstream of the W2A cluster. In another example, W2B is almost identical to W2A in sequence and restriction sites but none of seven W2B clones contained any part of W6. Instead, two W2B clones show additional bands for CH3/CH4 belonging to another ω cluster (Fig. 2, CH3-CH4 panel, arrows in lanes 18 and 45). When one of these two clones was fully characterized, the upstream cluster was determined to be W3-like with the BAC cloning site at the 3’ end of its CH2 exon. Figure 3A shows the linkage relationships determined in the 29 BAC clones.
Figure 2.
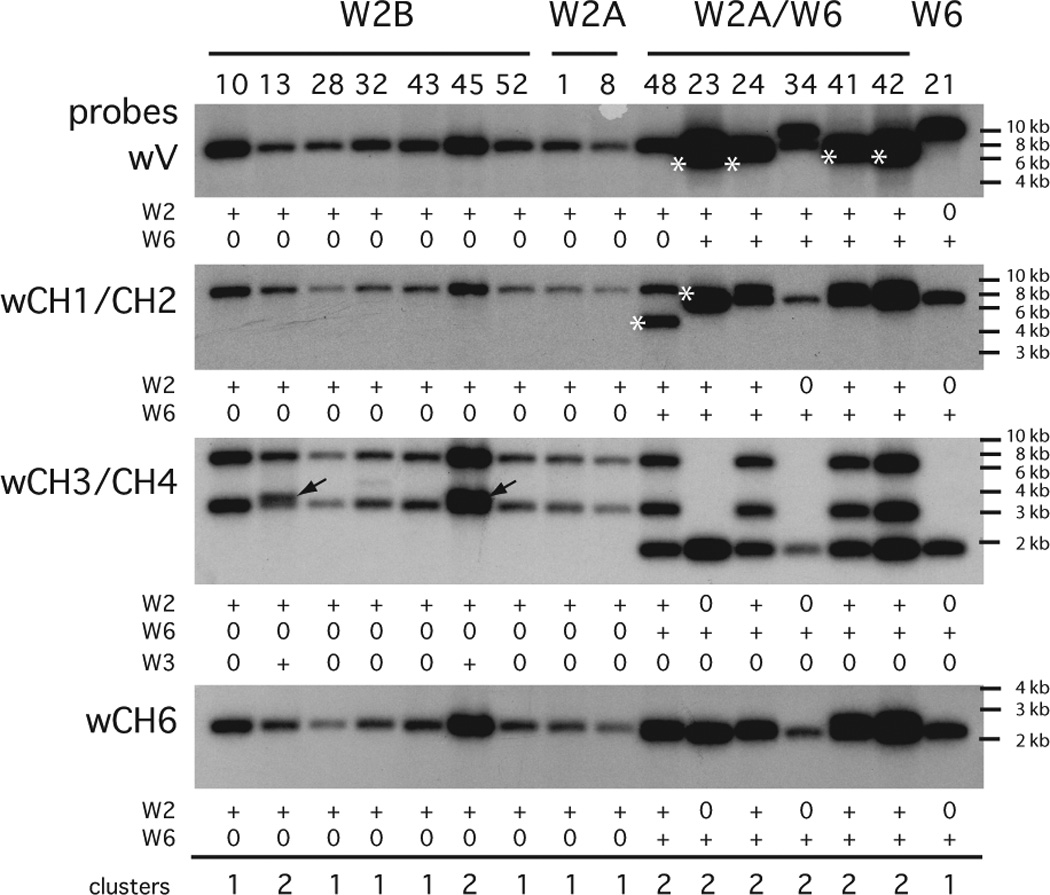
Analyses of BAC clones by Southern blotting. DNA samples from 16 BAC clones were digested with Nco I and electrophoresed on a 1% TAE agarose gel. Each lane is called by the clone name (see supplemental Table S1) and grouped according to whether they carried the VH PCR product of W2B, W2A, W2A and W6, or W6 as labeled. The DNA was transferred to a nylon filter and hybridized with wV probe, then stripped and hybridized with wCH1/CH2. The procedure was repeated with the wCH3/CH4 and wCH6 probes. The presence (+) or absence (0) of signal belonging to W2 or matching the electrophoretic mobility of the control W6 cluster (lane 21) is indicated below the panels. The asterisked bands indicate altered Nco I sites for fragments carrying W6 VH (lanes 23, 24, 41, 42), W6 CH1/CH2 (lane 48), and W2A CH1/CH2 (lane 23). These conclusions were verified by PCR and by additional Southern analyses using Pst I. With the wCH3/CH4 probe additional bands were found (arrows) and these belonged to W3. Molecular size markers are indicated at the right. The total number of ω clusters present is summarized at the bottom.
The insert size was determined for three BAC clones where both clusters were in close proximity to cloning sites, as deduced by altered restriction enzyme fragment sizes or absence of the 5’ or 3’ portions of the clusters. Long-extend PCR was performed using primers to the relevant gene segment and either BAC cloning site. The distance from cloning site to leader or to CH6/TM exons was measured and subtracted from the total insert size, which had been determined by pulse field electrophoresis (see figure legend). The intergenic distances for three different cluster pairs were 82–92 kb (Fig. 3B).
With the exceptions of W3GL/W3 and W3/W2B, the other BAC clones that carried two clusters all included W6. Although there are several W6 genes they are indistinguishable by restriction enzyme analysis or partial sequencing. Those W6 linked to W1GL, W1, W3GL, W2A are distinct, by deduction. However, BAC clones containing W3 or W6 with an upstream W3GL were also found, confounding attempts to construct a larger linkage group (Fig. 3). In shark-Y there are five subfamilies (W1-W6), eight ω isotypes and the total number of clusters is 11–12 (12 includes four W6 and two W3). It is not known whether some clusters are alleles.
C region organization
The organizations of two clusters, W2B and W6, are shown in Fig. 4A. The overall size of cluster W2B from leader to TM exon 2 is 26 kb, whereas that of W6 is about 24 kb.
W2B contains three rearranging gene segments (VD, D2, J), a J-C intron of 8 kb, eight C domain exons and two TM exons, TM2 consisting of 9 bp. Whereas the tail piece is contiguous with the ω CH6 exon similar to the μ CH4, there are two additional secretory exons, one (CH2sec) located downstream of CH2 and the other of CH4 (CH4sec). A poly(A) signal located within the coding sequence is the one used, as deduced by cDNA sequences (supplemental Fig. S1). The region containing exons CH3’ and CH4’ is a recent duplication of CH3 and CH4 but excluding the CH4sec.
The W6 cluster is similarly organized but lacks exons CH3’ and CH4’. Sequence 539 bp downstream of CH2 is homologous to CH2sec but nonfunctional due to stops (Fig. S1), and there is no CH4sec.
Expression
There are eight C domain exons in all ω clusters except for W6 (Fig. 4A). Furthermore the characterization of two clusters revealed not only additional splicing possibilities in W2B but restrictions for W6, suggesting a more complex pattern of expression than originally anticipated. cDNA clones were obtained from a neonatal spleen library, and RT-PCR experiments were performed to search for differentially spliced ω chains. Reverse PCR primers targeted the TM1 or the UT 3’ of CH6 to amplify the respective transmembrane (Tm) or secreted CH6 forms (Sec), while the forward primer was specific for W6 or for W1-W5. We found multiple forms of transcripts with all clusters.
For W6 there were three kinds of transcripts, two Tm and one Sec forms. Although the shorter Tm form was expected to be favored during PCR amplification, similar amounts were generated (not shown). Only the 6C-domain (six C region domains) Sec sequence was observed after blotting (not shown).
For the other clusters we found two Tm forms and three sizes of Sec forms using a reverse primer 3’ of CH6 (CH6-UT). The autoradiogram in Fig. 4B shows the W1-W5 species raised by RT-PCR. Probes for V, CH1-2, CH6 detected the same three CH6-UT fragments at ~3.1 kb, 2.5 kb and 1.8 kb (Fig. 4B). The first two contained VDJ with 8C-domains (Fig. 4B, CH6 UT, at 3 kb) and 6C-domains that lacked either the CH3/CH4 or else the CH3’/CH4’. The 1.8 kb species consisted of VDJ and CH1, CH2, CH5 and CH6 and proved to be an expanded clone in shark-JS, but was in trace amounts in shark-GR (Fig. 4B, CH6 UT panel, columns JS and GR); this will be referred to as “CH1256” below.
For the Tm fragments the CH6 probe was negative but V and CH1-2 hybridized to several bands. The upper bands (~2.3–2.6) also hybridized with a probe to the D2-J intersegmental region, and cloned sequences included partially rearranged or germline sequences such as leader spliced to VDD of W5, intersegmental sequence, and unrearranged J spliced to C region. Such sequences contained four C exons (CH1-4) but varied in size according to V gene segment configuration (brackets, Fig. 4B TM panel). The D2-J probe was negative for CH6-UT products. The two Tm fragments with VDJ (~1.3 kb, 1.8 kb) corresponded to structures previously reported [28].
Unexpected C region configurations
All splice variants isolated to date from cDNA libraries and by RT-PCR are illustrated in Fig. 4C, comparing the eight W1-W5 and four W6 forms found in this study and others (see Figure legend). Other than an out-of-frame VDJ rearrangement, all sequences appeared to be functional.
RT-PCR was performed on cDNA from a panel of organs some of which gave IgW signals (epigonal organ, thymus, kidney, pancreas) and others less (liver, rectal gland). Several different fragments were observed. The 3 kb 8C-domain Sec form was dominant in all tissues, although shorter fragments were expected to be favored during PCR amplification. One clearly variable other signal was the 1800 bp species which was in larger relative amounts in epigonal organ than in spleen and less so in kidney (not shown). This consisted of independent rearrangements CH1256, and in shark-JS they derived from the W1GL subfamily. The CH2 and CH5 exon junction contains unique sequence, so that it cannot derive from PCR in vitro recombination.
In the same PCR fragment pool we looked for W1 VDJ shared between the CH1256 sequences with those carrying 6C-domain (2500 bp) and 8C-domain (3100 bp). Of 15 6C-domain and 56 8C-domain sequences, none shared the same VDJ between C region isoforms, although a few dominant, mutated clones were present. The latter shared the same C region configuration among themselves. An equal number of the 6C-domain sequences carried CH3’CH4’ or CH3CH4.
Omega sequences without the VDJ were recently found in both spiny dogfish and the nurse shark [8]. Since those from nurse shark resemble transcripts of the W6 cluster, with the leader directly spliced to CH1, we used primers specifically targeting the leader and CH1 of W6. Although the VH-less fragment was predicted to be 360 bp and expected to have an amplification advantage over the ca. 720 bp fragment that contains leader, VDJ and CH1; only the latter was clearly visible by ethidium bromide. The trace VH-less fragment was cloned for verification (not shown). It was present in adult (shark-JS) and neonatal (shark-AQ) spleen, epigonal organ and liver but a distinct signal by blotting could not be ascertained in other adult tissues (thymus, PBL, pancreas).
Intergenic switching
The BAC library that defined W1GL and W6 linkage was constructed from shark-Y genomic DNA [22]. We screened an amplified cDNA library from shark-Y PBL [24], but there were few ω sequences; the animal is no longer alive. However, one cDNA contained a VDJ from W1GL gene switched to the C region of the W6 and is the first switching event found between ω clusters. In Fig. 5 cDNA clones 1 and 5 are compared to reference sequences from W1GL and W6. The VDJ of clones 1 and 5 are derived from rearrangement of W1GL gene segments. All shared nucleotides that differ from W6 are highlighted in yellow. There is rearrangement between VH and D1, demonstrating that clones 1 and 5 originated from W1GL, and a few unique sites identify the JH also as originating from W1GL. The C regions clearly differ between cDNA 1 and 5, showing the latter to resemble a isotype-switched H chain.
DISCUSSION
This work is the first extensive study on the organization of the ω genes in a cartilaginous fish. IgD/IgW is known to be structurally diverse among animals, even between teleost fish species [2], and this plasticity has been suggested to complement IgM function according to species’ needs [4]. However in nurse shark any one ω gene is by itself capable of producing a wide array of isoforms differing in length and domain combination. The significance of this domain shuffling on Ig-mediated function can only be speculated upon by comparison with other model systems and findings from antibody engineering.
The ω gene cluster C domains appear to produce three different secretory isoforms, two of which (CH2sec, CH4sec) are truncated versions of the third (CH6-tp). Similar forms of H chain "truncations" have been identified in ducks and turtles [29, 30]. For example, within the duck IgY CH gene there is a terminal exon 3’ of CH2 permitting a shorter form of the H chain to be transcribed [31]; in turtle the truncated form is separately encoded [32]. The presence of a 5.7S Ig in three vertebrate classes, independently evolved, suggests a size-dependent role. The duck 5.7S antibody does not perform the effector functions commonly associated with Fc such as complement fixation, opsonization or precipitation reactions; its part in the duck immune response is unknown and has been theorized to be immunomodulatory in nature [33]. It is thus likely that the three categories of secreted isoforms that we have observed also provide the immune system with discrete functions. Based on the examples in ducks and turtles, it is likely that the shorter, truncated forms will have similarly truncated effector functions.
It is less apparent if ω C regions of similar length but different domain content in nurse shark would have specific roles. However, recognition by effector molecules like complement or recruitment of effector cells via Fc receptors has been shown by Fc engineering not to be localized to one C-domain and to involve the oligosaccharide moiety [34, 35]. Activation of ω-mediated effector pathways could be controlled by expressing the relevant domain combinations. Even in the case of the closely related pairs CH3/CH4 and CH3’/CH4’, the replacement of serine in CH3 by leucine in the homologous position in CH3’ may affect glycosylation in the latter and so potentially differentiate function in the two kinds of 6C-domain H chains (Fig. 4C, f and g). A particular constellation of sites may be needed for optimal effector ligand recognition; or less obviously, interdomain relationships may influence conformation [34], for instance if CH2 is stabilized by CH3 contacts (Fig. 4C, c) but not by CH5 (Fig. 4C, e) then the latter structure has more mobility and permits greater access to ligand-binding sites.
In this way the various secreted ω isoforms could have different roles. However, the existence of two variant Tm forms [28] is surprising. Perhaps the interaction of the shark CD79 homolog with ω CH2 differs from that with CH4. Binding studies on different human Ig isotypes with the extracellular domains of the co-receptor showed preferential association for IgM, suggesting its enhancement of signaling [36]; possibly shark lymphocytes expressing the IgW 2C-domain receptor have different activation requirements from those with the 4C-domain form.
Genomic organization
Nurse shark [11, 28], spiny dogfish [8], and sandbar shark [12] ω cDNA sequences were reported to contain two, four or six C domains. In this work, a major eight C domain form in nurse shark is described, one encoded by most of its ω clusters. We defined eight ω genes in the shark-Y BAC library carrying distinct VH and C exons. Five of six previously defined VH subfamilies [27] (W1, W2, W3, W5, W6) were confirmed. Unlike the nurse shark μ clusters [16], some ω clusters carry germline-rearranged configurations (VD, VDD), an typical feature for elasmobranch Ig genes [for a review, see ref. 37]. Nonetheless, every nurse shark ω cluster must undergo somatic rearrangement in order to be expressed as B cell receptors.
The W6 cluster contained six C domain exons whereas the others carried eight C domain exons as well as two additional secretory exons. Characterization of the gene organizations clarified the relationship of four kinds of transcripts previously described in nurse shark [28], allowing us to look for and bring the total to eight and to show all eight could be generated from one cluster by RNA processing.
The 6C-domain cDNA in clearnose skate and other IgX/IgW sequences were subjected to a phylogenetic analysis that showed clustering of CH2 with CH4 and CH3 with CH5; the IgW CH6 clustered with IgM CH4 [10]. This result suggested an ancient organization of CH1-CH2-CH3-“CH6” where segmental duplication of CH2-CH3 generated CH4-CH5; this is borne out in the present studies, where similar Sec exons were found 3’ of CH2 and of CH4. Whereas in the nurse shark W6 gene the CH2sec became nonfunctional and the CH4sec deleted, making it the least organizationally complex, in other genes a subsequent segmental duplication of CH3 and CH4 produced what we designate here CH3’ and CH4’.
The Sec forms of IgW are generated from inclusion of one of three different secretory exons: CH2sec, CH4sec, or CH6-tp. The presence of a Sec exon 3’ of skate CH2 [13] suggests that this structure arose previous to divergence of sharks and Batoidea, >200 million years ago. Omega may have derived from an ancestral gene resembling μ that had a homologous secretory tail at its 3’-most C domain, like CH6, but two other secretory exons evolved thereafter. These acquisitions demonstrate selection for the ability to produce different secreted isoforms and suggest that the major forms each have a distinct function. The observation that W6 had once possessed one, if not both, interdomain secretory exons that were subsequently eliminated suggests an as yet unknown specialization for that Ig cluster.
Wide variety of isoforms
What is highly unusual in the IgW genes is the very extensive RNA processing that is possible. Although this hypothesis was put forward by Litman [37] and others [28], the source for varying forms of IgW was not clear until now, once the genomic organization was elucidated and the genes classified. Any cluster other than W6 is capable of producing at least eight isoforms. How is the RNA processing regulated?
Clearly, the plasma cell will be different from the resting B cell. There are three major secreted forms (2C, 4C, 6C/8C) as observed by northern blotting [28]; are these all produced at the same time in a plasma cell? Some clue may be gleaned from northern studies on organ distribution where in pancreas the dominant membrane form was 4C-domain but the only secreted form present was the 2C-domain mRNA. This result suggests that plasma cells can upregulate one kind of transcript. Such an interpretation is supported by a northern blot showing differentially expressed 2C-exon mRNA in spleens from four adult sharks, where it is absent in one sample and profuse in another [28]. This variation appears to reflect the on-going immune responses of the animals, which in some individuals included increased use of the 2C-exon isoform.
Our results on the upregulation of the CH1256 form in shark-JS but not shark-GR also suggests directed RNA processing, as does the unequal amounts of CH6-associated isoforms. There is currently no information on shark B cell subsets. The B cell production might rely on T cell direction, cognate or not, where cytokine uptake impacts on B cell differentiation and therefore gene expression at different levels. Since separate and independent secretory exons evolved 3’ of CH2 and CH4, it is not likely that the three major secreted forms are produced at random.
RNA Processing
IgM in all animals can be produced in two forms, a cell surface receptor and secreted molecule. The two H chain messages derive from a common precursor RNA, and there is a shift in expression of surface to secreted form from resting B cell to plasma cell. This is achieved largely by RNA processing, where the balance of factors effecting splicing and polyadenylation change with B cell activation [for a recent review, see ref. 38].
The regulation of alternative splicing in the IgW H chain is intriguingly even more complex due to the many components. There are two kinds of ω clusters, W6 and the general type represented by W2B (Fig. 6). The former is simpler for lacking the two secretory exons and produces three kinds of mRNA, predominantly the 4C-domain Tm and 6C-domain Sec forms, as observed in this study. The Tm transcript is the result of preferential splicing to the TM exons (Fig. 6, top, pink lines), and this is balanced by cleavage-polyadenylation 3' of the CH6-tp to produce the W6 secreted form (Fig, 6, top, arrow). In the resting B cell what is central to regulation is either inhibition of splicing of CH4 to CH5 or enhancement of splicing of CH4 to the TM1 exon. After B cell activation, changing levels of the trans-acting factors responsible for differential splicing could bring about preference for the W6 Sec form, wherein favoring of cleavage-polyadenylation at the secretory-specific poly(A) signal at CH6 (Fig. 6, top, arrow) would promote splicing of CH4 to CH5 instead.
Figure 6.
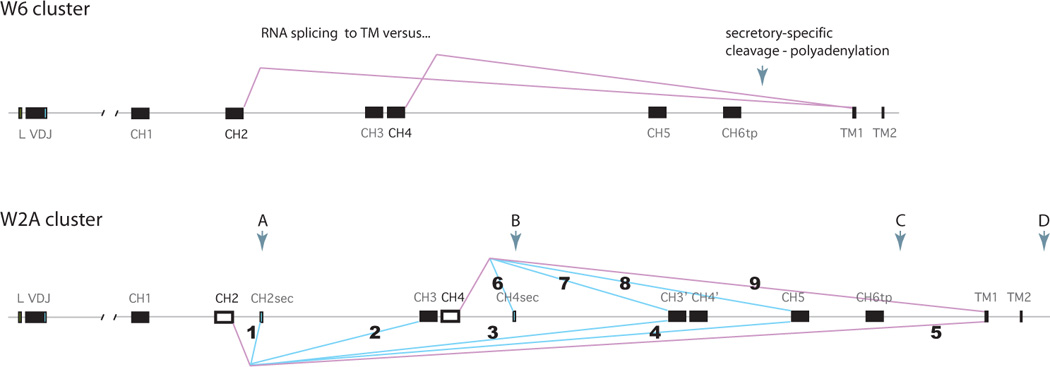
Alternative splicing patterns at two ω clusters. Top, W6 cluster. Leader, rearranged VDJ and C exons 1–6 and TM1-2 as labeled. Pink lines indicate splicing events, CH2 to TM1 and CH4 to TM1, generating the two kinds of Tm sequences observed for W6. The polyA signal is downstream of TM2, not indicated. When splicing only occurs to the adjacent downstream exons the 6C-domain transcript is generated and the polyA signal is 313 bp downstream from the CH6 exon, indicated by arrow. This map was based on genomic sequence and two cDNA 3’UT (Accession numbers KC920790, KC920797, KC920798).
Bottom, W2A cluster. Genetic elements described in legend to Fig. 4A. Exons CH2 and CH4 are shown as open boxes to distinguish them as the hypothesized key positions where differential splicing is determined. Pink lines (5 and 9) indicate splicing of CH2 and CH4 to TM1, as in the W6 cluster. Blue lines indicate alternative splicing patterns observed in cloned cDNA sequences, as follows. 1. CH2 to CH2sec. 2. CH2 to CH3/CH4 (followed by any of 6–9). 3. CH2 to CH3’/CH4’ (followed by splicing to CH5/CH6tp). 4. CH2 to CH5/CH6tp. 5. CH2 to TM1. 6. CH4 to CH4sec. 7. CH4 to CH3’/CH4’ (followed by splicing to CH5/CH6tp). 8. CH4 to CH5/CH6tp. 9. CH4 to TM1.
It is reasonable to speculate that similar regulation is present in the much more complex W2B gene (Fig. 6, bottom), in which case the resting B cell ω mRNA pool includes the 4C-domain Tm receptor and secreted 8C-domain molecule. The interesting question in a W2A gene is, once the balance is shifted away from the receptor form, how is the particular secreted isoform determined? Since there are four poly(A) signals present at the W2B gene (Fig. 6, bottom, arrows A-D), clearly there is differential recognition/utilization of these sites, probably regulated by levels of 3’ processing factors such as CstF64 [39] as well as differing strengths of the signals themselves [40]. We suggest that regulation is primarily determined at the CH2 and CH4 exons (open boxes), leading to eight kinds of transcripts through splicing events numbered 1–9 (Fig. 6, see legend). Splicing at CH4 determines splicing events 6–9 but splicing at CH2 directs events 1–5 and event 2 the downstream pathways from CH4; these account for all the different transcripts isolated.
Segmental duplication and evolution
Gene segment duplication is a frequently observed phenomenon that can serve to expand protein functionality through alternative splicing [41], and IgW presents an example where such possibilities are optimized in that (1) the tandemly duplicated exons are all in the same phase and (2) the autonomous Ig protein domains provide stability of structure and function in modular form. IgW and the putatively orthologous IgD in non-placental animals are unusually diverse in gene structure between species, compared to the evolutionarily stable IgM. On one hand this could suggest certain adaptability; on the other, what IgD and IgW have in common is that the C region can be expressed without a VDJ [6, 8]. In this context we suggest that participation in innate immune function [2] could also have influenced selection for the large diversity of IgW C isoforms.
Alternative splicing that involves exon skipping is commonly found in genes of the Ig superfamily of invertebrates and vertebrates [42, 43]. Less frequently encountered are forms generated by mutually exclusive alternative splicing as in the Dscam receptor of arthopods [44]. Among ω transcripts found in this study are 6C-domain sequences with CH3’/CH4’ or with CH3/CH4 but both splicing directly to CH5. These exon pairs could form competing cassettes managed through RNA secondary structures [45]; and possibly in absence of competition both are included. At the moment it is not clear whether the longer Sec forms are under tight regulation, and searches for DNA or RNA characteristics conferring splicing behavior are so far inconclusive. In the absence of novel regulator gene expression, control of RNA splicing may involve a balance of processing factor levels versus accessibility of the RNA. How this is directed undoubtedly will shed light not only on mammalian Ig processing but also how combinations of these pathways are utilized in a case of exceptional gene organization complexity.
Supplementary Material
Acknowledgments
We thank Helen Dooley for sharing her sequence data and Kathy Magor for comments.
Abbreviations
- Sec
secreted form of Ig
- Tm
transmembrane form of Ig
- nC-domain
sequence with n number of C region domains where n=1 to 8
Footnotes
This work was supported by National Institutes of Health grant GM068095.
REFERENCES
- 1.Flajnik MF, Du Pasquier L. Evolution of the Immune System. In: Paul WE, editor. Fundamental Immunology. 6th. Lippincott Williams & Wilkins; Philadelphia, PA: 2008. pp. 57–124. [Google Scholar]
- 2.Edholm E-S, Bengtén E, Wilson M. Insights into the function of IgD. Dev Comp Immunol. 2011;35:1309–1316. doi: 10.1016/j.dci.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Ota T, Rast JP, Litman GW, Amemiya CT. Lineage-restricted retention of a primitive immunoglobulin heavy chain isotype within the Dipnoi reveals an evolutionary paradox. Proc Natl Acad Sci U S A. 2003;100:2501–2506. doi: 10.1073/pnas.0538029100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohta Y, Flajnik MF. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc Natl Acad Sci U S A. 2006;103:10723–10728. doi: 10.1073/pnas.0601407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson M, Bengtén E, Miller NW, Clem LW, Du Pasquier L, Warr GW. A novel chimeric Ig heavy chain from a teleost fish shares similarities to IgD. Proc Natl Acad Sci U S A. 1997;94:4593–4597. doi: 10.1073/pnas.94.9.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen K, Xu W, Wilson M, He B, Miller NW, Bengtén E, Edholm ES, Santini PA, Rath P, Chiu A, Cattalini M, Litzman JB, Bussel J, Huang B, Meini A, Riesbeck K, Cunningham-Rundles C, Plebani A, Cerutti A. Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nat Immunol. 2009;10:889–898. doi: 10.1038/ni.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi K, Tomonaga S, Kajii T. A second class of immunoglobulin other than IgM present in the serum of a cartilaginous fish, the skate, Raja kenojei: isolation and characterization. Mol Immunol. 1984;21:397–404. doi: 10.1016/0161-5890(84)90037-3. [DOI] [PubMed] [Google Scholar]
- 8.Smith LE, Crouch K, Cao W, Müller MR, Wu L, Steven J, Lee M, Liang M, Flajnik MF, Shih HH, Barelle CJ, Paulsen J, Gill DS, Dooley H. Characterization of the immunoglobulin repertoire of the spiny dogfish (Squalus acanthias) Dev Comp Immunol. 2012;36:665–679. doi: 10.1016/j.dci.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Harding FA, Amemiya CT, Litman RT, Cohn N, Litman GW. Two distinct immunoglobulin heavy chain isotypes in a primitive, cartilaginous fish, Raja erinacea. Nuc. Acids Res. 1990;18:6369–6376. doi: 10.1093/nar/18.21.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson MK, Strong SJ, Litman RT, Luer CA, Amemiya CT, Rast JP, Litman GW. A long form of the skate IgX gene exhibits a striking resemblance to the new shark IgW and IgNARC genes. Immunogenetics. 1999;49:56–67. doi: 10.1007/s002510050463. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg AS, Hughes AL, Guo J, Avila D, McKinney EC, Flajnik MF. A novel "chimeric" antibody class in cartilaginous fish: IgM may not be the primordial immunoglobulin. Eur J Immunol. 1996;26:1123–1129. doi: 10.1002/eji.1830260525. [DOI] [PubMed] [Google Scholar]
- 12.Berstein RM, Schluter SF, Shen S, Marchalonis JJ. A new high molecular weight immunoglobulin class from the carcharhine shark: implications for the properties of the primordial immunoglobulin. Proc Natl Acad Sci U S A. 1996;93:3289–3293. doi: 10.1073/pnas.93.8.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson M, Amemiya C, Luer C, Litman R, Rast J, Nilmura Y, Litman G. Complete genomic sequence and patterns of transcription of a member of an unusual family of closely related, chromosomally dispersed Ig gene clusters in Raja. Int. Immunol. 1999;6:1661–1670. doi: 10.1093/intimm/6.11.1661. [DOI] [PubMed] [Google Scholar]
- 14.Hinds KR, Litman GW. Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature. 1986;320:546–549. doi: 10.1038/320546a0. [DOI] [PubMed] [Google Scholar]
- 15.Litman GW, Anderson MK, Rast JP. Evolution of antigen binding receptors. Annu Rev Immunol. 1999;7:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- 16.Lee V, Huang JL, Lui MF, Malecek K, Ohta Y, Mooers A, Hsu E. The evolution of multiple isotypic IgM heavy chains in the shark. J. Immunol. 2008;180:7461–7470. doi: 10.4049/jimmunol.180.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu C, Lee V, Finn A, Senger K, Zarrin AA, Du Pasquier L, Hsu E. Origin of immunoglobulin isotype switching. Curr. Biol. 2012;22:872–880. doi: 10.1016/j.cub.2012.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kokubu F, Hinds K, Litman R, Shamblott MJ, Litman GW. Extensive families of constant region genes in a phylogenetically primitive vertebrate indicate an additional level of immunoglobulin complexity. Proc Natl Acad Sci USA. 1987;84:5868–5872. doi: 10.1073/pnas.84.16.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luer CA, Walsh CJ, Bodine AB. The immune system of sharks, skates, and rays. In: Press CRC, Carrier JC, Musick JA, Heithaus MR, editors. Biology of sharks and their relatives. Boca Raton, FL: 2004. pp. 369–395. [Google Scholar]
- 20.Rumfelt LL, McKinney EC, Taylor E, Flajnik MF. The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand J Immunol. 2002;56:130–148. doi: 10.1046/j.1365-3083.2002.01116.x. [DOI] [PubMed] [Google Scholar]
- 21.Hackney JA, Misaghi S, Senger K, Garris C, Sun Y, Lorenzo MN, Zarrin AA. DNA targets of AID: evolutionary link between antibody somatic hypermutation and class switch recombination. Adv. Immunol. 2009;101:163–189. doi: 10.1016/S0065-2776(08)01005-5. [DOI] [PubMed] [Google Scholar]
- 22.Luo M, Kim H, Kudrna D, Sisneros NB, Lee S-J, Mueller C, Collura K, Zuccolo A, Buckingham EB, Grim SM, Yanagiya K, Inoko H, Shiina T, Flajnik MF, Wing RA, Ohta Y. Construction of a nurse shark (Ginglymostoma cirratum) bacterial artificial chromosome (BAC) library and a preliminary genome survey. BMC Genomics. 2006;7:106. doi: 10.1186/1471-2164-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rumfelt LL, Avila D, Diaz M, Bartl S, McKinney EC EC, Flajnik MF. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc Natl Acad Sci U S A. 2001;98:1775–1780. doi: 10.1073/pnas.98.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Criscitiello MF, Ohta Y, Saltis M, McKinney EC, Flajnik MF. Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J Immunol. 2010;184:6950–6960. doi: 10.4049/jimmunol.0902774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malecek K, Brandman J, Brodsky JE, Ohta Y, Flajnik MF, Hsu E. Somatic hypermutation and junctional diversification at Ig heavy chain loci in the nurse shark. J Immunol. 2005;175:8105–8115. doi: 10.4049/jimmunol.175.12.8105. [DOI] [PubMed] [Google Scholar]
- 26.Zhu C, Feng W, Weedon J, Hua P, Stepanov D, Ohta Y, Flajnik MF, Hsu E. The multiple shark immunoglobulin heavy chain genes rearrange and hypermutate autonomously. J. Immunol. 2011;187:2492–2501. doi: 10.4049/jimmunol.1101671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rumfelt LL, Lohr RL, Dooley H, Flajnik MF. Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 2004;5:8. doi: 10.1186/1471-2172-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rumfelt LL, Diaz M, Lohr RL, Mochon E, Flajnik MF. Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J Immunol. 2004;173:1129–1139. doi: 10.4049/jimmunol.173.2.1129. [DOI] [PubMed] [Google Scholar]
- 29.Grey HM. Duck immunoglobulins. I. Structural studies on a 5.7S and 7.8S gamma-globulin. J Immunol. 1967;98:811–819. [PubMed] [Google Scholar]
- 30.Chartrand SL, Litman GW, Lapointe N, Good RA RA, Frommel D. The evolution of the immune response. XII. The immunoglobulins of the turtle. Molecular requirements for biologic activity of 5.7S immunoglobulin. J Immunol. 1971;107:1–11. [PubMed] [Google Scholar]
- 31.Magor KE, Higgins DA, Middleton DL, Warr GW. One gene encodes the heavy chains for three different forms of IgY in the duck. J Immunol. 1994;153:5549–5555. [PubMed] [Google Scholar]
- 32.Li L, Wang T, Sun Y, Cheng G, Yang H, Wei Z, Wang P, Hu X, Ren L, Meng Q, Zhang R, Guo Y, Hammarström L, Li N, Zhao Y. Extensive diversification of IgD-, IgY-, and truncated IgY(ΔFc)-encoding genes in the red-eared turtle (Trachemys scripta elegans) J Immunol. 2012;189:3995–4004. doi: 10.4049/jimmunol.1200188. [DOI] [PubMed] [Google Scholar]
- 33.Magor KE. Immunoglobulin genetics and antibody responses to influenza in ducks. Dev Comp Immunol. 2011;35:1008–1016. doi: 10.1016/j.dci.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 34.Jefferis R, Lund J, Pound JD. IgG-Fc-mediated effector functions: molecular definition of interactions sites for effector ligands and the role of glycosylation. Immunol. Rev. 1998;163:59–76. doi: 10.1111/j.1600-065x.1998.tb01188.x. [DOI] [PubMed] [Google Scholar]
- 35.Lux A, Nimmerjahn F. Impact of differential glycosylation on IgG activity. Adv. Exp. Med. Biol. 2011;780:113–124. doi: 10.1007/978-1-4419-5632-3_10. [DOI] [PubMed] [Google Scholar]
- 36.Radaev S, Zou Z, Tolar P, Nguyen K, Nguyen A, Krueger PD, Stutzman N, Pierce S, Sun PD. Structural and functional studies of Igαβ and its assembly with the B cell antigen receptor. Structure. 2010;18:934–943. doi: 10.1016/j.str.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoder JA, Litman GW. Immune-type diversity in the absence of somatic rearrangement. Curr Top Microbiol Immunol. 2000;248:271–282. doi: 10.1007/978-3-642-59674-2_12. [DOI] [PubMed] [Google Scholar]
- 38.Peterson ML. Immunoglobulin heavy chain gene regulation through polyadenylation and splicing competition. Wiley Interdiscip Rev RNA. 2011;2:92–105. doi: 10.1002/wrna.36. [DOI] [PubMed] [Google Scholar]
- 39.Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1996;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 40.Peterson ML, Perry RP. The regulated production of μm and μs mRNA is dependent on the relative efficiencies of μs poly(A) site usage and the Cμ4-to-M1 splice. Mol Cell Biol. 1989;9:726–738. doi: 10.1128/mcb.9.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I, Copley RR, Bork P. Common exon duplication in animals and its role in alternative splicing. Hum Mol Genet. 2002;11:1561–1567. doi: 10.1093/hmg/11.13.1561. [DOI] [PubMed] [Google Scholar]
- 42.Brites D, McTaggart S, Morris K, Anderson J, Thomas K K, Colson I, Fabbro T, Little TJ, Ebert D, Du Pasquier L. The Dscam homologue of the crustacean Daphnia is diversified by alternative splicing like in insects. Mol Biol Evol. 2008;25:1429–1439. doi: 10.1093/molbev/msn087. [DOI] [PubMed] [Google Scholar]
- 43.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 44.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Li G, Yang Y, Wang W, Zhang W, Pan H, Zhang P, Yue Y, Lin H, Liu B, Bi J, Shi F, Mao J, Meng Y, Zhan L, Jin Y. An RNA architectural locus control region involved in Dscam mutually exclusive splicing. Nat Commun. 2012;3:1255. doi: 10.1038/ncomms2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.