Abstract
We report on a competitive electrochemical detection system that is free of wash-steps and enables the real-time monitoring of adenosine triphosphate (ATP) in a quantitative manner over a five-log concentration range. The system utilizes a recognition surface based on ATP aptamer (ATPA) capture probes pre-bound to electro-active Flavin adenine dinucleotide (FAD) molecules, and a signaling surface utilizing graphene (Gr) and gold nanoparticle (AuNP) modified carbon paste electrode (Gr-AuNP-CPE) that is optimized to enhance electron transfer kinetics and signal sensitivity. Binding of ATP to ATPA at the recognition surface causes the release of an equivalent concentration of FAD that can be quantitatively monitored in real-time at the signaling surface, thereby enabling a wide linear working range (1.14 ×10−10–3.0×10−5M), a low detection limit (2.01×10−11M using graphene and AuNP modified glassy carbon), and fast target binding kinetics (steady-state signal within 12 minutes at detection limit). Unlike assays based on capture probe-immobilized electrodes, this double-surface competitive assay offers the ability to speed-up target binding kinetics by increasing the capture probe concentration, with no limitations due to intermolecular coulombic interactions and non-specific binding. We utilize the real-time monitoring capability to compute kinetic parameters for target binding and to make quantitative distinctions on degree of base-pair mismatch through monitoring target binding kinetics over a wide concentration range. Based on simplicity of the assay chemistry and the quantitative detection of ATP within fruit and serum media, as demonstrated by comparison of ATP levels against those determined using a standard HPLC-UV absorbance method, we envision a versatile detection platform for applications requiring real-time monitoring over a wide target concentration range.
INTRODUCTION
Clinical diagnostic systems seek highly sensitive, quantitative and selective detection platforms for the real-time multiplexed monitoring of target analytes.1 To realize this vision, it is preferable for the assays to have minimal chemical complexity, contain no wash steps and enable a wide detection range for multiple targets.2 Electrochemical detection can be seamlessly integrated towards this need for real-time monitoring of analytes without requiring wash-steps, since the electron transfer event is triggered only when the electro-active species is proximal to the signaling electrode.3 Usually, this occurs through the capture of diffusing target analytes onto capture probes immobilized on the electrode for localization of the electro-active reporter species to within a nanometer of the same electrode.4 While this scheme can result in highly sensitive detection of the captured target analytes,5 it is limited by the slow assay time,6 especially for target levels close to the sensor detection limit.7 This is attributed to: (a) the slow settling time for target species onto the sensor surface due to mass transport limitations arising from the lack of 3D diffusion conditions at the proximity of the sensor surface;8 and (b) the poor target binding kinetics due to steric hindrance from neighboring capture probe/target complexes on the sensor surface.9 Miniaturization of the sensor, which can enhance detection sensitivity due to the reduced background, acts only to accentuate these mass transport limitations due to inability of hydrodynamic means for stirring in proximity of the diffusion layer where the mass transport limitations are most adverse.10 While electrokinetic methods on microfluidic device platforms for target enrichment in proximity of the capture probes using dielectrophoresis11 or isotachophoresis12 are capable of addressing this bottleneck, the ensuing device complexity hampers their consideration towards point-of-care clinical diagnostics.
De-coupling of the target recognition function from the signal transduction function is one method to address this limitation. In this so-called double surface technique,13,14, the target recognition is performed at a different surface from that enabling signal transduction; thereby allowing for their independent optimization. The recognition surface is usually a micro or nano-sized bead of high surface area that is capable of immobilizing single capture probe molecules up to an extremely high concentration.15 The ensuing free 3D diffusion conditions and high capture probe concentration at the recognition surface ensure fast target binding kinetics. On the other hand, the signaling electrode and transduction technique can be separately optimized, perhaps through surface modification or nanostructuring to enhance sensitivity.3d,16 However, prior applications of this double-surface technique have involved a wash step to release the target or signal reporter species. Herein, we are able to eliminate the wash step through the application of a DNA aptamer-based competitive assay that can enable the real-time electrochemical monitoring of the target species, without the need for target labeling.17 Additionally, through graphene (Gr) modification of the signaling electrode, we are able to reduce detection limits and obtain signal linearity over an unprecedentedly wide concentration range. We illustrate this detection system by using a relatively weak affinity assay for Adenosine triphosphate (ATP) as the target and its aptamer (ATPA) as the capture probe [Kd = (6 ± 3) ×10−6M].18 The recognition surface is composed of the ATPA capture probe immobilized on ca. 1µm sized beads prebound with a weaker affinity to electro-active flavin adenine dinucleotide (FAD).17 The displacement assay is based on the release of FAD, due to preferential binding of ATP to the immobilized ATPA, since the aptamer was pre-selected to bind this target. Due to 3D diffusion conditions at the recognition surface and the ability to inordinately raise concentration of beads with the capture probe to enhance collision frequency with the target species, the binding kinetics can be sped-up without raising nonspecific binding. This is because, unlike ELISA-type sandwich assays,19 the current assay does not require a secondary label and the electrochemical signal is triggered only when the diffusing FAD species are within a nanometer of the detection electrode. Additionally, steric limitations from intermolecular interactions between neighboring target complexes do not affect target binding kinetics, because of the smaller size of DNA aptamer bound target complexes versus immuno-assay schemes, and due to the prevalence of Poisson statistics for excess beads under low target levels,20 so that a majority of beads are bound to either single or zero target molecules. The released FAD can be electrochemically monitored in real-time at the signaling surface, using Gr and gold nanoparticle (AuNP) modified carbon paste (Gr-AuNP-CPE) or glassy carbon electrodes (Gr-AuNP-GCE) that are optimized for fast electron transfer and adsorption kinetics. Using adsorptive stripping square wave voltammetry (AdS-SWV), highly sensitive (detection limit of 2.01×10−11M ATP using Gr-AuNP-GCE) and quantitative target detection can be achieved, with a wide range for signal linearity (3×10−5 – 1.14 ×10−10 M), which would be especially useful for the detection of targets such as microRNA's that are either down regulated or overexpressed in different cancer types.21 Finally, we illustrate the utility of the capability for real-time monitoring of the target binding kinetics by applying the detection system to compute the kinetic (kon and koff) and affinity (Kd) parameters for the binding of ATP to ATPA, as well as by applying it towards the quantitative distinction of various degrees of DNA base pair mismatch by monitoring target binding kinetics and signal saturation level over a wide concentration range.
MATERIALS AND METHODS
Chemicals
All chemicals were of A. R. grade and used as received without any further purification, as per the list provided in the Supporting Information (Section S-1.1). The capture probe was prepared based on prior methods.17 A typical experiment used for the recognition technique is detailed in the Supporting Information (Section S-1.2). The information about the nucleic acid sequences used in the present work is given in Table S-1.
Instrumentation and preparation of modified electrodes
Detailed information about the instruments and electrodes used in the present work are described in Supporting Information (Sections S-1.3 and S-1.4). The working electrodes used herein were fabricated based on procedures in prior work.22
Determination of FAD and characterization of electrode surface
The AdS-SWV of FAD was carried out by taking an appropriate quantity of the analyte into a 25 mL standard volumetric flask and then making up to the mark with pH 7.0 citrate-phosphate buffer (0.1 M). The solution was then transferred into a micro-electrochemical cell where the measurements were performed. A magnetic stirrer (SCILOGEX MS-H-Pro) with a stirring bar was used to enhance the convective transport of the analyte during its accumulation onto the carbon paste electrode surface. During the pre-concentration step, an accumulation potential (Eacc) of: −0.7 V was applied for an accumulation time (tacc) ranging from 30 s for kinetics measurements to 240 s for steady-state signal measurements, within the solution stirred at 400 rpm using the magnetic stirrer. Following a 10 s rest period for the solution to become quiescent, the SWV signal was recorded by scanning the electrode potential toward the positive direction from −0.7 V to +0.2 V, using a frequency (f) of 100 Hz. The Cyclic voltammetric experiments were carried out by scanning the potential between −0.75 and −0.15 V. Electrochemical impedance spectroscopy was carried out under 10 to 106 Hz frequency range and 5mV amplitude. Chronocoulometry was carried out using an interval time of 5 s from −0.20 to −0.7 V for FAD vs. Ag / AgCl, 3M KCl. TEM images and Raman spectra of the Gr and AuNP modified surface are provided in Supporting Information (Figures S8–S10).
ATP and complementary DNA target binding studies
To determine the dissociation constant of target ATP binding with ATPA capture probe, the target at different concentration levels [(5–25)×10−6M] was mixed at time t=0 with 4.5×10−5M of capture probe (ATPA). The AdS-SWV electrochemical signal due to release of pre-bound FAD was studied every 30 seconds until 3 minutes, beyond which the measurements were carried out once every minute. Similar kinetic studies for release of FAD were conducted using ATPA complementary target DNA strands with differing levels of base-pair mismatch, to study variations in time to obtain signal saturation and the saturation signal level.
Determination of target within biological media
Analysis of ATP was carried out in blood serum samples. Analysis of ATP was also carried out in pericarp tissue of litchi, banana and tomato seed samples according to the reported literature procedure.23 The detailed experimental procedure has been given in the Supporting Information (Section S-1.5).
RESULTS AND DISCUSSION
Displacement Assay for detection of target ATP
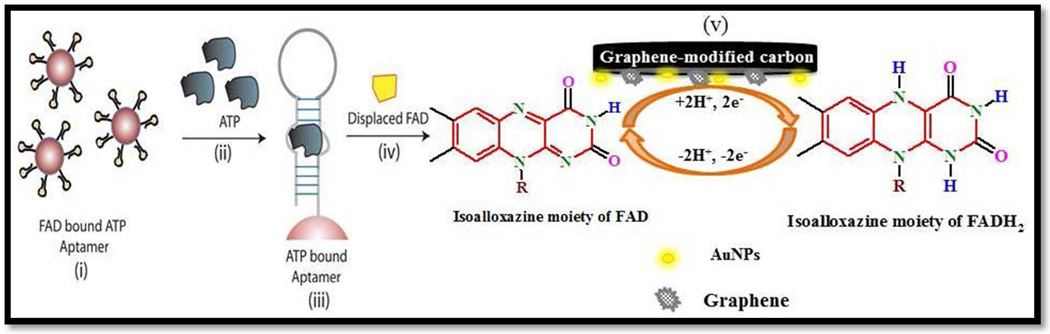
The competitive assay for ATP is shown schematically in Figure 1. Herein, ATP aptamer (ATPA) is non-covalently bound to FAD and is biotinylated at the 5' end for linking to avidin coated beads to serve as the capture probe. Since ATPA binds to adenosine and adenosine monophosphate groups of ATP, it binds specifically to FAD, which also consists of an identical sugar and nucleobase region. The presence of target species, such as ATP (or complementary DNA) within an appropriately buffered supporting electrolyte, causes the preferential binding of the aptamer to the higher affinity target, thereby releasing an equivalent quantity of FAD as per Figure 1.17 The redox reaction of FAD (Figure 1) is quantified electrochemically by AdSSWV at Gr-AuNP-CPE surfaces. Since the electrochemical signal is only caused by displaced FAD, with no contribution from FAD species originally bound to the DNA aptamer, the signal correlates linearly to the concentration of ATP. Furthermore, since there is no intervening wash-step, the electrochemical signal can be monitored in real-time every 30 seconds (time for the AdS-SWV measurement) to quantify the target binding kinetics. The same assay was applied on target DNA containing a perfect match (CS-DNA), single-base mismatch (SBM-DNA) and double-base mismatch (DBM-DNA) to ATPA, respectively, in order to measure the differences in binding kinetics and saturation signal levels. A buffered citrate-phosphate supporting electrolyte is utilized for simultaneously carrying out the competitive assay and electrochemical detection steps. The optimization of its pH and composition is described in the Supporting Information (Section S-2).
Figure 1.
Design of an ATP detection system with FAD displaced signaling via AdS-SWV. (i) FAD as a suboptimal target is bound to the ATP aptamer (ATPA) pre-conjugated to streptavidin coated magnetic beads (sphere). (ii) The presence of ATP, which is the native target for ATPA, displaces the pre-bound FAD (iii). The displaced electroactive FAD (iv) is measured via AdS-SWV [employing Gr-AuNP-CPE (v)] to generate a measurable signal.
Characterization of the Gr and AuNP modified electrochemical detection system
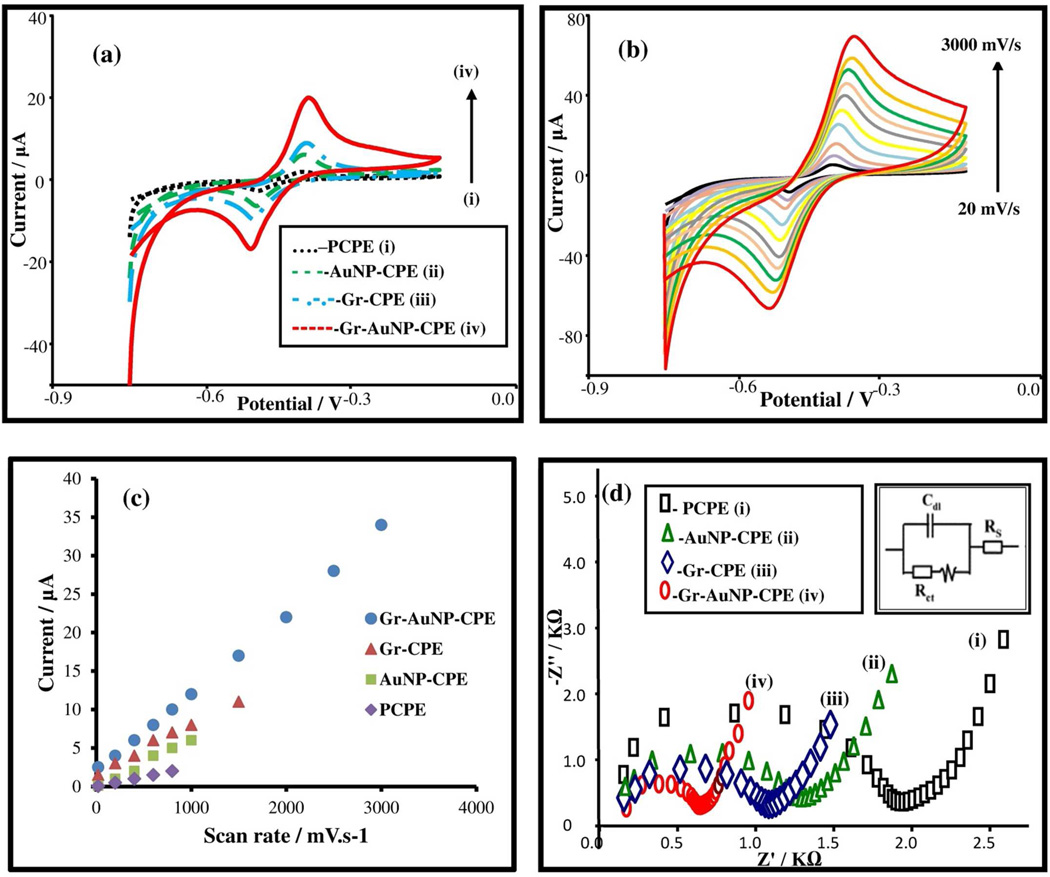
Plain and glassy carbon electrodes are well-suited for the electrochemical detection of organic molecules,24 due to their ease of synthesis, wide potential window and low background currents. Additionally, they can be combined with modifiers, such as graphene,25,26 nanomaterials,27,28 macrocycles,29 and metal complexes,30,31 to enhance the sensitivity and selectivity for determination of various analytes of interest. By utilizing SWV signals for standard FAD in the supporting electrolyte, we optimized the percentage of Gr and AuNP modifiers within the Gr-AuNP-CPE at 3% and 4%, respectively, as shown in Figures S-2 and S-3. Next, the accumulation potential and accumulation time for stripping voltammetry and frequency for SWV were optimized (Figure S-4). The cyclic voltammograms of FAD (4.5×10−6 M) at the four investigated electrode surfaces are shown in Figure 2 (a). In comparison to plain carbon paste electrodes [PCPE (i)], it is apparent that the anodic and cathodic peak currents rise continuously upon electrode modification with AuNPs (ii), Gr (iii) and Gr-AuNPs (iv); suggesting that the redox reaction of FAD becomes facile on Gr-AuNP-CPE. The reversible redox reaction (Figure 1) takes place at its isoalloxazine moiety, wherein FAD is reduced to FADH2. In order to better understand the underlying mechanism for signal enhancement at the Gr-AuNP-CPE surface, the CV for FAD was acquired at scan rates varying from 20 to 3000 mV/s, for all the respective electrode surfaces. Based on the CVs [for Gr-AuNP-CPE in Figure 2 (b)] and the linear rise in peak current with scan rate in Figure 2 (c), we infer that the process is adsorption controlled. Furthermore, based on the higher slope of the linear region for Gr-AuNP-CPE in comparison to that for PCPE, AuNP-CPE and Gr-CPE, we infer the likely role of improved adsorption kinetics on the observed signal enhancement with Gr-AuNP-CPE, under conditions of AdS-SWV. Based on the successively higher surface coverage (Γ) values for FAD (determined from the CV data) of 1.405 × 10−10 moles/cm2 for PCPE, 2.568 × 10−10 moles/cm2 for AuNP-CPE, 3.115×10−10 moles/cm2 for Gr-CPE, and 4.213 × 10−10 moles/cm2 for Gr-AuNP-CPE, we infer an increase in the active surface area upon surface modification. This is also apparent within the CV data (Figure S-5) comparing PCPE to that after Gr and AuNP modification (PCPE vs. Gr-AuNPCPE). While the background capacitance due to increased surface area rises 20-fold upon modification, the faradaic signal rises 60-fold. Hence, the net gain in signal to background after modification is ca. 3-fold for CV signals (Figure S-5) and ca.10-fold for AdS-SWV signals (Figure S-6). The scan rate dependent CV data was utilized with Laviron’s equation32 to confirm that while the charge transfer coefficient (α) was unchanged upon Gr and AuNP modification of PCPE, the electrode reaction rate constant (ks) successively increases from 0.17 s−1 for PCPE; to 0.46 s−1 for AuNP-CPE, 0.55 s−1 for Gr- CPE and 1.2 s−1 for Gr-AuNP-CPE. The data from chronocoulometric characterization of the Gr and AuNP modified CPEs (Table S-2) shows that PCPE modification enhances the values of slope, charge (Qads) and surface coverage (Γ0) of Gr-AuNP-CPE as compared to PCPE, as computed from Anson equation33. This indicates that modification of PCPE with Gr and AuNPs enables more effective accumulation of FAD onto the surface. Furthermore, based on the successive reduction in impedance at the electrode surface upon Gr and AuNP modification of CPEs, as apparent from the decreasing charge transfer resistance (Rct) in Figure 2(d) (PCPE=1.623 kΩ, AuNP−CPE=1.073 kΩ; Gr−CPE=0.923 kΩ and Gr−AuNP−CPE=0.453 kΩ), we infer the successive enhancement of electron transfer kinetics at the Gr-AuNP-CPE surface. In summary, the electrochemical characterization data confirm that the modification of the CPE with Gr and AuNPs causes a net enhancement in signal sensitivity due to the enhanced adsorption and electron transfer kinetics of FAD at the modified electrode surface.
Figure 2.
(a) Cyclic voltammograms for 4.5 × 10−6 M FAD at (i) PCPE (- - -), (ii) AuNP-CPE ( ), (iii) Gr-CPE (
), (iii) Gr-CPE ( ) and (iv) Gr-AuNP-CPE (
) and (iv) Gr-AuNP-CPE ( ) in citrate-phosphate buffer solution (pH 7.0) and at scan rate of 100 mV s−1. (b) Cyclic voltammograms of FAD (4.5 × 10−6 M) obtained in citrate-phosphate buffer solution (pH 7.0) employing varying scan rates (mV s−1): 20, 200, 400, 600, 800, 1000, 1500, 2000, 2500 and 3000 mV s−1 by scanning electrode potential between −0.75 and −0.15V. (c) Comparative plots of Ip vs scan rate for FAD at PCPE, AuNP-CPE, Gr-CPE and Gr-AuNP-CPE. (d) Nyquist plots for EIS measurements (1 × 10−3 M K3[Fe(CN)6]) at (i) PCPE (□□□), (ii) AuNP-CPE (
) in citrate-phosphate buffer solution (pH 7.0) and at scan rate of 100 mV s−1. (b) Cyclic voltammograms of FAD (4.5 × 10−6 M) obtained in citrate-phosphate buffer solution (pH 7.0) employing varying scan rates (mV s−1): 20, 200, 400, 600, 800, 1000, 1500, 2000, 2500 and 3000 mV s−1 by scanning electrode potential between −0.75 and −0.15V. (c) Comparative plots of Ip vs scan rate for FAD at PCPE, AuNP-CPE, Gr-CPE and Gr-AuNP-CPE. (d) Nyquist plots for EIS measurements (1 × 10−3 M K3[Fe(CN)6]) at (i) PCPE (□□□), (ii) AuNP-CPE ( ), (iii) Gr-CPE (
), (iii) Gr-CPE ( ) and (iv) Gr-AuNP-CPE (
) and (iv) Gr-AuNP-CPE ( ). Inset: Box on the right upper side shows the equivalent circuit used for data fitting.
). Inset: Box on the right upper side shows the equivalent circuit used for data fitting.
ATP detection sensitivity, selectivity and real-time monitoring capability
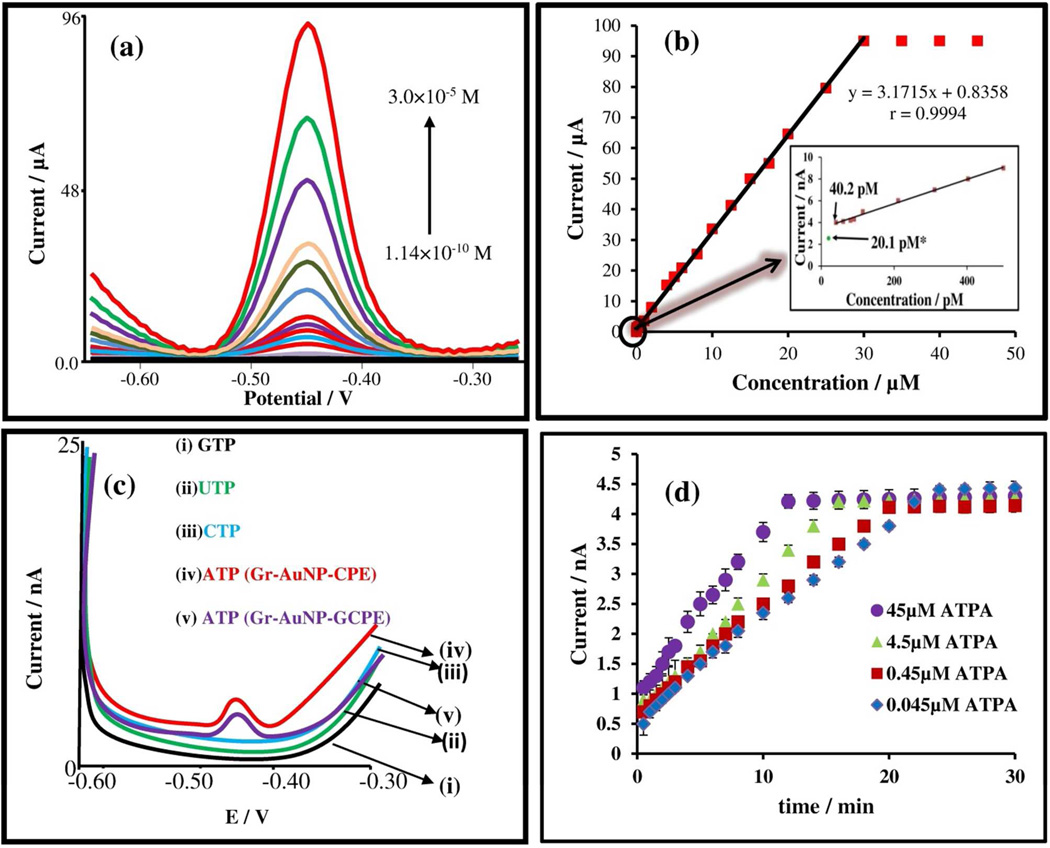
The displacement assay, as per Figure 1, was applied towards the detection of ATP by real-time monitoring of the released FAD at the Gr-AuNP-CPE electrode by utilizing AdS-SWV. The linearity range of the signal for ATP detection in the pico-molar to micro-molar concentration range is shown in Figure 3, utilizing a capture probe concentration of 4.5×10−5 M ATPA in pH 7 citrate-phosphate buffer. The raw data of Figure 3(a) are summarized as peak current versus the ATP concentration in Figure 3(b). As can be seen from the Figure 3(b), a wide linear working range of 3.0×10−5 to 1.14 ×10−10 M ATP is obtained, with a detection limit of: 4.02×10−11 M with Gr-AuNP-CPE and 2.01×10−11 M with Gr-AuNP-GCE detection surfaces. The raw AdS-SWV scan at the ATP detection limit in Figure 3(c), shows an easily distinguishable signal in comparison to the near-zero non-specific signals from common ATP analogs such as GTP, UTP and CTP. It is apparent from Figure 3(c) that the slower rise in capacitive background beyond the signal for the Gr-AuNP-GCE versus the Gr-AuNP-CPE detection surface enables higher signal sensitivity and a two-fold lower detection limit for the former. A key strength of this detection platform is the capability for real-time monitoring of the ATP target and the ability to speed-up target binding kinetics by modulating the capture probe concentration over a wide range. Assays based on capture probes immobilized at the signaling surface exhibit poor kinetics due to limitations imposed by diffusion and steric hindrance at the electrode surface.7, 9 Within the double-surface scheme, as per Figure 1, the unhindered 3D diffusion conditions at the recognition surface cause fast binding of the target molecules to the immobilized capture probes. Subsequently, free diffusion of the FAD released due to the displacement assay can be sensitively monitored at the nanostructured signaling surface. Furthermore, since the ATPA capture probe concentration can be inordinately increased over a wide range to enhance its collision frequency with the ATP target molecules, without any steric hindrance from neighboring bound molecules, the binding kinetics for ATPA with ATP can be sped-up within this particular assay. As apparent from Figure 3(d), the binding kinetics for ATP target with ATPA capture probes can be sped-up at the target detection limit by increasing the ATPA concentration in the range of 4.5×10−8M to 4.5×10−5 M. In fact, in this manner, 4.02×10−11M of ATP can be detected within 30 seconds and a saturation signal for the binding kinetics can be obtained within 12 minutes by utilizing an ATPA capture probe concentration of 4.5×10−5 M.
Figure 3.
(a) AdSSW voltammograms of ATP (indirect determination by release of FAD) in a concentration range of 1.14 ×10−10 to 3.0×10−5 M at Gr-AuNP-CPE after hybridization with 45×10−6 M ATPA in pH 7.0 citrate-phosphate buffer. (b) Linear calibration curve for the I vs E data. Inset : Magnification of signal near the detection limit: 4.02×10−11M for Gr-AuNP-CPE and 2.01×10−11M for Gr-AuNP-GCPE. (c) Comparative AdSSWV at 4.02×10−11 M for various targets: (i) GTP; (ii) UTP; (iii) CTP and (iv) ATP (at Gr-AuNP-CPE) and (v) ATP (at Gr-AuNP-GCPE). (d) Effect of binding between 4×10−11 M ATP and various concentrations of the capture probe.
Effect of base-pair mismatch on binding kinetics
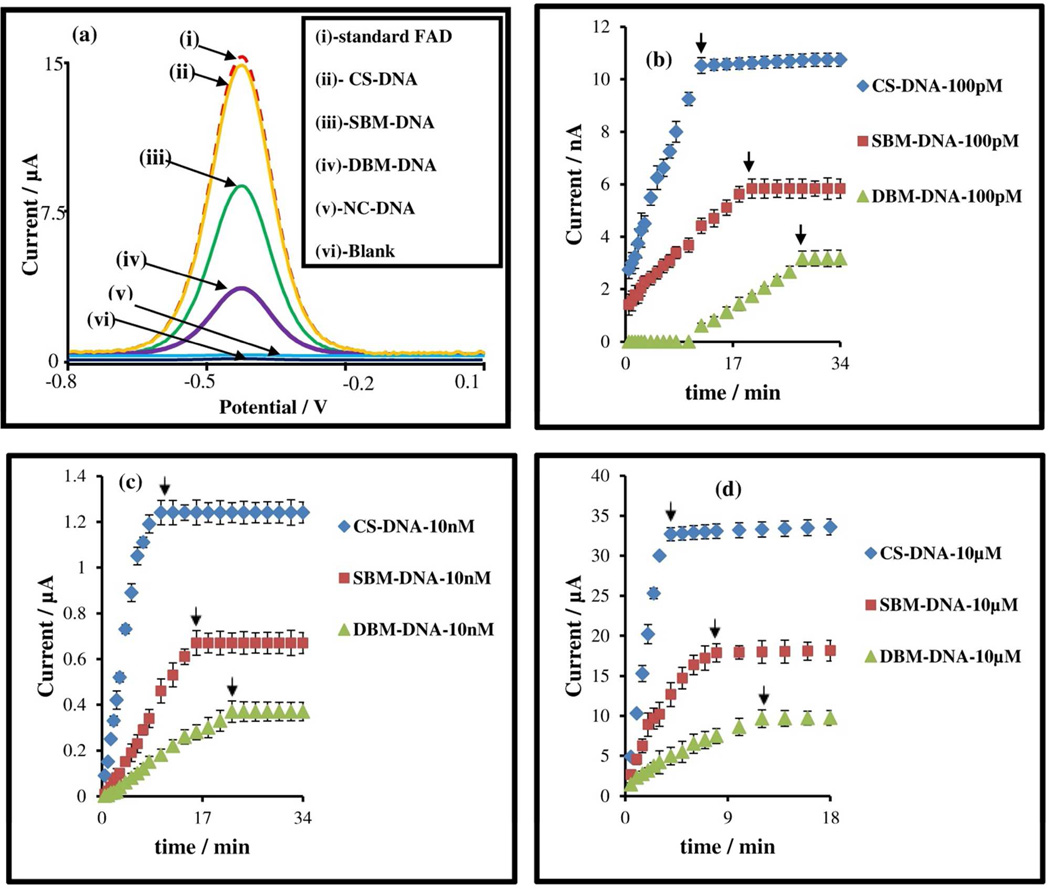
The binding of target DNA composed of bases with varying degrees of mismatch to the ATPA capture probe sequence was studied using real-time detection to probe variations in hybridization kinetics and steady-state signal level. Three types of target DNA strands were used; viz., complementary strand with perfect-match (CS-DNA), single-base mismatch (SBM-DNA), and double-base mismatch (DBM-DNA). Prior investigations have studied the influence of the disrupted π stacking, due to base-pair mismatch, on the signal level at steady-state.34 Given the capability of the displacement assay towards real-time electrochemical monitoring, we have focused instead on studying the influence of base-pair mismatch on DNA hybridization kinetics. As per Figure 4, base-pair mismatches cause differences, not only in signal level at steady-state [Figure 4(a) and Table S-3] but also in binding kinetics of the target, as quantified by the time needed to reach signal saturation [Figure 4(b–d)]. One prior study has utilized surface plasmon fluorescence spectroscopy to demonstrate differences in target binding kinetics arising from varying degrees of base-pair mismatches.9 However, these differences were only apparent within a small target concentration range. This was due to the limits imposed on capture probe concentration within the surface immobilized assay towards avoiding intermolecular columbic interactions. Within the current work, since capture probe concentration can be increased without limitations from steric hindrance and due to the inherently faster DNA hybridization in solution versus on the signaling surface,6 we are able to quantitatively monitor distinct differences in target binding kinetics with varying degrees of base-pair mismatch, over a wide target concentration range 1.14×10−10 M to 1×10−5 M. By utilizing a high ATPA concentration (4.5×10−5 M), the time required by 1×10−5 M target at signal saturation is 4, 8, and 14 minutes for CS-DNA, SBM-DNA and DBMDNA, respectively; whereas at 1.14×10−10 M target it is 12, 20, and 28 minutes for CS-DNA, SBM-DNA and DBM-DNA, respectively. Furthermore the signal level at saturation follows a distinct selectivity ratio of ~0.55 for SBM-DNA to CS-DNA and ~0.3 for DBM-DNA to CS-DNA, irrespective of the target concentration. All of these characteristics demonstrate the quantitative nature of this electrochemical detection system for kinetic and steady-state measurements.
Figure 4.
(a) AdS-SWV voltammograms of 2×10−6M of different targets (CS-DNA, SBM-DNA, DBMDNA and NC-DNA) when hybridized with 45×10−6 M ATPA in pH 7.0 citrate-phosphate buffer. (b) A comparative Ip vs time plot to study the effect of hybridization process between 1×10−10 M of target (CSDNA, SBM-DNA and DBM-DNA) and 45×10−6 M of ATPA. (c) A comparative Ip vs time plot to study the effect of hybridization process between 1×10−8 M of target (CS-DNA, SBM-DNA and DBM-DNA) and 45×10−6 M of ATPA. (d) A comparative Ip vs time plot to study the effect of hybridization process between 1×10−5 M of target (CS-DNA, SBM-DNA and DBM-DNA) and 45×10−6 M of ATPA.
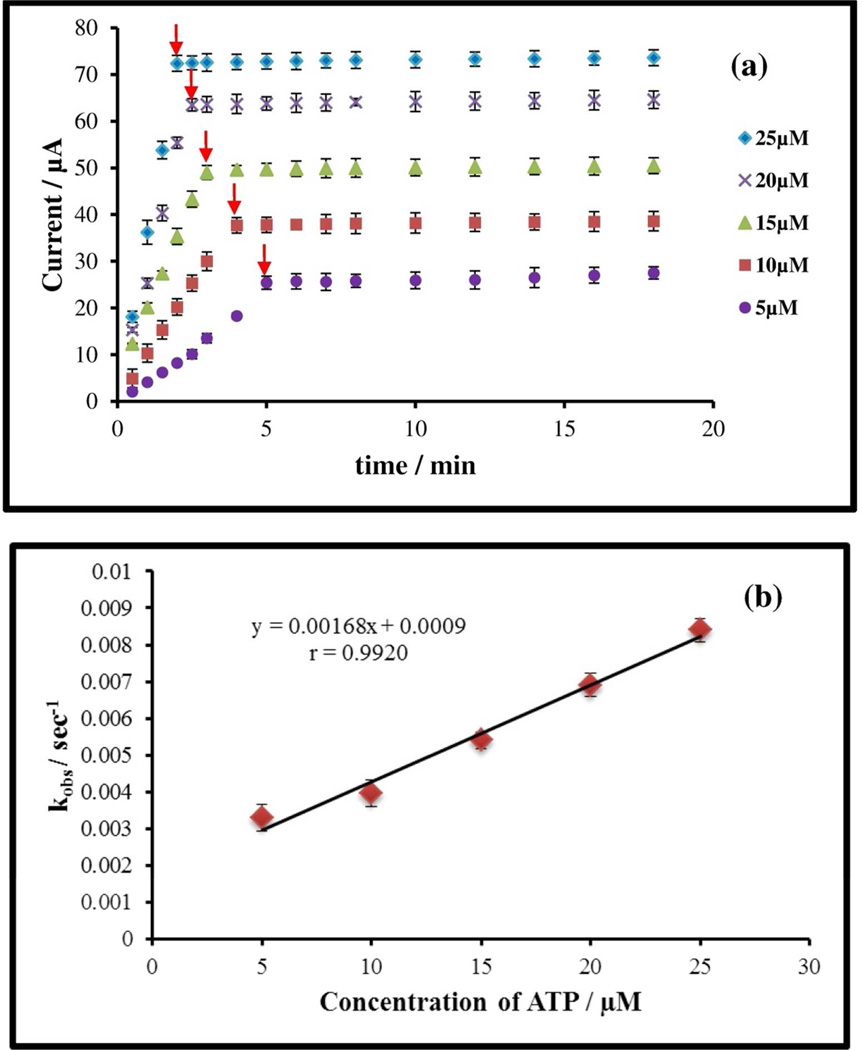
Determination of kinetic and affinity parameters (kobs, kon, koff and Kd)
To further illustrate the utility of this highly quantitative assay for kinetic measurements, we used the ATP binding kinetics plot to compute a few important parameters viz., kobs (apparent rate constant), kon (rate constant for the association reaction), koff (rate constant for the dissociation reaction) and Kd (dissociation constant) for interaction with ATPA capture probes. Using a fixed capture probe concentration of 4.5×10−5 M, the target ATP concentration was varied in the range of 5×10−6 –25×10−6 M. As apparent from the plot of Ip vs time [Figure 5 (a)], by increasing the target concentration, the peak current reaches steady state at earlier time points due to the higher slope for rise towards steady state. This is due to the higher collision frequency between the target and capture probe species, thereby reaching steady-state more quickly. Based on the time required to reach signal saturation, kobs can be determined for each target concentration of interest and this is plotted for varying ATP concentrations (Figure 5(b)). The straight line can be fit by the equation (1):35
| (1) |
Based on this, kon can be determined from the slope (M−1s−1), and koff can be determined from the intercept (s−1) of the line. The dissociation constant, Kd, can be determined as the ratio of koff to kon.36 These kinetic and affinity parameters for ATP were computed as: kon=168 M−1s−1, koff=0.0009 s−1 and Kd=5.36×10−6M. The Kd value for the ATP binding with ATPA (5.36×10−6 M) is in accordance with the literature value of 6×10−6 M.18 Moreover, there are no prior reports on determination of the kon and koff values of ATP with the ATPA.
Figure 5.
(a) A comparative Ip vs time plot to study the effect of variation in hybridization kinetics between ATPA (45×10−6 M) and ATP on varying the concentration of ATP in a range of 5×10−6 M to 25×10−6 M. (b) Plot of kobs vs concentration of ATP from the data obtained in (a) to calculate kinetic parameters. Calculated values of kinetic binding parameters: kon (M−1.s−1) =168; koff (s−1) = 0.0009; Kd (GM) = 5.36. Arrows in Figure 5 (a) above indicate the value for 1/kobs.
Quantitative detection within various media
Further validation of the detection platform was performed by measuring repeatability, reproducibility, precision and accuracy of analysis using five replicate measurements for the detection of 7.12 × 10−9 M ATP over a single day (intra assay) (n=5) and for five days over a period of one week (inter-assay). Satisfactory mean percentage recoveries (% R) and relative standard deviations (% RSD) were obtained [Table S-4 (a)]. The robustness of the detection system was examined by studying the effect of small variations in pH, accumulation potential (Eacc) accumulation time(tacc), and SWV frequency(f), on the recovery of ATP. As apparent from Table S-4 (b), the high % R for ATP (within a range of 98–101 %) with limited change in signal upon variations in important detection conditions (viz., pH, Eacc, tacc and f) suggest a robust and quantitative detection system. Recovery tests for ATP were carried out in samples of litchi, banana, tomato seeds and blood serum. The results (Table 1) show % R in the range of 98.0–102 % for ATP, demonstrating the reliability of the detection system within various matrices. As required for analytical applications, the amount of ATP within a sample has been determined by the standard addition method. The amount of ATP obtained in litchi, banana and tomato seed samples by our detection system agrees well with that determined using the HPLC-UV absorbance method (Table S-5). Paired F- and t-tests showed an agreement at the 95% confidence level, demonstrating that the electrochemical method reported herein corresponded quantitatively with samples measured by the HPLC method. Thus determination of ATP can be carried out quantitatively in various matrices by the method reported herein. The stability and reproducibility of Gr-AuNP-CPE is described in the Supporting Information (Section S-8).
Table 1.
Recovery test for ATP in blood serum, litchi, banana and tomato seed samples.
| a | b | c | d | |
|---|---|---|---|---|
| Human serum type AB (male) | --- | ND | --- | 98.7 ± 0.51 |
| 1.45 | 1.43 | 98.6 | ||
| 2.90 | 2.85 | 98.3 | ||
| 4.35 | 4.32 | 99.3 | ||
| Human serum (from platelet poor human plasma) | --- | ND | --- | 99.0 ± 0.41 |
| 2.90 | 2.86 | 98.6 | ||
| 5.80 | 5.75 | 99.1 | ||
| 8.70 | 8.65 | 99.4 | ||
| Litchi | --- | 0.73 | --- | 98.6 ± 0.21 |
| 1.18 | 1.88 | 98.4 | ||
| 2.36 | 3.05 | 98.7 | ||
| 3.54 | 4.22 | 98.8 | ||
| Banana | --- | 1.28 | --- | 98.9 ± 0.40 |
| 2.05 | 3.28 | 98.5 | ||
| 4.10 | 5.32 | 98.9 | ||
| 6.15 | 7.38 | 99.3 | ||
| Tomato seeds | --- | 3.13 | --- | 99.3 ± 0.39 |
| 1.42 | 4.50 | 98.9 | ||
| 2.84 | 5.93 | 99.3 | ||
| 4.26 | 7.37 | 99.7 |
ND : Not detected
: ATP spiked (10−10 M)
: ATP recovered (10−10 M)
: Recovery (%) (n=5)
: % Average recovery ± %RSD (n = 5)
Comparison of the current ATP detection system against prior methods
Finally, we compare the current ATP detection system versus prior methods,37 to identify the possible analytical applications that are best suited to the current method. Based on sensitivity levels, the current method is at the state-of-the-art, with only one other recent study reporting better sensitivities.37a In this report, the target binding reaction sets-off a cascade generating multiple signaling moieties per target binding event, whereas our detection method only generates a single FAD molecule per two ATP molecules bound to the aptamer, which we will seek to address in future work. On the other hand, a unique strength of the current detection methodology is the relatively fast target binding kinetics and the ability for real-time target monitoring without an intervening wash step. Additionally, the ability to carry out the assay within fruit and serum media, as well as its ability for quantitative detection of ATP, as demonstrated by its correlation to the levels obtained by other methods, demonstrate a versatile detection system.
CONCLUSIONS
We have reported here on a detection system for ATP that is based on a competitive assay utilizing an ATP-aptamer (ATPA) that is pre-bound to electro-active FAD molecules. Binding of ATP to ATPA caused the release of an equivalent concentration of FAD that can be quantitatively detected by electrochemical adsorption stripping square-wave voltammetry (AdS-SWV) at graphene (Gr) and gold nanoparticle (AuNP) modified carbon paste electrode (Gr-AuNP-CPE). The Gr and AuNP modified CPE surface enhanced adsorption and electron transfer kinetics versus the plain CPE, thereby enhancing the net signal sensitivity. As a result, the ATP detection system exhibited a wide linear working range (1.14×10−10 to 3.0×10−5M), low detection limit (4.02×10−11M with Gr-AuNP-CPE and 2.01×10−11M with Gr-AuNP modified glassy carbon), and extremely fast target binding kinetics at the detection limit, as apparent from signal detection within 30 seconds and signal saturation within 12 minutes. A unique strength of the reported competitive assay that utilizes a different surface for target recognition versus signaling was the ability to increase the capture probe concentration, with no limitation arising from intermolecular columbic interactions, to enable relatively fast kinetics due to target binding in solution rather than on an immobilized electrode surface. Furthermore, the lack of an intervening wash-step permits real-time monitoring of the target. This capability was applied to compute kinetic parameters of the assay viz., kon (association rate), koff (dissociation rate), and Kd (dissociation constant). The calculated Kd values agree with prior reports, while determinations of the kon and koff values for the binding of ATP and ATPA have not been previously reported. Additionally, the degree of base-pair mismatch of complementary target DNA with the ATPA capture probe was demonstrated to quantitatively alter both, the target binding kinetics, as apparent from time to achieve a steady-state signal, and the number of bound target molecules, as apparent from the steady-state signal level. Well-modulated differences in binding kinetics were apparent for single and double-base mismatch versus perfect-match over a wide target concentration range (µM-pM). Finally, the simplicity of the assay chemistry enables its straightforward modification towards various target species, and the electrochemical detection platform enables reliable quantification of ATP within litchi, banana, tomato seeds and serum media, as demonstrated by its comparison to that obtained using HPLC methods.
Supplementary Material
ACKNOWLEDGEMENT
NSS acknowledges support from the Asian Office Aerospace Research & Development (grant #114083). M.F.A. is supported by the LCRC and the NIH-RCMI grant #8G12MD007595-04 and by the RCSA Cottrell College Science Program. S.S. is supported by the NSF PREM (Award Number DMR-0934111).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Experimental procedure for analysis in biological matrices (S1), optimization and characterization of supporting electrolyte (S2) and electrode materials (S3–S6), and stability and reproducibility of the detection system (S8 & S9) are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Martinez A, Phillips S, Whitesides GM, Carrilho E. Anal. Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 2.Von Lode P. Clin. Biochem. 2005;38:591–606. doi: 10.1016/j.clinbiochem.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 3.(a) Swami N, Chou CF, Terberueggen R. Langmuir. 2005;21:1937–1941. doi: 10.1021/la0493767. [DOI] [PubMed] [Google Scholar]; (b) Drummond TG, Hill MG, Barton JK. Nat. Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]; (c) Wang J. Analyst. 2005;130:421–426. doi: 10.1039/b414248a. [DOI] [PubMed] [Google Scholar]; (d) Wang J. Electroanalysis. 2007;19:769–776. [Google Scholar]
- 4.Abi A, Ferapontova EE. J. Am. Chem. Soc. 2012;134:14499–14507. doi: 10.1021/ja304864w. [DOI] [PubMed] [Google Scholar]
- 5.(a) Hu J, Wang T, Kim J, Shannon C, Easley CJ. J. Am. Chem. Soc. 2012;134:7066–7072. doi: 10.1021/ja3000485. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Angew. Chem. 2005;117:5592–5595. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 6.Gao Y, Wolf LK, Georgiadis RM. Nucleic Acids Res. 2006;34:3370–3377. doi: 10.1093/nar/gkl422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei C-W, Cheng J-Y, Huang C-T, Yen M-H, Young T-H. Nucleic Acids Res. 2005;33:e78. doi: 10.1093/nar/gni078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nair PR, Alam MA. Appl. Phys. Lett. 2006;88:233120:1–233120:3. [Google Scholar]
- 9.Tawa K, Knoll W. Nucleic Acids Res. 2004;32:2372–2377. doi: 10.1093/nar/gkh572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheehan PE, Whitman LJ. Nano Lett. 2005;5:803–807. doi: 10.1021/nl050298x. [DOI] [PubMed] [Google Scholar]
- 11.(a) Swami N, Chou CF, Ramamurthy V, Chaurey V. Lab Chip. 2009;9:3212–3220. doi: 10.1039/b910598k. [DOI] [PubMed] [Google Scholar]; (b) Liao KT, Tsegaye M, Chaurey V, Chou CF, Swami NS. Electrophoresis. 2012;33:1958–1966. doi: 10.1002/elps.201100707. [DOI] [PubMed] [Google Scholar]; (c) Chaurey V, Polanco C, Chou CF, Swami NS. Biomicrofluidics. 2012;6:012806. doi: 10.1063/1.3676069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bercovici M, Han CM, Liao JC, Santiago JG. Proc. Natl. Acad. Sci. 2012;109:11127–11132. doi: 10.1073/pnas.1205004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palecek E, Bartosik M. Chem. Rev. 2012;112:3427–3481. doi: 10.1021/cr200303p. [DOI] [PubMed] [Google Scholar]
- 14.Palecek E, Fojta M. Talanta. 2007;74:276–290. doi: 10.1016/j.talanta.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 15.(a) Pedrero M, Campuzano S, Pingarron JM. Electroanalysis. 2012;24:470–482. doi: 10.1002/elan.201100528. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fojta M, Kostecka P, Trefulka MR, Havran L, Palecek E. Anal. Chem. 2007;79:1022–1029. doi: 10.1021/ac0616299. [DOI] [PubMed] [Google Scholar]; (c) Hason S, Vetterl V. Anal. Chem. 2006;78:5179–5183. doi: 10.1021/ac052227o. [DOI] [PubMed] [Google Scholar]
- 16.Yu FL, Li G, Qu B, Cao W. Biosens. Bioelectron. 2010;26:1114–1117. doi: 10.1016/j.bios.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 17.Sitaula S, Branch SD, Ali MF. Chem. Commun. 2012;48:9284–9286. doi: 10.1039/c2cc34279k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huizenga DE, Szostak JW. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 19.Song L, Shan D, Zhao M, Pink BA, Minnehan KA, York L, Gardel M, Sullivan S, Phillips AF, Hayman RB, Walt DR, Duffy DC. Anal. Chem. 2013;85:1932–1939. doi: 10.1021/ac303426b. [DOI] [PubMed] [Google Scholar]
- 20.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Duffy DC. Nat. Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. Nat. Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 22.Svancara I, Vytras K, Kalcher K, Walcarius A, Wang J. Electroanalysis. 2009;21:7–28. [Google Scholar]
- 23.(a) Liu H, Jiang Y, Luo Y, Jiang W. Food Technol. Biotechnol. 44:531–534. [Google Scholar]; (b) Spoelstra P, Joosen RVL, van der Plas LHW, Hilhorst HWM. Seed Sci. Res. 2002;12:231–238. [Google Scholar]
- 24.Zima J, Svancara I, Barek J, Vytras K. Crit. Rev. Anal. Chem. 2009;39:204–227. [Google Scholar]
- 25.Kochmann S, Hirsch T, Wolfbeis OS. TRAC-Trend. Anal. Chem. 2012;39:87–113. [Google Scholar]
- 26.Kuila T, Bose S, Khanra P, Mishra AK, Kim NH, Lee JH. Biosens. Bioelectron. 2011;26:4637–4648. doi: 10.1016/j.bios.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs CB, Peairs MJ, Venton BJ. Anal. Chim. Acta. 2010;662:105–127. doi: 10.1016/j.aca.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Sanghavi BJ, Hirsch G, Karna SP, Srivastava AK. Anal. Chim. Acta. 2012;735:37–45. doi: 10.1016/j.aca.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 29.Gadhari NS, Sanghavi BJ, Srivastava AK. Anal. Chim. Acta. 2011;703:31–40. doi: 10.1016/j.aca.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 30.Sanghavi BJ, Mobin SM, Mathur P, Lahiri GK, Srivastava AK. Biosens. Bioelectron. 2013;39:124–132. doi: 10.1016/j.bios.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 31.Mobin SM, Sanghavi BJ, Srivastava AK, Mathur P, Lahiri GK. Anal. Chem. 2010;82:5983–5992. doi: 10.1021/ac1004037. [DOI] [PubMed] [Google Scholar]
- 32.Laviron E. J. Electroanal. Chem. 1979;101:19. [Google Scholar]
- 33.Anson FC. Anal. Chem. 1966;38:54–57. [Google Scholar]
- 34.Ahangar LE, Mehrgardi MA. Biosens. Bioelectron. 2012;38:252–257. doi: 10.1016/j.bios.2012.05.040. [DOI] [PubMed] [Google Scholar]
- 35.Smirnova I, Kasho V, Sugihara J, Choe JY, Kaback HR. Biochemistry. 2009;48:8852–8860. doi: 10.1021/bi9011918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braisted AC, Wells JA. Proc. Natl. Acad. Sci. 1996;93:5688–5692. doi: 10.1073/pnas.93.12.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.(a) Yin BC, Guan YM, Ye BC. Chem. Commun. 2012;48:4208–4210. doi: 10.1039/c2cc30997a. [DOI] [PubMed] [Google Scholar]; (b) Ross AE, Venton BJ. Analyst. 2012;137:3045–3051. doi: 10.1039/c2an35297d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Xu YD, Venton BJ. Electroanalysis. 2010;22:1167–1174. [Google Scholar]; (d) Zhang HF, Han YJ, Guo YJ, Dong C. J. Mater. Chem. 2012;22:23900–23905. [Google Scholar]; (e) Zuo X, Song S, Zhang J, Pan D, Wang L, Fan C. J. Am. Chem. Soc. 2007;129:1042–1043. doi: 10.1021/ja067024b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.