Abstract
This review is focused on current findings implicating miRNAs in the polycystic liver diseases, which we categorized as cholangiociliopathies. Our recent data suggest that deregulation of miRNA pathways is emerging as a novel mechanism in the development of cholangiociliopathies. Experimental evidence demonstrates that miRNAs (i.e., miR-15a) influence hepatic cyst growth by affecting the expression of the cell cycle regulator, Cdc25A. Given that abnormalities in many cellular processes (i.e., cell cycle regulation, cell proliferation, cAMP and calcium signaling, the EGF-stimulated mitogen-activated protein kinase (MAPK) pathway and fluid secretion) contribute to the hepatic cystogenesis, the potential role of miRNAs in regulation of these processes is discussed.
Keywords: miRNA, cholangiociliopathies, miR-15a, Cdc25A, cholangiocyte, proliferation
MicroRNAs: Biogenesis, Action, Function
Initially discovered in C. elegans in 1993 as essential regulators for the timing of larval development, microRNAs [(miRNAs); i.e., small, non-coding RNAs ~20–22 nucleotides in length)] have been identified in many different organisms including vertebrates, flies, plants and viruses. To date, the list of miRNAs consists of more than 6,000 entries.1 MiRNAs are encoded by ~2% of all known human genes; at same time ~30% of genes are predicted to be regulated by miRNAs.2–5 A single miRNA has the potential to interact with many mRNA transcripts whereas a single mRNA can serve as a target of multiple miRNAs.6–8 However, only limited miRNA targets have been validated experimentally; many of them are still predicted based on different computational programs.9 To confirm the validity of miRNA targets, several criteria have been recommended: (1) a miRNA/mRNA interaction needs to be verified; (2) the miRNA and its target mRNA should co-expressed in the tissue under study; (3) the miRNA should affect protein expression; and (4) a functional role of the miRNA/mRNA complex should be proven.10,11
The processing of miRNAs involves both nuclear and cytoplasmic events. The biogenesis of miRNAs begins in nucleus were they transcribed by RNA polymerase II2 or RNA polymerase III12,13 as a long primary miRNA (pri-miRNA, contains at least several hundred base pairs) and cleaved by the nuclear RNase III enzyme, Drosha, and its associate DGCRG8 (DiGeorge syndrome critical region gene 8) into shorter (~60–70 nucleotides in length) precursor miRNA (pre-miRNA).14,15 The pre-miRNA is transported from nucleus to the cell cytoplasm via the exportin-5 that works in conjunction with Ran-guanosine triphosphate.16 The pre-miRNA is further cleaved by cytoplasmic RNase III, Dicer, and its partner TRBP [transactivating region (Tar) RNA-binding protein] and PACT [interferon-inducible double-stranded-RNA-dependent protein kinase (PKR) activator] to the mature miRNA duplex of 20–25 nucleotides.15,17–19 This miRNA duplex is included into a RISC complex (RNA-induced silencing complex) which consists of Dicer, TRBP and Ago 2 (Argonaute 2).2,17,18 One of the miRNA strands is degraded based on thermodynamic properties of the duplex and the mature miRNA which has the weakest thermodynamic stability at its 5′ terminus is intended to target the mRNA.2,19
MiRNAs bind to the 3′ UTR region of their specific target mRNA relying mainly on sequences at the 5′ end of miRNA (i.e., “seed” region). Depending on the efficiency of binding, miRNA may restrain the expression of mRNA or induce degradation. Perfect complementarity results in mRNA degradation while imperfect binding blocks translation of the target mRNA.20 The translationally silenced mRNA then can be sequestered into P-bodies (or GW bodies) where it is stored or degraded.2 While the majority of miRNAs downregulate the expression of their targets, upregulation by miRNAs has also been reported. Indeed, miR-369-3 binds to the 3′UTR of TGFα mRNA leading to its activation.21 Also miR-10a positively affects protein synthesis as a result of its direct binding to 5′UTR of ribosomal mRNA.22
MiRNAs in Diseases
MiRNAs play an important role in both normal and pathological conditions controlling cell differentiation, proliferation, apoptosis, cell cycle progression and angiogenesis. They are implicated in the pathogenesis of a wide spectrum of human disorders including cardiovascular and neurological diseases, viral infections and different cancers.9,20,23–28 The first connection between a miRNA and cancer was established in patients with chronic lymphocytic leukemia (CLL). In this tumor, the region of chromosome 13 that contains two miRNA genes, miR-15 and miR-16, is often deleted; as a result miR-15 and miR-16 levels are significantly downregulated leading to malignant transformation.29 Still then, numerous miRNAs have been identify as oncogenic regulators including the miR-17-92 cluster,30 miR-145,31,32 miR-155,33 miR-21,32 miR-200 and miR-222.34
Molecular Pathways in Cholangiociliopathies and miRNA
The polycystic liver diseases (which we recently categorized as cholangiociliopathies)35,36 are a group of pleiotropic hereditary disorders that includes (but is not limited to) Autosomal Dominant Polycystic Kidney Disease (ADPKD), Autosomal Recessive PKD (ARPKD), Meckel-Gruber Syndrome (MKS), Bardet-Biedl Syndrome (BBS), Nephronophthisis (NPHP) and Joubert Syndrome (JBTS). In cholangiociliopathies, mutations in respective genes result in hepatic cytogenesis and/or hepatic fibrosis. Liver cysts are originated from cholangiocytes (i.e., bile duct epithelial cells) and their progressive expansion is coupled to at least three cellular mechanisms: (1) cell proliferation and cell cycle progression; (2) cell-matrix interactions and (3) fluid secretion. A diverse range of hormones, growth factors and cytokines impact these processes acting individually or in combination.35,37
We recently found that besides these three cellular processes, deregulation of the miRNA pathways is involved in the cholangiociliopathies.38 Given that expression of many miRNAs is significantly affected in pathological conditions, we examined miRNA signatures in normal and PCK rats (an animal model of one of the cholangiociliopathies, ARPKD) and identified by microarray analysis 126 miRNAs in both normal and cystic cholangiocytes. Out of these 126 miRNAs, four (i.e., miR-25, miR-28, miR-99b and miR-121) were exclusively present in normal rat cholangiocytes and 46 miRNAs were exclusively expressed in cystic cholangiocytes. These 46 miRNAs include miRNAs previously implicated in different pathological conditions and cellular processes such as cancer (miR-17-92 family, miR-107, miR-181, miR-208 and miR-221),9,26,39,40 cardiovascular diseases (miR-133),2 hedgehog signaling (miR-324-5p),41 EGF-stimulated mitogen-activated protein kinase (MAPK) signaling (miR-7),42 cell differentiation (miR-142, miR-181)2 and cell cycle progression (miR-221, miR-210).40,43
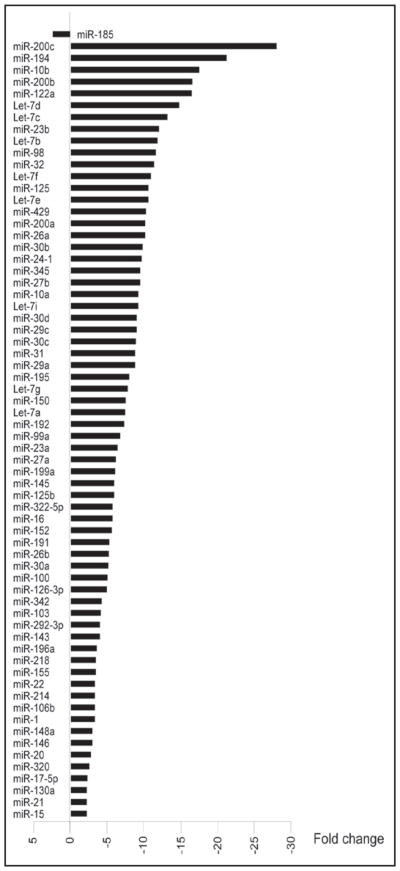
We further analyzed the expression of 76 miRNAs which were present in both normal and cystic cholangiocytes. Consistent with previously made observations in cancer tissues, miRNA levels in cystic cholangiocytes were significantly (more than 2 fold) altered with majority of them (67 miRNAs or 91%) being downregulated (Fig. 1). One miRNA was upregulated, i.e., miR-185 (Fig. 1), and expression of 6 miRNAs (miR-19b, miR-29b, miR-93, miR-106a, miR-223 and miR-483) was not changed (less than 2-fold).
Figure 1.
MiRNA profiles in cystic cholangiocytes of the PCK rat. Sixty-eight miRNAs are differentially expressed in cystic cholangiocytes with the majority of them being downregulated as determined by microarray analysis.
Considering the global changes in miRNA signatures in the cholangiociliopathy, as well as the ability of miRNAs to control the expression of many genes involved in different cellular pathways,44 we clustered miRNA targets around several pathological and functional categories including genes related to polycystic liver and kidney diseases, cell cycle deregulation, cAMP and calcium intracellular signaling, and fluid secretion. Computational analysis (using Target Scan, miRBase and PicTar) revealed that 25% of miRNAs present in cystic cholangiocytes target genes mutated in the cholangiociliopathies; 29% of miRNAs target genes related to the cAMP signaling pathway, 5% to the intracellular calcium signaling, 29% to the cell cycle progression; and 12% to fluid secretion (Fig. 2).
Figure 2.
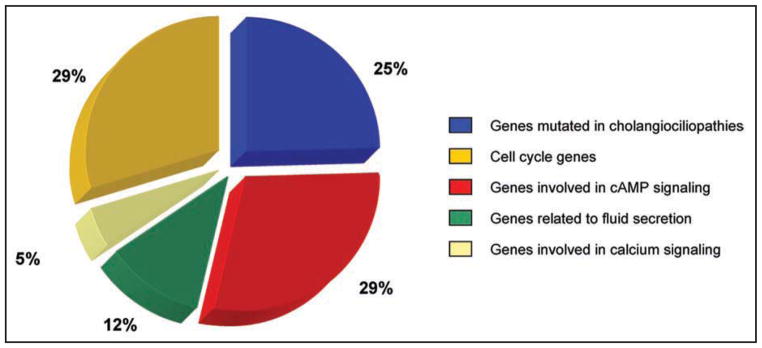
MiRNA targets. Potential miRNA targets were grouped as genes mutated in cholangiociliopathies and genes related to cellular pathways involved in hepatic cystogenesis.
Cholangiociliopathies result from mutations in respective genes following by deregulation of multiple cellular functions (Fig. 3). Hyper-proliferation of cholangiocytes associated with cell cycle dysregulation are considered to be major contributing mechanisms in hepatic cyst growth.37,45 Indeed, substantial evidence demonstrates that an increased rate of cholangiocyte proliferation is triggered by activation of the EGF-stimulated mitogen-activated protein kinase (MAPK) pathway,46 upregulation of the cAMP signaling,45 downregulation of the calcium signaling,45,47 and deregulation of the cell cycle38 (Fig. 3). Importantly, many downstream components (i.e., MAPK, ERK1/2, PKA, EPAC, AKT and Cdc25A) of these signaling pathways are upregulated in cystic cholangiocytes.45–47 Accelerated fluid secretion also known to contribute to cyst expansion is associated with overexpression of the water channel AQP-1, the anion exchanger AE2, and the chloride channel CFTR48 (Fig. 3). Thus, because the expression of many proteins implicated in hepatic cystogenesis is significantly upregulated in cystic cholangiocytes it is tempting to speculate that the downregulation of miRNAs might be responsible for the observed changes. Indeed, based on computational analysis of the miRNA gene targets, multiple hypothetical miRNA/mRNA interactions (predicted by at least one of three program used—Target Scan, miRBase and PicTar) were identified (Fig. 3). Even though detailed experimental functional evidence is required to examine the physiological and/or pathophysiological importance of particular miRNA/gene target relationships, it appears that miRNAs may function in concert with each other affecting critical cellular functions and thus influencing liver cyst growth. Indeed, combinatorial effects of several miRNAs (i.e., miR-16, miR-34a and miR-106b) on cell cycle progression has been recently shown.49
Figure 3.
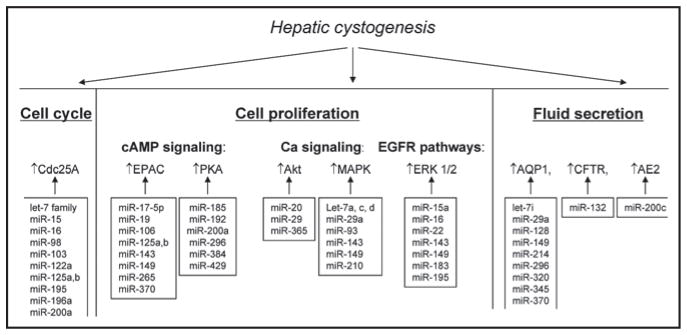
The potential role of miRNAs in regulation of down-stream components of the intracellular pathways which contribute to hepatic cystogenesis. Please note that the scheme includes only proteins expression of which in cystic cholangiocytes has been verified experimentally.
To begin to elucidate the potential involvement of miRNA pathways in the pathogenesis of polycystic liver and kidney diseases, we found that in the PCK rat overexpression of the cell cycle regulator, Cdc25A, is strongly associated with downregulation of miR-15a.38 A detailed experimental analysis revealed that miR-15a directly binds to Cdc25A mRNA downregulating the expression of transcribed Cdc25A protein. The physiological consequences of miR-15a/Cdc25A interaction lead to cell cycle deregulation and an increased rate of cell proliferation accelerating hepatic cystogenesis.38
Although these data provide evidence of the importance of the miR15a/Cdc25a complex in cyst growth,38 it seems likely (based on the nature of miRNA) that collective or concerted regulation of multiple targets related to different cellular pathways by miR-15a should contribute to the disease phenotype. As detected by computational analysis of miR-15a binding partner’s, miR-15a could also target other cell cycle genes—Cdk6 and Cdc25B. Indeed a recent study showed that experimental modulation of Cdk6 levels by miR-16 affects cell cycle progression and proliferation.50 We also identified more potential targets of miR-15a including down-stream effectors (i.e., ERK and MAP kinases, AKT and PIK3) of the EGFR, intracellular calcium and cAMP signaling pathways. These data suggest that miR-15a alone may affect multiple signaling pathways regulating hepatic cystogenesis at different cellular levels.
Polycystic Kidney Disease and miRNAs
The cholangiociliopathies occur in conjunction with polycystic kidney disease. A recent study has also incriminated miRNA pathways in renal cystogenesis.51 In the Han:Sprague-Dawley (SPRD)-cy rat, an animal model of PKD, thirty miRNAs were differentially expressed and their predicted targets might be involved in regulation of genetic switches in PKD. Interestingly, 43% of all miRNAs detected in polycystic kidneys of the Han:Sprague-Dawley (SPRD)-cy rat51 are identical to miRNAs detected in cystic cholangiocytes of the PCK rat.38 These data suggest that similar cellular pathways controlled by miRNAs might be implicated in the pathogenesis of polycystic liver and kidney diseases.
Mechanisms of miRNA Deregulation
The microarray analysis of miRNAs in cystic cholangiocytes revealed widespread changes in profiles demonstrating: (1) the majority of miRNAs were downregulated; (2) one miRNA was upregulated; (3) four miRNAs present in normal cholangiocytes were not detected and (4) the expression of 46 miRNAs was induced. Why miRNAs undergo such global changes in pathological conditions is unknown, but these multidirectional changes suggest that several mechanisms could be involved.
Based on the complexity of miRNA biogenesis, miRNA deregulation observed in pathological conditions in general and in cystic cholangiocytes in particular might reflect abnormalities at different stages of miRNA processing machinery. Indeed, mice with disrupted miRNA biogenesis exhibit higher susceptibility to cancer.52 Dysregulation of mature miRNAs at the levels of Drosha and/or Dicer processing could also be involved.9 Consistent with this, overexpression and downregulation of Dicer has been observed in prostate cancer53 and Burkitt’s lymphoma,54 respectively.
Several other mechanisms including regulation by transcriptional and epigenetic factors and by other miRNAs have been proposed.5,55 Indeed, the miR-17-92 cluster is directly regulated by the oncogene, Myc.56 In contrast, the levels of miR-127, miR-124a and let-7a-3 depend on histone acetylation and DNA methylation.26,57
Recent studies demonstrate that regulation of gene expression by miRNAs is cell cycle dependent.58 In the G0 phase of the cell cycle (i.e., quiescent phase), miRNAs activate translation of their target genes while in S/G2 phase, they may act as repressors of translation.58
Our data showed that cystic cholangiocytes expressed 46 miRNAs not found in normal cholangiocytes. We speculate that, beside the above mentioned mechanisms, miRNA expression might also be influenced by intracellular second messengers, particularly intracellular calcium. Indeed, experimental data suggest that in the presence of calcium, the formation of Dicer is enhanced.59 Another potential candidate is intracellular cAMP, levels of which are significantly increased in cystic cholangiocytes and its overexpression is associated with an accelerated rate of cyst growth.45 cAMP phosphorylates CREB (cAMP response element-binding protein) via two downstream effectors, PKA and EPAC, that are also upregulated in cystic cholangiocytes.47 Current evidence implicates that CREB controls the expression of several miRNAs, in particular miR-132.60,61 Interestingly, we found that miR-132 is present in cystic cholangiocytes but not in normal cells. However, it remains to be proven experimentally whether cAMP or calcium or both might trigger expression of miRNAs under pathological conditions.
Conclusions and Perspectives
To date, only a few studies38,51 have focused on the possible contributions of miRNAs in the pathogenesis of polycystic liver and kidney diseases. Indeed, only one report showed experimental evidence of miRNA involvement in hepatic cystogenesis.38 Defining miRNAs as specific regulators of affected intracellular pathways in polycystic liver and kidney will help to further understand the mechanism of disease progression and help to design targeted therapies. The following questions need answer: (1) what miRNAs are involved in regulation of affected intracellular signaling pathways? (2) Why do miRNAs undergo global changes in cystic cholangiocytes? (3) What mechanisms underlie the multi-directional changes of miRNA expression? (4) What are the miRNA profiles in different forms of polycystic liver and kidney diseases? (5) Are the miRNA signatures different in polycystic liver and polycystic kidney? and (6) Could miRNAs be therapeutic targets and if yes, which ones?
MiRNAs might indeed represent promising therapeutic targets—overexpressing of downregulated miRNAs or knock down of overexpressed miRNAs may potentially help reverse many abnormal cellular functions.62 Animal studies addressing this possibility have began to emerge.26 The first attempt to use miRNA as a therapeutic target showed a promising outcome—i.e., intravenous administration of antagomirs (i.e., modified antisense oligonucleotides) against miR-16, miR-122, miR-192 and miR-194 lead to their inhibition in vivo.63 Since in many pathological conditions miRNAs are significantly downregulated, interventions to increase their levels are required. So far no data showing successful and safe delivery of miRNA vectors have been published.27,64
References
- 1.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erson AE, Petty EM. MicroRNAs in development and disease. Clin Genet. 2008;74:296–306. doi: 10.1111/j.1399-0004.2008.01076.x. [DOI] [PubMed] [Google Scholar]
- 3.Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–8. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 5.McDaneld TG. MicroRNA: mechanism of gene regulation and application to livestock. J Anim Sci. 2008 doi: 10.2527/jas.2008-1303. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 7.Hagan JP, Croce CM. MicroRNAs in carcinogenesis. Cytogenet Genome Res. 2007;118:252–9. doi: 10.1159/000108308. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Rane S, Sayed D, Abdellatif M. MicroRNA with a MacroFunction. Cell Cycle. 2007;6:1850–5. doi: 10.4161/cc.6.15.4551. [DOI] [PubMed] [Google Scholar]
- 10.Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen K, Song F, Calin GA, Wei Q, Hao X, Zhang W. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29:1306–11. doi: 10.1093/carcin/bgn116. [DOI] [PubMed] [Google Scholar]
- 12.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. Rna. 2004;10:1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microR-NAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 14.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 15.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 16.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microR-NA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 18.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarnow P, Jopling CL, Norman KL, Schutz S, Wehner KA. MicroRNAs: expression, avoidance and subversion by vertebrate viruses. Nat Rev Microbiol. 2006;4:651–9. doi: 10.1038/nrmicro1473. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can upregulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 22.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–71. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–69. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Skawran B, Steinemann D, Becker T, Buurman R, Flik J, Wiese B, et al. Loss of 13q is associated with genes involved in cell cycle and proliferation in dedifferentiated hepato-cellular carcinoma. Mod Pathol. 2008;21:1479–89. doi: 10.1038/modpathol.2008.147. [DOI] [PubMed] [Google Scholar]
- 25.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–96. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 26.Blenkiron C, Miska EA. miRNAs in cancer: approaches, aetiology, diagnostics and therapy. Hum Mol Genet. 2007;16:106–13. doi: 10.1093/hmg/ddm056. [DOI] [PubMed] [Google Scholar]
- 27.Hennessy E, O’Driscoll L. Molecular medicine of microRNAs: structure, function and implications for diabetes. Expert Rev Mol Med. 2008;10:24. doi: 10.1017/S1462399408000781. [DOI] [PubMed] [Google Scholar]
- 28.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–63. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 29.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and downregulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 32.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 33.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–9. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 34.Toloubeydokhti T, Bukulmez O, Chegini N. Potential regulatory functions of microR-NAs in the ovary. Semin Reprod Med. 2008;26:469–78. doi: 10.1055/s-0028-1096127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masyuk AI, Masyuk TV, LaRusso NF. Cholangiocyte primary cilia in liver health and disease. Dev Dyn. 2008;237:2007–12. doi: 10.1002/dvdy.21530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masyuk T, Masyuk A, LaRusso N. Cholangiociliopathies: Mechanisms of development and therapeutic targets. Transworld Research Network; 2008. [Google Scholar]
- 37.Masyuk T, LaRusso N. Polycystic liver disease: new insights into disease pathogenesis. Hepatology. 2006;43:906–8. doi: 10.1002/hep.21199. [DOI] [PubMed] [Google Scholar]
- 38.Lee SO, Masyuk T, Splinter P, Banales JM, Masyuk A, Stroope A, et al. MicroRNA15a modulates expression of the cell cycle regulator Cdc25A and affects hepatic cystogenesis in a rat model of polycystic kidney disease. J Clin Invest. 2008;118:3714–24. doi: 10.1172/JCI34922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, Marsh CB. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17–92, E2F and Myc. Proc Natl Acad Sci USA. 2008;105:19678–83. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams BD, Guttilla IK, White BA. Involvement of microRNAs in breast cancer. Semin Reprod Med. 2008;26:522–36. doi: 10.1055/s-0028-1096132. [DOI] [PubMed] [Google Scholar]
- 41.Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27:2616–27. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem. 2008 doi: 10.1074/jbc.M804280200. [DOI] [PubMed] [Google Scholar]
- 43.Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, et al. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–80. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ivanovska I, Cleary MA. Combinatorial microRNAs: working together to make a difference. Cell Cycle. 2008;7:3137–42. doi: 10.4161/cc.7.20.6923. [DOI] [PubMed] [Google Scholar]
- 45.Masyuk TV, Masyuk AI, Torres VE, Harris PC, Larusso NF. Octreotide inhibits hepatic cystogenesis in a rodent model of polycystic liver disease by reducing cholangiocyte adenosine 3′,5′-cyclic monophosphate. Gastroenterology. 2007;132:1104–16. doi: 10.1053/j.gastro.2006.12.039. [DOI] [PubMed] [Google Scholar]
- 46.Sato Y, Harada K, Furubo S, Kizawa K, Sanzen T, Yasoshima M, et al. Inhibition of intrahepatic bile duct dilation of the polycystic kidney rat with a novel tyrosine kinase inhibitor gefitinib. Am J Pathol. 2006;169:1238–50. doi: 10.2353/ajpath.2006.051136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banales JM, Masyuk TV, Gradilone SA, Masyuk AI, Medina JF, LaRusso NF. The cAMP effectors Epac and protein kinase a (PKA) are involved in the hepatic cystogenesis of an animal model of autosomal recessive polycystic kidney disease (ARPKD) Hepatology. 2009;49:160–74. doi: 10.1002/hep.22636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banales JM, Masyuk TV, Bogert PS, Huang BQ, Gradilone SA, Lee SO, et al. Hepatic cystogenesis is associated with abnormal expression and location of ion transporters and water channels in an animal model of autosomal recessive polycystic kidney disease. Am J Pathol. 2008;173:1637–46. doi: 10.2353/ajpath.2008.080125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishikawa H, Kubo A, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol. 2005;7:517–24. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- 50.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–52. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pandey P, Brors B, Srivastava PK, Bott A, Boehn SN, Groene HJ, et al. Microarray-based approach identifies microRNAs and their target functional patterns in polycystic kidney disease. BMC Genomics. 2008;9:624. doi: 10.1186/1471-2164-9-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 53.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, et al. Upregulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–20. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaul D, Sikand K. Defective RNA-mediated c-myc gene silencing pathway in Burkitt’s lymphoma. Biochem Biophys Res Commun. 2004;313:552–4. doi: 10.1016/j.bbrc.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 55.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. Rna. 2006;12:1161–7. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microR-NAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 57.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 58.Vasudevan S, Tong Y, Steitz JA. Cell cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–9. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- 60.Cheng HY, Obrietan K. Revealing a role of microRNAs in the regulation of the biological clock. Cell Cycle. 2007;6:3034–5. doi: 10.4161/cc.6.24.5106. [DOI] [PubMed] [Google Scholar]
- 61.Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–4. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 62.Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–55. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 63.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 64.Esau CC, Monia BP. Therapeutic potential for microRNAs. Adv Drug Deliv Rev. 2007;59:101–14. doi: 10.1016/j.addr.2007.03.007. [DOI] [PubMed] [Google Scholar]